Abstract
The Italian Toscano cigar production includes a fermentation step that starts when dark fire-cured tobacco leaves are moistened and mixed with ca. 20% prefermented tobacco to form a 500-kg bulk. The dynamics of the process, lasting ca. 18 days, has never been investigated in detail, and limited information is available on microbiota involved. Here we show that Toscano fermentation is invariably associated with the following: (i) an increase in temperature, pH, and total microbial population; (ii) a decrease in reducing sugars, citric and malic acids, and nitrate content; and (iii) an increase in oxalic acid, nitrite, and tobacco-specific nitrosamine content. The microbial community structure and dynamics were investigated by culture-based and culture-independent approaches, including denaturing gradient gel electrophoresis and single-strand conformational polymorphism. Results demonstrate that fermentation is assisted by a complex microbial community, changing in structure and composition during the process. During the early phase, the moderately acidic and mesophilic environment supports the rapid growth of a yeast population predominated by Debaryomyces hansenii. At this stage, Staphylococcaceae (Jeotgalicoccus and Staphylococcus) and Lactobacillales (Aerococcus, Lactobacillus, and Weissella) are the most commonly detected bacteria. When temperature and pH increase, endospore-forming low-G+C content gram-positive bacilli (Bacillus spp.) become evident. This leads to a further pH increase and promotes growth of moderately halotolerant and alkaliphilic Actinomycetales (Corynebacterium and Yania) during the late phase. To postulate a functional role for individual microbial species assisting the fermentation process, a preliminary physiological and biochemical characterization of representative isolates was performed.
The habit of smoking cigars can be traced back to American natives, and after the discovery of the New World cigar smoking spread throughout Europe (56). In Italy, tobacco fermentation for cigar production dates back to the end of the 18th century (54). The Italian Toscano cigar differs from other world-famous cigars because the filler tobacco undergoes a fermentation process which is shorter (weeks, versus months for other cigars) and is characterized by higher moisture content of the fermenting leaves (50 to 55% versus 20 to 30% for other cigars). This fermentation can be compared to that of composting (15) or other alkaline pH tobacco products, such as snuff and chewing tobacco (60), and differentiates the final aroma from that of other cigars. Toscano cigars are smoked by the majority of Italian cigar smokers, although production and distribution of cigars similar to Toscano now extend to many other countries. The total cigar market in Italy in 2005 was 646,700 kg, corresponding to 129,337,000 pieces. Toscano cigar total sales were 535,000 kg (107,000,000 pieces), corresponding to an 82.8% market share with a turnover of approximately 60,000,000€.
Filler solid-state fermentation is carried out with dark fire-cured tobacco leaves adjusted to a moisture content of 50 to 55%. Leaves are placed into partially insulated tanks containing up to 500 kg of moist tobacco that, at the beginning of fermentation, has a moderately acidic pH. Under these conditions, fermentation starts and a considerable amount of heat is generated as a function of the metabolic activities of indigenous microorganisms. Heat is not easily dissipated due to the poor thermal conductivity of the tank and of the tobacco bulk itself. Temperatures to 65°C can be reached. During a 2- to 3-week fermentation, periodic shuffling of the tobacco bulk is performed in order to improve bulk homogeneity and avoid overheating. Concomitantly, high ammonia levels are generated, likely by proteolysis and/or other processes, contributing to the characteristic alkaline smoke of Toscano cigars. Undesirable fermentation by-products, such as tobacco-specific nitrosamines (TSNA) (5, 20), whose increase in concentration parallels that of nitrite, are generated during fermentation. An additional unpleasant by-product of organic acid metabolism is oxalic acid, which negatively influences the tobacco taste (34). Minimizing the generation of nitrite and TSNA represents a major goal of tobacco fermentation technology. It has become mandatory to define the microbial species that play a role in this process with regard to their impact on carbon and nitrogen metabolism to improve the quality of the end product (13) and, possibly, its safety.
Even if the fermentation operational process and macroscopic effects (pH and temperature increase) are well known and have been handed down for two centuries, extensive biological and chemical investigations of the process have never been performed. The only microbiological study on Toscano cigar fermentation dates back to the 1940s, when Giovannozzi (17) isolated a predominant yeast belonging to the genus Debaryomyces and several bacteria. Thereafter, no other investigation has been carried out, and most of the current knowledge can be inferred from seminal studies on fermentation of other alkaline pH tobacco products which have demonstrated the biotic nature of leaf fermentation, the colonization of broadleaf tobacco by thermotolerant microorganisms (Micrococcus sensibilis, Micrococcus bicolor, Bacillus sphaericus, Bacillus mycoides, Phytomonas mellea, Bacillus cereus, and Staphylococcus spp.) (24), and the possibility for association of bacterial strains or species with definite desirable qualities, such as aroma and mildness (13). In this study we have defined the physical-chemical profile of dark fire-cured tobacco fermentation and have investigated the structure and dynamics of the microbial community assisting the process by both culture-independent and culture-based approaches.
MATERIALS AND METHODS
Fermented tobacco technology and sampling procedure.
Fermented cigar tobacco filler was produced by BAT Italia S.p.a. according to the traditional technology in Lucca and Cava de' Tirreni plants (Italy). Blends of Italian dark fire-cured tobacco whole leaves were first wet with tap water to reach an average moisture content of 55% and then transferred into partially insulated cubic tanks (open on the top and with a grating bottom) of about 2 m3 containing up to 500 kg of moistened tobacco. Fermented tobacco from previous cycles was added to the bulk at approximately 20% (wt/wt) to act as a starter. Fermentation was carried out for approximately 18 days, and aseptic sampling was performed each day of fermentation from the middle of the bulk.
Chemical assays.
pH was measured in a water suspension of 2% tobacco. All the chemical assays were performed on dried milled tobacco. Nitrite was estimated by using the Griess reagent (α-naphthylamine/sulfanilic acid), using a standard curve obtained with known quantities of NaNO2 (18). TSNA were evaluated by gas chromatography-thermal energy analysis, after extraction with methylene chloride and solid-phase extraction clean-up. TSNA were reported as the sum of N-nitrosonornicotine, N-nitrosoanatabine, N-nitrosoanabasine, and 4-methyl-N-nitrosoamino-1-(3-pyridyl)-1-butanone. Nitrates and reducing sugars were analyzed by continuous flow analysis according to Coresta recommended methods (http://www.coresta.org). Ammonia determination was performed colorimetrically as described by Collins et al. (9). Organic acids were extracted with 1.0 N H2SO4 and analyzed by high-performance liquid chromatography as described by Qiu (46). Total carbon and nitrogen contents were determined by using a Leco TruSpec elemental analyzer.
Isolation of microorganisms from fermenting tobacco leaves.
Potato dextrose agar (PDA; Oxoid), tryptic soy agar (TSA; Difco), de Man, Rogosa, and Sharpe (MRS) agar (Oxoid), pseudomonas isolation agar (PIA; Difco), violet red bile lactose agar (VRBLA; Oxoid), plate count agar (PCA; Oxoid), and a tobacco-based (TB) medium were used as culture media. TB medium was prepared from a fire-cured tobacco water extract. In detail, 5 g of dried milled dark fire-cured tobacco collected at the start of fermentation was suspended in 100 ml of distilled water, adjusted to pH 7.0, and shaken for 15 min at room temperature (RT). A tobacco extract was obtained by serial passages through Whatman no. 1 paper and a 0.22-μm nitrocellulose filter (Millipore). This extract was used as a liquid medium or diluted 1:2 with distilled water supplemented with 15 g/liter of bacteriological agar (Difco) to obtain solid TB medium. In some experiments, the pH of the TB medium was adjusted at 5.2 to facilitate yeast growth.
For microbial isolation, 20 g of fermenting tobacco was collected at the indicated times and, under sterile conditions, suspended in a 200-ml final volume of sterile saline (0.9% NaCl) and homogenized with a BagMixer (Interscience) for 2 min. Decimal dilutions of the homogenized samples were plated onto suitable medium. PDA or TB medium plates supplemented with 100 μg/ml chloramphenicol (Sigma) were used for total yeast number determination, after incubation at 30°C under aerobic conditions for up to 96 h. Total aerobic bacterial counting and colony isolation were performed by using TSA or TB medium plates containing 20 μg/ml nystatin (Sigma) and 30 μg/ml amphotericin B (Sigma) incubated aerobically at 37°C for up to 48 h. The number of cultivable spore-forming bacteria was determined aerobically at 37°C for up to 48 h on TSA or TB medium plates supplemented with nystatin and amphotericin B, after 10 min of heating at 80°C. Coliforms and pseudomonads were determined by plating on VRBLA and PIA media, respectively, after aerobic incubation at 35°C for up to 48 h. The spread plating method was used for the above media. Lactic acid bacteria were counted and isolated by pour plating on MRS agar and incubated at 30°C for 48 h under aerobic conditions. All platings were performed in triplicate, and dilutions yielding 30 to 600 CFU/plate were considered for bacterial enumeration and further isolation.
Identification of microorganisms.
A total of 200 apparently different microbial isolates were collected: 15 a day from PDA plates on days 1, 3 and 5, 15 a day from TSA plates on days 1, 3, 5, 8, 10, 12, 15, and 17, and 5 a day from MRS agar plates on days 1, 3, 5, 8, 10, 12, and 15.
Phenotypic identification of bacterial strains was performed by conventional microbiological methods. A number of phenotypic criteria were considered: (i) colony morphology (size, shape, and pigmentation); (ii) cell morphology (Gram stain, size, shape, and presence of endospore) and motility; (iii) oxidase and catalase tests; (iv) oxygen requirement for growth; (v) salt tolerance (growth in the presence of up to 20% NaCl); (vi) pH range for growth (from 3 to 9); and (vii) temperature tolerance (up to 65°C) (3). Metabolic tests were performed by using the API 50 CHB/E and API Coryne systems (Bio Mérieux). Yeast colonies were classified by color, size, shape, texture, margin or edge, and elevation (59). Carbon requirement tests were carried out by using the API 20C AUX system (Bio Mérieux). A number of biochemical characteristics were also determined for both bacterial and yeast isolates, as outlined below.
Molecular identification of microbial strains was performed by 16S or 18S rRNA gene sequence analysis. For this purpose, bacteria or yeast cells were grown under appropriate conditions to late logarithmic phase. After centrifugation, high-molecular-weight genomic DNA was extracted by using the Ultra Clean DNA extraction kit (MO BIO Laboratories) according to the manufacturer's instructions. Genomic DNA was used in PCR experiments to amplify the full-length 16S rRNA gene or the NS1-NS8 region of the 18S rRNA gene with primer pairs 27-F/1492-R or NS1/NS8, respectively. Primer annealing temperatures in PCRs are listed in Table 1. All PCRs were performed as follows: 5 min initial denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 45 s, annealing for 30 s at the indicated temperature (Table 1), and extension at 72°C for 1 to 2 min, depending on amplicon length. The final extension was set at 72°C for 10 min. Reactions were carried out with Pfu DNA polymerase (Stratagene) according to the manufacturer's instructions in a Perkin-Elmer Gene Amp 2400 PCR system. PCR products were separated by electrophoresis through 1% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer (48) and recovered by using the Qiaex II DNA purification kit (QIAGEN). PCR products were cloned into the pGEM-T Easy vector (Promega) or directly subjected to DNA sequencing of both strands as a service by the PHRED quality custom sequencing service (MWG Biotech). For this purpose, primers 16SE20-42-F, 16SEB20-42-F, 16SEB683-R, 16SEB786-F, 16SEB1488-R, DS-F, and DS-R were designed a posteriori on the basis of available sequences (DDBJ, EMBL, RDP, and GenBank databases) to improve matches with 16S or 18S rRNA gene sequences of the tobacco isolates preliminarily identified on the basis of metabolic tests and were used for sequencing of internal amplicon regions (Table 1).
TABLE 1.
Oligonucleotide primers used in this study
| Primer name | Target gene | Use | Sequencea | Annealing temp (°C) in PCR | Positionb | Reference |
|---|---|---|---|---|---|---|
| 27-F | 16S rRNA | Broad-range PCR | 5′-AGAGTTTGATCMTGGCTC-3′ | 55 | 10-27 | 29 |
| 1492-R | 16S rRNA | Broad-range PCR | 5′-GGTTACCTTGTTACGACTT-3′ | 55 | 1510-1492 | 29 |
| NS1 | 18S rRNA | Broad-range PCR | 5′-GTAGTCATATGCTTGTCTC-3′ | 55 | 19-38 | 62 |
| NS8 | 18S rRNA | Broad-range PCR | 5′-TCCGCAGGTTCACCTACGGA-3′ | 55 | 1789-1769 | 62 |
| Com1-F | 16S rRNA (V4 region) | SSCP-RFLP | 5′-CAGCAGCCGCGGTAATAC-3′ | 50 | 519-536 | 30 |
| Com2-R | 16S rRNA (V5 region) | SSCP-RFLP | 5′-CCGTCAATTCCTTTGAGTTT-3′ | 50 | 927-908 | 30 |
| F243-Fc | 16S rRNA (V3 region) | RFLP | 5′-GGATGAGCCCGCGGCCTA-3′ | 65 | 209-226 | 42 |
| R531-Rc | 16S rRNA (V3 region) | RFLP | 5′-CGGCCGCGGCTGCTGGCACGTA-3′ | 65 | 503-482 | 42 |
| FUN18S1-F | 18S rRNA | SSCP-RFLP | 5′-CCATGCATGTCTAAGTATAA-3′ | 47 | 49-68 | 33 |
| FUN18S2-R | 18S rRNA | SSCP-RFLP | 5′-GCTGGCACCAGACTTGCCCTCC-3′ | 47 | 569-548 | 33 |
| 341-FGCd | 16S rRNA (V3 region) | DGGE | 5′-CCTACGGGAGGCAGCAG-3′ | 55 | 341-357 | 37 |
| 518-R | 16S rRNA (V3 region) | DGGE | 5′-ATTACCGCGGCTGCTGG-3′ | 55 | 534-518 | 37 |
| 16SE20-42-F | 16S rRNA | Sequencing | 5′-TGGCTCAGATTGAACGCTGGCGG-3′ | 55 | 20-42 | This study |
| 16SEB20-42-F | 16S rRNA | Sequencing | 5′-TGGCTCAGGAYGAACGCTGGCGG-3′ | 58 | 20-42 | This study |
| 16SEB683-R | 16S rRNA | Sequencing | 5′-CTACGCATTTCACCGCTACAC-3′ | 58 | 703-683 | This study |
| 16SEB786-F | 16S rRNA | Sequencing | 5′-GATTAGATACCCTGGTAGTCCACG-3′ | 58 | 786-809 | This study |
| 16SEB1488-R | 16S rRNA | Sequencing | 5′-TACCTTGTTACGACTTCACC-3′ | 58 | 1507-1488 | This study |
| DS-F | 18S rRNA | Sequencing | 5′-AAACGGCTACCACATCCAAG-3′ | 60 | 399-418 | This study |
| DS-R | 18S rRNA | Sequencing | 5′-GTACAAAGGGCAGGGACGTA-3′ | 60 | 1630-1611 | This study |
Y = C or T; M = A or C.
Numbering is relative to the Escherichia coli 16S rRNA gene or to the Saccharomyces cerevisiae 18S rRNA gene and in a 5′-to-3′ orientation relative to the coding strand.
Specifically designed for Actinobacteria.
This primer has the following GC clamp at its 5′ end: CGCCCGCCGCGCGCGCGCGGGCGGGGCGGGGGCACGGGGGG.
Biochemical characterization of microorganisms.
Starch and casein hydrolysis, urease activity, and nitrate reduction were analyzed according to the methods of Cappuccino and Sherman (6). Pectinolytic activity was detected according to the methods of Tamburini et al. (53). To determine cellulose hydrolysis, cellulose agar, composed of nutrient agar (Difco) supplemented with 0.5 g/liter carboxymethylcellulose (CMC; Sigma), was used. Cellulolytic activity was detected after a 24-h incubation by staining with 1.0 g/liter Congo red (Sigma) and fixing with 1.0 M NaCl. The ability of bacterial strains to utilize nitrate, nitrite, or ammonium as nitrogen sources was tested in a chemically defined medium containing 0.5 g/liter K2HPO4, 0.5 g/liter MgSO4·7H2O, 0.3 g/liter NaCl, 0.02 g/liter MnSO4, and 5% (wt/vol) glycerol, alternatively supplemented with 1.0 g/liter KNO3, 1.0 g/liter NaNO2, or 0.5 g/liter (NH4)2SO4 as nitrogen source. For evaluation of yeast organic acid metabolism, cells were grown in TB medium at 30°C with orbital shaking (200 rpm). At fixed times, 2-ml samples were centrifuged and the supernatant was used for the determination of the organic acid concentration by high-performance liquid chromatography (46). The organic acid concentration in the neutral TB medium was 43 ppm oxalic acid, 1,050 ppm citric acid, and 4,200 ppm malic acid, equivalent to 1.6%, 83%, and 93% recovery from cured tobacco leaves (see Table 2, below). In agreement with a previous report (45), the poor recovery of oxalic acid at neutral pH indicates its presence in the tobacco matrix as water-insoluble species (e.g., calcium salt).
TABLE 2.
Physical and chemical parameters during dark fire-cured tobacco fermentationa
| Day of fermentationb | pH | Temp (°C) | H2O (%) | C/N ratio | Reducing sugars (g/kg) | Malic acid (g/kg) | Citric acid (g/kg) | Oxalic acid (g/kg) | Ammonium (g/kg) | Nitrate (g/kg) | Nitrite (mg/kg) | TSNA (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 6.0 | 20 | 54.7 | 10.5 | 15.6 | 41.0 | 11.5 | 23.8 | 6.5 | 11.3 | 33.7 | 31.2 |
| 5 | 6.8 | 38 | 57.3 | 11.0 | 12.4 | 19.5 | 7.9 | 25.0 | 6.9 | 11.8 | 176.4 | 56.0 |
| 10 | 8.0 | 65 | 51.9 | 11.5 | 12.1 | 13.2 | 7.4 | 42.3 | 6.2 | 8.6 | 895.8 | 95.0 |
| 15 | 8.7 | 62 | 49.7 | 11.2 | 9.6 | 3.4 | 7.2 | 62.3 | 5.3 | 3.9 | 617.4 | 73.3 |
| 18 | 8.8 | 65 | 49.1 | 11.9 | NDc | NDc | 4.9 | 61.2 | 3.8 | 5.5 | 381.6 | 89.0 |
Values for chemical assays are reported on a dried milled tobacco basis.
Samples corresponding to days 5, 10, and 15 were taken before shuffling of the tobacco bulk.
ND, not detectable.
DNA extraction from the microbial community of tobacco.
In order to obtain total microbial DNA from dark fire-cured fermenting tobacco samples, 7 to 10 g of tobacco was suspended under sterile conditions in 100 ml of 0.9% NaCl, 0.05% Tween X-100 for 1.5 h with rotary shaking (150 rpm) at RT. This suspension was passed through sterile gauze and centrifuged at 4°C for 2 min at 2,200 × g to remove particulate matter. The supernatant was centrifuged at 4°C for 60 min at 3,000 × g. The pellet was suspended in 1.7 ml SET buffer (75 mM NaCl, 25 mM EDTA, 20 mM Tris-Cl [pH 7.5]) and incubated in the presence of 1 mg/ml lysozyme (Sigma) for 30 min at 37°C. Samples were briefly sonicated (sonifier model 250/240; Branson Ultrasonic Corporation) for 10 s and incubated in the presence of 0.5 mg/ml proteinase K (Sigma) and 1% sodium dodecyl sulfate for 2 h at 55°C. Nucleic acids were extracted by phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]), and 15 μg/ml RNase A was used to remove RNA. After additional phenol-chloroform-isoamyl alcohol extraction, high-molecular-weight DNA was ethanol precipitated, collected by spooling using Shepherd's crooks, analyzed by 1% agarose gel electrophoresis in 1× TAE buffer, and diluted for PCR analysis.
DGGE analysis of the V3 region of the 16S rRNA gene.
Two μl (10 to 30 ng of DNA) of each DNA sample was subjected to PCR amplification of the 16S rRNA gene. The primers used were the domain Bacteria-specific 27-F and 1492-R primers (Table 1). PCRs were performed as reported above, with LA Taq DNA polymerase (TaKaRa) and a 1-min 40-s extension time. The amplified products were checked by electrophoresis in agarose gel in 1× TAE buffer, purified by use of the Qiaex II DNA purification kit (QIAGEN), and then subjected to nested PCR. Fragments of approximately 200 bp, corresponding to the V3 region of 16S rRNA genes (37), were amplified with Platinum Taq DNA polymerase (Invitrogen) by the use of primers 341-FGC and 518-R and a 40-s elongation time (Table 1). Fragments were purified as described above and resolved by denaturing gradient gel electrophoresis (DGGE) in a DCode universal mutation detection system (Bio-Rad). An 8% polyacrylamide (acrylamide:N,N-methylenebisacrylamide, 37.5:1) gel with a 30 to 55% parallel denaturing gradient was prepared with a model 475 gradient delivery system (Bio-Rad). A 100% denaturing solution contained 7 M urea and 40% deionized formamide. The gels were polymerized for at least 12 h. Approximately 400 ng of purified PCR products was loaded in each well. The gels were run for 5 h at 200 mV in 1× TAE buffer at 60°C, stained with 1 μg/ml ethidium bromide for 20 min, and destained in water.
Generation of PCR-SSCP genetic profiles.
For single-strand conformational polymorphism (SSCP) analysis, different primer pairs, namely, Com1-F/Com2-R and FUN18S1-F/FUN18S2-R (Table 1), were used to amplify 16S or 18S rRNA genes from total DNA of the tobacco microbial community. Each PCR was performed with Ampli Taq DNA polymerase (Applied Biosystems) and a 1-min elongation time. PCR products were separated by electrophoresis in a 1% agarose gel in 1× TAE buffer and purified by using a Qiaex II DNA purification kit. Nine μl of purified PCR sample was denatured by adding 12 μl of formamide loading buffer (80% [wt/vol] deionized formamide, 10 mM EDTA [pH 8], 1 mg/ml xylene cyanol FF, 1 mg/ml bromophenol blue) at 95°C for 5 min and then resolved by 10% polyacrylamide (29:1) gel electrophoresis in 0.8× Tris-borate-EDTA (TBE) buffer at 25 mA and 4°C. Gels were silver stained according to standard procedures (48).
Generation of libraries and RFLP fingerprinting.
Eight independent eubacterial 16S rRNA gene libraries were generated, including four with pooled PCR products extracted from four SSCP gel slices for fermentation days 15 and 18 (gels were cut horizontally to pool similarly positioned bands for both days), and the other four included amplicons generated for the same days with SSCP primers (Com1-F/Com2-R) prior to electrophoretic separation or with Actinobacteria-specific primers (F243-F/R531-R). Two more libraries were obtained with Eukarya-specific primers (FUN18S1-F/FUN18S2-R) using DNA samples extracted on days 1 and 5. DNA fragments (see primer positions in Table 1 for length estimates) were cloned into the pGEM-T Easy vector according to the manufacturer's instructions and used to transform Escherichia coli DH5αF′ (48). For restriction fragment length polymorphism (RFLP) analysis, pGEM-T Easy inserts were amplified with M13 forward and reverse primers (M13F and M13R; Promega), and 500 ng DNA from each independent clone was subjected to restriction analysis with 2 to 5 U of CfoI and/or HinfI endonucleases, in a total volume of 20 μl. Aliquots (5 μl) of each digested product were resolved by electrophoresis through a nondenaturing 6% polyacrylamide (29:1) gel containing 2.5% glycerol in 1× TBE buffer (48), and RFLP patterns were visualized by the silver staining method. For each library, ca. 50 independent clones were analyzed and grouped on the basis of their restriction profile, and 1 prototypic clone for each RFLP pattern was sequenced with the M13F and M13R primers.
Sequence analysis of single products from PCR-SSCP and DGGE microbial community profiles.
Bands identified in DGGE or SSCP polyacrylamide gels were excised with a razor blade. Blocks of gel were transferred in 50 μl of sterile water, and the DNA was allowed to diffuse overnight at 4°C. The eluted DNA was reamplified with the same primers and PCR conditions described for DGGE or PCR-SSCP analysis. The PCR products were tested by DGGE or SSCP for purity and identity with the original bands in the community profiles and then cloned in pGEM-T Easy. Sequencing was performed as described above, using commercial M13F/M13R primers.
Phylogenetic analysis of 16S rRNA gene sequences.
For a preliminary similarity assessment, 1,100- to 1,450-nucleotide (nt)-long 16S rRNA gene sequences were used, in most cases encompassing positions 60 to 1450 relative to the E. coli rrnB gene. Sequence similarity searches were performed using the BLAST network service of the NCBI database and Seqmatch tool of the Ribosomal Database Project II (RDP) (http://www.ncbi.nlm.nih.gov/BLAST/ and http://rdp.cme.msu.edu/, respectively). For phylogenetic analysis, 16S rRNA gene sequences were aligned in conjunction with representative sequences deposited in the RDP database by using the alignment software Clustal W (55). The 16S rRNA gene sequences considered for tree construction encompassed nt 123 to 1333 for cultured Bacillaceae, nt 341 to 534 for DGGE-derived DNA fragments, and nt 519 to 927 for SCCP-derived DNA fragments. Phylogenetic trees were constructed using the MEGA 3 program (28) with the Kimura two-parameter algorithm and the neighbor-joining method. The robustness of the inferred phylogenies was determined by bootstrap analysis based on 1,000 resamplings of data.
RESULTS
Physical-chemical profile of dark fire-cured tobacco fermentation.
The tobacco fermentation process was monitored for 10 cycles lasting approximately 18 days each, with periodic shuffling. The physical-chemical profile and the microbial community structure and dynamics for a representative cycle, with shuffling on days 5, 10, and 15, are reported below.
The evolution of major physical and chemical parameters is shown in Table 2. Before fermentation, dark fire-cured tobacco leaves have an average pH of 5.5, though inoculation of the bulk with fermented leaves as a starter raised the pH value to 6.0. The pH was in the neutral to mild acidic range during early fermentation (days 1 to 5) and then shifted to more basic values (pH ≥ 8) during the late stages. The temperature rose progressively from RT to 65°C on day 10, retaining high values until the end of fermentation. Concomitantly, the moisture content progressively decreased from ca. 55% to less than 50%, and a huge ammonia evolution occurred. The C/N ratio moderately increased from 10.5 on day 1 to 11.9 on day 18 (Table 2). The content of reducing sugars decreased with time from 15.6 g/kg to an undetectable level. Malic, citric, and oxalic acids were the prevalent acid-extractable organic acids in the fermenting bulk. The content of the first two compounds progressively decreased during the process, while oxalic acid concentration almost tripled. Acetic and lactic acids were undetectable.
Direct measurement of the ammonium concentration in tobacco samples showed nearly stable levels with a moderate decrease in the second part of the fermentation (days 15 to 18), likely resulting from loss by volatilization as gaseous ammonia at alkaline pH and high temperature. Nitrate concentration declined to about half of the initial value, while nitrite concentration substantially increased to a maximum value of 895.8 mg/kg on day 10, attaining final values 10-fold higher than those present in the starting bulk. An increase in TSNA levels paralleled nitrite peaking and tripled during the course of fermentation.
Estimation of major microbial groups in bulk of fermenting tobacco.
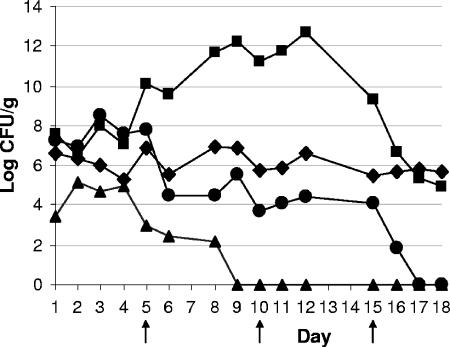
A quantitative estimation of the major microbial groups in the fermenting tobacco bulk was preliminary to any attempt to identify and/or isolate individual species and assess their role in the process. Selective media and/or growth conditions were used to determine the relative abundance of total aerobic bacteria, endospore-forming bacteria, lactic acid bacteria, and yeasts (Fig. 1). Results demonstrated an almost-6-log increase in the number of total aerobic bacteria during 12 days (from ca. 107 to ca. 1013 CFU/g) and a dramatic decrease in the following 6 days, ending with ca. 105 CFU/g on day 18. Endospore-forming bacteria showed moderate variation during the entire process with values between 5.5 × 105 and 107 CFU/g. The number of lactic acid bacteria increased to ca. 108 CFU/g during the first 3 days and then declined to ca. 104 during days 6 to 15 and to undetectable levels on day 18. The yeast population peaked during the second day (ca. 105 CFU/g) and then progressively decreased and became undetectable after day 9 (Fig. 1). Gram-negative bacteria, including coliform bacteria and pseudomonads, were sporadically recovered at low density (ca. 102 CFU/g) during the early fermentation and became undetectable after day 5 (data not shown).
FIG. 1.
Microbiological profile of tobacco fermentation as determined by plate counts of major microbial groups. Symbols: ▪, total aerobic bacteria; ⧫, endospore-forming bacteria; •, lactic acid bacteria; ▴, yeasts. Shuffling of the tobacco bulk was performed on days 5, 10, and 15, as shown by arrows.
Analysis of microbial community of fermenting tobacco by culture-independent methods.
To investigate the species composition and the temporal dynamics of the tobacco bacterial community during an 18-day fermentation cycle, a time course profiling of partial domains of the eubacterial 16S rRNA gene was performed by DGGE and SSCP. Both the DGGE and the SSCP profiles with the Bacteria-specific primers revealed a relatively low complexity, with the predominance of a few species (Fig. 2 and 3A). Qualitatively, the banding profiles denoted a bacterial community that evolved from day 1 (start) to day 5, becoming stable thereafter and exhibiting only minor differences compared with the fermentation end-point (day 18). This finding was consistent with the chemical-physical parameters (Table 2) and with the population density profile of the major microbial groups associated with the process (Fig. 1). The most evident variation of DGGE profiles was observed on days 5, 10, and 15, concomitant with tobacco shuffling.
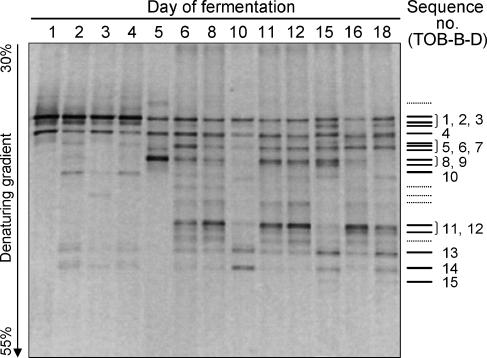
FIG. 2.
DGGE analysis of the eubacterial community during tobacco fermentation. DGGE fingerprints are of the V3 region of the 16S rRNA gene amplified with primers 341-FGC and 518-R from DNA samples extracted at different times, as indicated. The formamide-urea denaturing gradient ranged from 30% to 55%. Bands excised and successfully identified by sequencing (Table 3) are reported as continuous lines and sequentially numbered from top to bottom. Dotted lines show the bands for which the reamplification step failed. Shuffling of the tobacco bulk was performed on days 5, 10, and 15.
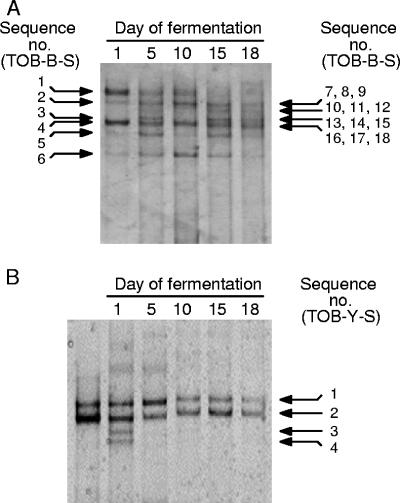
FIG. 3.
SSCP analysis of the microbial community during tobacco fermentation. (A) Profile with the Bacteria-specific primer pair Com1-F/Com2-R. (B) Profile with the Eukarya-specific primer pair FUN18S1-F/FUN18S2-R. The day of sampling is indicated above each lane. Arrows mark the position of sequentially numbered bands excised and identified by DNA sequencing (Table 3). The four groups of bands TOB-B-S7 to -9, TOB-B-S10 to -12, TOB-B-S13 to -15, and TOB-B-S16 to -18 (A, right) could not be resolved by SSCP, and the corresponding sequences were determined by RFLP analysis as detailed in the text. B, left lane, shows the profile obtained with DNA extracted from D. hansenii TOB-Y7 cultures (Table 5), used as an internal control.
The relatively low complexity of partial 16S rRNA gene profiles prompted an attempt to characterize individual DNA bands. The nucleotide sequence for 15 dominant DGGE bands (Fig. 2, TOB-B-D1 to TOB-B-D15) was determined. All DGGE-derived sequences were assigned to the group of gram-positive bacteria based on a combination of BLAST searches and phylogenetic analysis of sequences (Table 3; see also Fig. S1 in the supplemental material). Eight sequences were referred to members of the genera Aerococcus, Facklamia, Weissella, Alkalibacterium, and Lactobacillus (order Lactobacillales), and three were referred to Bacillus and Salinicoccus (order Bacillales) within the low-G+C group of gram-positive bacteria (Firmicutes). One sequence (TOB-B-D6) was similar to the uncharacterized Firmicutes sp. oral clone BX005 (41) and phylogenetically close to the genus Marinilactibacillus (see Fig. S1 in the supplemental material). The remaining three sequences matched the 16S rRNA gene sequences of the genera Corynebacterium and Yania within the high-G+C gram-positive bacteria (Actinobacteria). Some DGGE bands were observed during the entire tobacco fermentation cycle, while others showed a definite timing. TOB-B-D1, TOB-B-D2, and TOB-B-D4 (Aerococcus- and Weissella-related organisms) characterized the early stages of the fermentation process, while TOB-B-D7 to TOB-B-D9 (Lactobacillus-related organisms) appeared between day 5 and day 15, becoming undetectable on day 18. On the other hand, TOB-B-D10 to TOB-B-D12 (thermotolerant microorganisms belonging to the Bacillaceae family) and, to a lesser extent, TOB-B-D13 to TOB-B-D15 (corynebacteria) were more easily detectable during later stages of tobacco fermentation (Fig. 2).
TABLE 3.
Sequence similarities between rRNA gene sequences of uncultured tobacco fermentation-associated microorganisms and those of the closest relatives in the NCBI database
| Sequence | Closest relative(s) | Accession no. | Identity (%) | E value |
|---|---|---|---|---|
| TOB-B-D1 | Aerococcus viridans | M58797 | 100 | 1e-103 |
| TOB-B-D2 | A. viridans | M58797 | 97.9 | 5e-94 |
| TOB-B-D3 | Facklamia tabacinasalis | Y17820 | 99.5 | 3e-101 |
| TOB-B-D4 | Weissella hellenica | AB015642 | 99.5 | 3e-101 |
| TOB-B-D5 | Alkalibacterium iburiense | AB188091 | 97.4 | 1e-91 |
| TOB-B-D6 | Firmicutes sp. oral clone BX005 | AY005049 | 96.9 | 3e-89 |
| TOB-B-D7 | Lactobacillus sp. | AF305930 | 98.9 | 8e-99 |
| TOB-B-D8 | Lactobacillus sp. | AF316587 | 99.0 | 5e-97 |
| TOB-B-D9 | Lactobacillus sp. | AF305930 | 97.4 | 5e-94 |
| TOB-B-D10 | Salinicoccus sp. | AY536543 | 99.5 | 3e-101 |
| TOB-B-D11 | Bacillus pseudofirmus | AB201799 | 97.9 | 1e-94 |
| TOB-B-D12 | Bacillus pumilus | AF526907 | 97.4 | 1e-91 |
| TOB-B-D13 | Yania flava | AY684123 | 97.7 | 7e-87 |
| TOB-B-D14 | Corynebacterium ammoniagenes | X84440 | 99.4 | 2e-93 |
| TOB-B-D15 | Y. flava | AY684123 | 98.9 | 1e-91 |
| TOB-B-S1 | A. viridans | M58797 | 100 | 0.0 |
| TOB-B-S2 | C. ammoniagenes | X84440 | 100 | 0.0 |
| TOB-B-S3 | Weissella cibaria | AJ295989 | 98.8 | 0.0 |
| Weissella confusa | AB023241 | 98.8 | 0.0 | |
| TOB-B-S4 | A. viridans | M58797 | 100 | 0.0 |
| TOB-B-S5 | W. cibaria | AJ295989 | 98.8 | 0.0 |
| W. confusa | AB023241 | 98.8 | 0.0 | |
| TOB-B-S6 | C. ammoniagenes | X84440 | 100 | 0.0 |
| TOB-B-S7 | Bacillus subtilis | AY030331 | 100 | 0.0 |
| TOB-B-S8 | B. pumilus | AB098578 | 100 | 0.0 |
| TOB-B-S9 | Uncultured bacterium B5 | AF317373 | 100 | 0.0 |
| TOB-B-S10 | B. subtilis | AJ830709 | 100 | 0.0 |
| Bacillus mojavensis | AY030339 | 100 | 0.0 | |
| TOB-B-S11 | Bacillus cereus | AY138271 | 100 | 0.0 |
| Bacillus thuringiensis | DQ328632 | 100 | 0.0 | |
| TOB-B-S12 | Bacillus licheniformis | AF234859 | 100 | 0.0 |
| TOB-B-S13 | B. subtilis | AY030331 | 100 | 0.0 |
| TOB-B-S14 | B. pumilus | AB098578 | 100 | 0.0 |
| TOB-B-S15 | Uncultured bacterium B5 | AF317373 | 100 | 0.0 |
| TOB-B-S16 | B. subtilis | AJ830709 | 100 | 0.0 |
| B. mojavensis | AY030339 | 100 | 0.0 | |
| TOB-B-S17 | B. cereus | AY138271 | 100 | 0.0 |
| B. thuringiensis | DQ328632 | 100 | 0.0 | |
| TOB-B-S18 | Bacillus licheniformis | AF234859 | 100 | 0.0 |
| TOB-Y-S1 | Debaryomyces hansenii | AB106349 | 100 | 0.0 |
| Debaryomyces vanrijiae | AB054267 | 100 | 0.0 | |
| Debaryomyces prosopidis | AB054277 | 100 | 0.0 | |
| Debaryomyces maramus | AB054261 | 100 | 0.0 | |
| TOB-Y-S2 | D. hansenii | AB106349 | 100 | 0.0 |
| D. vanrijiae | AB054267 | 100 | 0.0 | |
| D. prosopidis | AB054277 | 100 | 0.0 | |
| D. maramus | AB054261 | 100 | 0.0 | |
| TOB-Y-S3 | Trichosporon cutaneum | AB001753 | 99.8 | 0.0 |
| TOB-Y-S4 | T. cutaneum | AB001753 | 99.8 | 0.0 |
| TOB-B-R1 | Uncultured bacterium 13-20:1 | AB029412 | 99.2 | 0.0 |
| TOB-B-R2 | Uncultured bacterium 4-49 | AB034713 | 98.9 | 0.0 |
| TOB-B-R3 | Uncultured bacterium pHS3 | AF493257 | 99.0 | 0.0 |
| TOB-B-R4 | Corynebacterium casei | DQ361022 | 99.3 | 7e-156 |
| TOB-B-R5 | Brevibacterium stationis | AJ620367 | 98.6 | 3e-152 |
| TOB-Y-R1 | Candida etchellsii | AB018142 | 98.0 | 5e-110 |
The same sequence-based strategy was applied to identify major SSCP bands. In SSCP experiments the Bacteria-specific primers amplify a 16S rRNA gene region (ca. 400 bp) that is larger than in DGGE experiments (ca. 200 bp), generally increasing the probability of taxonomic assignation (Table 3; compare E-values). SSCP worked successfully for samples representative of earlier fermentation stages (days 1 to 10) (Fig. 3A). However, for samples from later stages (days 15 and 18), which generated more complex patterns, groups of bands had to be excised and cloned to generate libraries according to the strategy described in Materials and Methods. The data from this combined approach are summarized in Table 3. In agreement with DGGE results (Fig. 2), the SSCP analysis demonstrated the presence of 16S rRNA gene sequences related to the lactic acid bacterium Aerococcus viridans, particularly evident during earlier fermentation stages, and to Corynebacterium ammoniagenes, whose signal increased progressively during fermentation up to day 10. An additional lactic acid bacterium, taxonomically related to the genus Weissella (Weissella confusa or Weissella cibaria), also characterized the SSCP patterns. During later fermentation stages (days 15 to 18) the SSCP analysis confirmed the presence of thermotolerant microorganisms belonging to Bacillaceae (related to Bacillus subtilis and/or Bacillus mojavensis, Bacillus pumilus, Bacillus cereus/Bacillus thuringiensis, and Bacillus licheniformis). In addition, 16S rRNA gene sequences (TOB-B-S9 and TOB-B-S15) closely related to that of an uncultured microorganism from feedlot manure, belonging to the Enterococcaceae or Carnobacteriaceae families of Lactobacillales (Table 3; see also Fig. S2 in the supplemental material) were detected.
The SSCP profiles with Eukarya-specific primers exhibited very low complexity (Fig. 3B). DNA bands could be directly excised from the gel and subjected to cloning and nucleotide sequence analysis. The results revealed the presence of spoilage yeast(s) belonging to the genus Debaryomyces, which was particularly abundant during the early stage of fermentation (days 1 to 5). A band referable to Trichosporon cutaneum could also be detected in the starting sample (Fig. 3B).
Because of the complexity and poor resolution of SSCP patterns from late fermentation samples, a library of pooled PCR products obtained with Bacteria-specific SSCP primers from days 15 and 18 was generated, and ca. 100 clones were screened by RFLP. This additional approach not only confirmed the presence of all DNA sequences previously detected by SSCP, but also led to the discovery of a few additional ones (Table 3). Two 16S rRNA gene sequences (TOB-B-R1 and TOB-B-R2), taxonomically related to those of two uncultured microorganisms from compost (23), were found (see Fig. S2 in the supplemental material). Another sequence (TOB-B-R3) was similar to that of an uncultured bacterium from the maize phyllosphere (26), taxonomically close to W. confusa or W. cibaria (see Fig. S2 in the supplemental material). In addition, the same library-based approach with Actinobacteria- or Eukarya-specific primers (Table 1) led to the detection of additional clones taxonomically related to Actinomycetales (best matches with Corynebacterium casei and Brevibacterium stationis) and to the yeast Candida etchellsii, respectively (Table 3).
Isolation and identification of microorganisms from fermenting tobacco leaves by culture-based methods.
TSA, MRS, and PDA media were used to isolate and identify individual members of the microbial community by culture-based methods. A mesophilic temperature was chosen for microbial isolation (30 to 37°C for bacteria and 30°C for yeasts), as suggested from optimum growth temperatures of taxa detected by culture-independent methods. A total of 200 apparently different colonies derived from samples taken during the whole course of fermentation were examined. The microbial isolates were tentatively assigned to 69 preliminary phenons by morphology, motility, staining, growth, and basic biochemical properties and then identified by combining specific phenotypic tests, including the API systems and 16S or 18S rRNA gene sequencing. Multiple isolates showing identical phenotype and 16S or 18S rRNA gene sequences were considered only once, lowering the number of distinct isolates to 41. Definitive identification was assigned when molecular data were fully consistent with phenotypic traits, while presumptive identification denotes partial inconsistency between molecular and phenotypic data and/or lack of informative biochemical results. On this basis, distinct isolates were referred to a total of 17 taxa, including 5 definitive and 8 presumptive bacterial species (10 belonging to Firmicutes, 2 to Actinobacteria, and 1 to Proteobacteria) plus 3 definitive and 1 presumptive yeast species (Tables 4 and 5 and text below). No mold was detected during the whole fermentation.
TABLE 4.
Growth and biochemical properties of representative bacterial isolates from fermenting tobaccoa
| Isolate | 16S rRNA gene sequenceb
|
API identificationc
|
Growthd
|
Hydrolysis of:
|
Nitrate reduction | Utilization of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Highest similarity (accession no.) | Identity (%) | Species | Identity (%) | pH range | Tmax (°C) | Starch | Pectin | Casein | CMC | Urea | Nitrate | Nitrite | Ammonium | ||
| TOB-B1 | B. subtilis (AY881640) | 99.3 | B. subtilis | 97.4 | 4.7-8.7 | 60 | ++ | ++ | ++ | ++ | − | + | + | − | + |
| TOB-B5 | B. subtilis (AY030331) | 99.7 | B. subtilis | 91.8 | 4.7-8.7 | 60 | + | + | ++ | + | − | + | − | + | + |
| TOB-B6 | B. subtilis (AY030331) | 99.8 | B. subtilis | 95.3 | 4.7-8.7 | 60 | ++ | + | ++ | + | − | − | + | − | + |
| TOB-B12 | B. subtilis (AY881640) | 99.5 | B. pumilus | 99.9 | 5.7-8.7 | 55 | − | − | ++ | + | − | + | + | − | + |
| TOB-B14 | B. subtilis (AY162133) | 98.0 | B. subtilis | 96.0 | 4.7-8.7 | 60 | + | + | ++ | + | − | + | − | − | + |
| TOB-B15 | B. subtilis (AB018486) | 100 | B. subtilis | 94.6 | 4.7-8.7 | 60 | + | + | ++ | + | − | + | − | − | ± |
| TOB-B30 | B. subtilis (AY030331) | 99.9 | B. subtilis | 96.9 | 4.7-8.7 | 50 | + | + | ++ | + | − | + | − | + | + |
| TOB-B36 | B. subtilis (AB016721) | 98.9 | B. subtilis | 96.9 | 4.7-8.7 | 60 | + | + | ++ | + | − | + | − | − | + |
| TOB-B39 | B. subtilis (DQ779883) | 99.6 | B. subtilis | 88.8 | 4.7-8.7 | 60 | − | + | + | + | − | + | − | − | + |
| TOB-B42 | B. subtilis (AY881640) | 99.2 | B. subtilis | 95.3 | 5.7-8.7 | 60 | + | + | ++ | + | − | + | − | − | ± |
| TOB-B60 | B. subtilis (AY030331) | 100 | B. subtilis | 95.3 | 5.7-8.7 | 60 | + | ± | ++ | + | − | + | + | + | + |
| TOB-B61 | B. subtilis (AJ830709) | 100 | B. subtilis | 96.9 | 5.7-8.7 | 60 | + | + | ++ | + | − | + | + | − | + |
| TOB-B3 | B. pumilus (AY919667) | 99.4 | B. pumilus | 99.7 | 4.7-8.7 | 60 | − | − | + | − | − | − | ± | ± | ± |
| TOB-B4 | B. pumilus (AB098578) | 99.9 | B. pumilus | 99.9 | 4.7-8.7 | 60 | − | − | + | − | − | − | + | ± | + |
| TOB-B7 | B. pumilus (AY030327) | 99.5 | B. pumilus | 97.3 | 4.7-8.7 | 60 | − | − | + | + | − | − | − | − | + |
| TOB-B9 | B. pumilus (AY188840) | 99.5 | B. pumilus | 99.9 | 5.7-8.7 | 55 | − | − | + | + | − | − | − | − | + |
| TOB-B11 | B. pumilus (AY030327) | 98.8 | B. pumilus | 99.9 | 4.7-8.7 | 60 | − | − | ++ | + | − | − | − | − | + |
| TOB-B38 | B. pumilus (AY167882) | 99.5 | B. pumilus | 97.1 | 4.7-8.7 | 60 | − | + | + | − | − | − | − | − | + |
| TOB-B40 | B. pumilus (DQ232736) | 99.3 | B. licheniformis | 97.5 | 4.7-8.7 | 60 | − | − | − | − | − | + | − | − | − |
| TOB-B41 | B. pumilus (AB098578) | 99.9 | B. pumilus | 99.9 | 4.7-8.7 | 60 | − | − | + | − | − | + | − | − | ± |
| TOB-B63 | B. pumilus (AB098578) | 100 | B. pumilus | 99.9 | 5.7-8.7 | 60 | − | − | + | − | − | − | − | ± | − |
| TOB-B17 | B. licheniformis (AY786999) | 99.9 | B. licheniformis | 99.9 | 5.7-8.7 | 60 | + | − | + | ++ | − | + | + | − | + |
| TOB-B19 | B. licheniformis (X68416) | 100 | B. licheniformis | 98.6 | 5.7-8.7 | 50 | + | − | + | ++ | − | + | + | ± | + |
| TOB-B24 | B. licheniformis (AJ586340) | 99.7 | B. licheniformis | 86.0 | 5.7-8.7 | 60 | + | − | + | + | − | + | ++ | + | ± |
| TOB-B27 | B. licheniformis (DQ238044) | 98.5 | B. licheniformis | 98.6 | 5.7-8.7 | 50 | + | − | + | + | − | + | ++ | + | − |
| TOB-B2 | B. mojavensis (AB021191) | 99.7 | B. subtilise | 97.0 | 4.7-8.7 | 60 | ++ | ++ | ++ | ++ | − | − | − | − | + |
| TOB-B64 | B. thuringiensis (AY741718) | 99.9 | B. cereuse | 93.2 | 4.7-8.7 | 60 | − | − | − | − | − | − | − | − | − |
| TOB-B65 | B. thuringiensis (AF290545) | 99.9 | B. licheniformise | 98.8 | 5.7-8.7 | 60 | − | ± | + | + | − | + | + | ± | + |
| TOB-B28 | Bacillus sp. (AY553066) | 97.6 | B. subtilis | 87.1 | 6.7-8.7 | 55 | − | − | ± | − | − | + | − | − | − |
| TOB-B70 | Staphylococcus cohnii (AY395015) | 99.8 | NT | NT | 5.7-8.7 | 60 | − | − | − | − | + | + | − | − | − |
| TOB-B71 | Jeotgalicoccus halotolerans (AY028925) | 97.1 | NT | NT | 6.7-8.7 | 60 | − | − | − | − | − | − | − | − | − |
| TOB-B67 | A. viridans (AY707779) | 99.7 | NT | NT | 6.7-8.7 | 60 | − | − | − | − | − | − | − | − | − |
| TOB-B31 | C. ammoniagenes (X84440) | 99.7 | Corynebacterium pseudodiphtheriticume | 97.9 | 6.7-8.7 | 50 | − | − | − | − | + | + | − | − | − |
| TOB-B32 | C. ammoniagenes (X84440) | 97.8 | Corynebacterium propinquume | 96.9 | 6.7-8.7 | 50 | − | − | − | − | − | + | − | − | − |
Symbols and abbreviations: −, negative; ±, weakly positive; +, positive; ++, strongly positive; NT, not tested; Tmax, maximum growth temperature.
NCBI-BLAST values. These were fully consistent with those determined by the Seqmatch tool of the RDP (data not shown).
Tests were performed using the API 50CHB/E and API Coryne systems.
Determined in TSA medium.
The species identified by 16S rRNA gene sequencing is not included in the API 50CHB/E and API Coryne systems.
TABLE 5.
Growth and biochemical properties of yeasts isolated from fermenting tobacco
| Isolate | 18S rRNA gene sequence
|
API identificationa
|
NaCl (% wt/vol) toleranceb | Tmax (°C)b | pH rangeb | Generation time (h)c | ||
|---|---|---|---|---|---|---|---|---|
| Closest relative (accession no.) | Identity (%) | Species (code) | Identity (%) | |||||
| TOB-Y4 | C. parapsilosis (AY055857) | 99.8 | C. parapsilosis (6756171) | 92.2 | ≤15 | 35 | 3.0-9.0 | 4.7 |
| TOB-Y5 | T. ovoides (AB001765) | 99.6 | C. laurentii (6777773) | 46.2 | ≤9 | 30 | 5.0-9.0 | 28.0 |
| TOB-Y6 | C. rugosa (AB013502) | 99.3 | C. rugosa (6442100) | 98.1 | ≤12 | 45 | 3.0-9.0 | 4.7 |
| TOB-Y7 | D. hansenii (AB070854) | 100 | D. hansenii (6776771) | 99.8 | ≤15 | 30 | 5.0-9.0 | 5.8 |
Tests were performed using the API 20C AUX system; because of the absence of a T. ovoides code in the database, the identity value refers to the best-fitting species, Cryptococcus laurentii.
Determined in PDA medium.
Calculated by A600 measurements during the exponential phase (6 to 24 h) after inoculation in TB medium at pH 5.2 (see Materials and Methods).
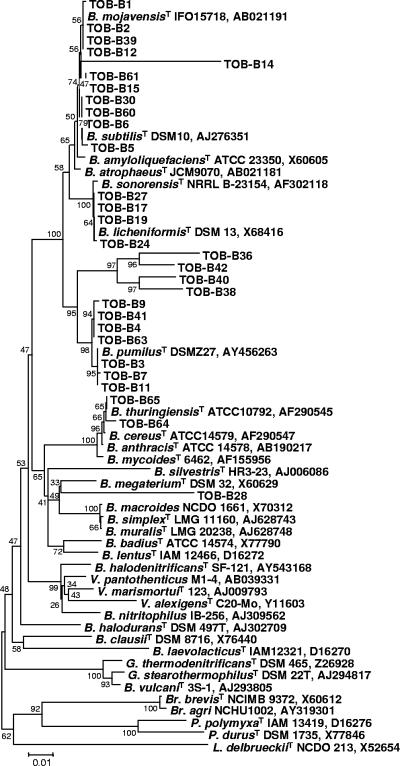
Among bacterial isolates, there was a clear prevalence (29/37) of Bacillaceae, and 23 of these could definitively be identified at the species level (11 as B. subtilis, 8 as B. pumilus, and 4 as B. licheniformis). Phylogenetic analysis confirmed the taxonomic allocation of Bacillus spp., with few ambiguities. This was the case for TOB-B36 and TOB-B42 (B. subtilis), which belonged to a separate cluster sharing a common node with the B. pumilus group (Fig. 4). Moreover, TOB-B1 and TOB-B39 (B. subtilis) branched in the B. mojavensis lineage, although they also appeared to be closely related to the B. subtilis type strain. Discordant results between the API50CHB/E system and 16S rRNA gene-based identification were observed for TOB-B12 and TOB-B40 isolates. Based on phylogenetic analysis, TOB-B12 can most likely be assigned to the B. subtilis-B. mojavensis group, while TOB-B40 fell into a distinct cluster, proximal to B. pumilus (Fig. 4). Moreover, starch hydrolysis and nitrate reduction tests, which are discriminatory for these species, provided ambiguous results (Table 4). Such discordance was also observed for the isolate TOB-B28, identified with low (87.1%) probability as B. subtilis by the API system but showing the highest 16S rRNA gene sequence homology with a still-uncharacterized Bacillus sp. strain. The unique phenotypic traits and phylogenetic position of TOB-B28 do not allow any confident identification for this isolate (Table 4). We also noted that the API system failed to identify or provided false readings for isolates that do not belong to any of the species for which the method was developed. This is true for those isolates which showed the highest 16S rRNA gene sequence similarity with B. mojavensis (TOB-B2), B. thuringiensis (TOB-B64 and TOB-B65), and C. ammoniagenes (TOB-B31 and TOB-B32). Indeed, two isolates were referable to C. ammoniagenes based on 16S rRNA gene sequence, but only for TOB-B31 was the identification confirmed by a urease-positive reaction (Table 4). Additional isolates were recovered from tobacco samples and presumptively assigned by 16S rRNA gene sequencing as being similar to A. viridans, Staphylococcus cohnii, and Jeotgalicoccus halotolerans (Bacillales), Pediococcus pentosaceus (Lactobacillales), Brevibacterium sp. (Actynomycetales), and Brevundimonas sp. (Alphaproteobacteria, Caulobacterales), in accordance with preliminary phenotypic tests (Table 4 and data not shown).
FIG. 4.
Phylogenetic relationships inferred from the alignment of partial 16S rRNA gene sequences (nt 123 to 1333 relative to the E. coli rrnB gene; see Materials and Methods for details) of 29 Bacillaceae isolated from fermenting tobacco and 34 reference strains of representative species belonging to the genera Bacillus, Brevibacillus, Geobacillus, Paenibacillus, Virgibacillus, and Lactobacillus. GenBank accession numbers of reference sequences are reported. The T superscript stands for type strain. The tree was constructed using the Kimura two-parameter algorithm and the neighbor-joining method. The bar indicates 1% sequence divergence. Bootstrap values (expressed as percentages of 1,000 replications) are reported at each node. Species are abbreviated as follows: B., Bacillus; Br., Brevibacillus; G., Geobacillus; P., Paenibacillus; V., Virgibacillus; L., Lactobacillus.
Based on identification by conventional methods and 18S rRNA gene sequence analysis, the yeast isolates were attributed to the species Debaryomyces hansenii, Candida parapsilosis, Candida rugosa and, presumptively, Trichosporon ovoides (Table 5).
Growth and biochemical properties of microorganisms involved in tobacco fermentation.
To investigate the possible role of individual microbial isolates in tobacco fermentation, several growth and biochemical properties were evaluated, including the effects of pH, temperature, and salt concentration on growth and the ability to hydrolyze urea, casein, and polysaccharides or to utilize various nitrogen sources (Table 4).
The results demonstrated the overall ability of Bacillus isolates to grow over a wide range of pHs (4.7 to 8.7) and temperatures (up to 60°C). In general, B. licheniformis and B. subtilis strains shared the ability to reduce nitrate with and without N2 gas production, respectively, while B. pumilus-related isolates did not. B. subtilis isolates except TOB-B12 (recognized as B. pumilus by the API system) were able to hydrolyze the three polysaccharides and showed strong proteolytic activity on casein. Both B. pumilus and B. licheniformis lacked pectinolytic activity and showed moderate casein hydrolysis. B. pumilus-related strains revealed a heterogeneous behavior with regard to CMC hydrolysis. Growth on nitrate, nitrite, or ammonium as the only nitrogen source was highly variable within individual Bacillus species, with B. licheniformis growing on nearly all tested substrates and B. subtilis and B. pumilus primarily utilizing ammonium and, rarely, other nitrogen sources. Except for nitrate reduction, the single B. mojavensis-related isolate (TOB-B2) showed the same metabolic pattern as most B. subtilis strains, in agreement with assimilative and biochemical properties in the API 50CHB/E system. The two isolates which showed highest similarity with B. thuringiensis by 16S rRNA gene sequencing (TOB-B64 and TOB-B65) were endowed with unexpectedly different hydrolytic patterns and nitrogen metabolism, in spite of their close phylogenetic relatedness with the B. thuringiensis type strain (Fig. 4).
Among Staphylococcaceae, no relevant metabolic data were obtained for S. cohnii and a J. halotolerans-related isolate, although the urease test related the former to S. cohnii subsp. urealyticum.
Coryneform bacteria, including C. ammoniagenes-related species, were able to grow under neutral or alkaline conditions (pH range of 6.7 to 8.7) at temperatures of ≤50°C. None of them was able to degrade starch, pectin, casein, or CMC or to grow on nitrite, nitrate, or ammonium as a nitrogen source, while both reduced nitrate without further evolution, resulting in nitrite accumulation. Only TOB-B31 exhibited urease activity, which represents a discriminatory trait for C. ammoniagenes (25) (Table 4).
All bacterial isolates belonging to the order Bacillales (genera Bacillus, Staphylococcus, and Jeotgalicoccus) grew in TSA medium supplemented with up to 10% (wt/vol) NaCl, while isolates related to A. viridans and C. ammoniagenes failed to grow at >7% salt concentrations (data not shown). Growth and metabolic tests were not performed with isolates presumptively identified as P. pentosaceus, Brevibacterium sp., and Brevundimonas sp.
Growth properties of yeast isolates, including NaCl tolerance, temperature, pH, and generation time in TB medium were determined (Table 5). D. hansenii, C. parapsilosis, and C. rugosa showed rapid growth, with generation times ranging from 4.7 to 5.8 h, while T. ovoides grew very slowly in TB medium. All strains well tolerated the pH range typical of the fermentation process (from 5 to 9), with Candida spp. growing at even more acidic pH. The maximum growth temperature was 30 to 35°C with the exception of C. rugosa, which grew up to 45°C. Moreover, C. parapsilosis and D. hansenii were endowed with the highest halotolerance, although all isolates showed a remarkable ability to grow in the presence of up to 9% (wt/vol) NaCl (Table 5).
Assuming that (i) the acidic pH at the beginning of fermentation fosters the development of yeasts and (ii) consumption of available organic acids by yeast cells contributes to the pH rise during fermentation, we investigated the role of D. hansenii, C. parapsilosis, and C. rugosa in metabolism (production or utilization) of oxalic, citric, and malic acids using TB medium as a tobacco surrogate. T. ovoides was not tested due to its high generation time in TB medium (Table 5). Both citric and malic acids were consumed by all three yeast species, although to a highly variable extent. C. rugosa utilized almost exclusively malic acid, while C. parapsilosis and D. hansenii utilized both citric and malic acids, with some preference of C. parapsilosis for citric acid and of D. hansenii for malic acid. Conversely, oxalic acid showed a modest increase during yeast growth (see Fig. S3 in the supplemental material). Remarkably, the pH of TB medium rose progressively from 5.2 to ca. 7.5 during the first 24 h growth of all three yeasts and remained stable thereafter (data not shown).
DISCUSSION
Toscano cigars are a traditional product whose fermentation methods and sensory characteristics greatly differ from other commercial cigars and could be assimilated to fermented smokeless products (60). Although consumed by the vast majority of Italian cigar smokers, the microbiological aspects of its traditional fermentation are still largely unknown (17). In the present study, the microbial community of Toscano fermentation was studied for the first time by using a combination of classical and molecular techniques.
The traditional technology of Toscano cigar tobacco fermentation is based on a semicontinuous process whose physical-chemical profile suggests a model resembling that of the well-known solid waste composting process (11, 23, 39, 51, 52), being characterized by intrinsic variation of chemical and physical parameters. Major sources of variability are the local heterogeneity of the bulk, the seasonal fluctuation of temperature, and inconstant shuffling times, all reflecting the small-scale, semiartisanal, nonstandardized nature of the tobacco fermentation process. Nevertheless, a general trend can be inferred by the analysis of 10 fermentation cycles, including increase of pH, temperature, nitrite, and TSNA and decrease of reducing sugar, nitrate, ammonia, and moisture content (see Table S1 in the supplemental material). As shown for a representative fermentation cycle (Table 2), moisture content (ranging from 55 to 50%) and shifts in temperature (from RT to 65°C) and pH (from 6.0 to 8.8) are in the same ranges as those observed in composting. Distinctive features include a low C/N ratio (considering 25 as the optimal value as suggested for composting [15]), high levels of nitrate and ammonia, low levels of reducing sugars, and high levels of malic and citric acids, in agreement with previously reported data (56). A composite microbial population that increases up to 6 orders of magnitude (from 107 to 1013 CFU/g) sustains this process (Fig. 1). Microbiological investigations of the fermenting tobacco bulk by both culture-independent and culture-based analyses support this view.
SSCP and RFLP analyses of libraries revealed a simple yeast population composed of a few species. Sequencing of the partial 18S rRNA gene identified three yeast genera (Candida, Debaryomyces, and Trichosporon). Species belonging to these genera, namely D. hansenii, C. rugosa, C. parapsilosis and, presumptively, T. ovoides, were the only yeasts recovered from fermenting tobacco by culture-based analysis, highlighting the complete superimposition of cultural and molecular results for the tobacco-associated yeast population.
Preliminary microbial count data (Fig. 1) suggested that the Bacteria community of fermenting tobacco was considerably more diverse than the yeast community. It is commonly held that the use of different molecular techniques and/or different “universal” primer sets affect the community profile detectable by PCR-based methods (22, 49). Therefore, we approached the analysis of the structure and dynamics of the eubacterial community by combining DGGE with SSCP. The V3 region of the 16S rRNA gene was selected to enhance the range of DGGE resolution (63), while longer DNA fragments encompassing the V4 to V5 region were generated to improve the phylogenetic inference through SSCP analysis.
Bacterial species most frequently detected by culture were closely related to taxa previously identified by culture-independent methods (Table 4). Interestingly, 16S rRNA gene sequencing of individual isolates revealed remarkable intraspecific diversity among Bacillaceae which was otherwise undetectable by DGGE and/or SSCP. For most Bacillus isolates, differences in metabolic profiles were consistent with sequence heterogeneity, suggesting that the observed diversity was not biased by sequence polymorphisms of high-copy-number rrn operons in Bacillus species (1).
As a whole, microbial diversity detected on laboratory media was in good agreement with the community structure inferred by culture-independent methods. Both approaches depicted a bacterial population consisting of gram-positive bacteria with a prevalence of species belonging to Bacillaceae, Aerococcaceae (Firmicutes), and Corynebacteriaceae (Actinobacteria). Nevertheless, some differences were seen between the different methods. Firstly, DGGE resolved a group of sequences whose phylogenetic allocation indicates the presence of Lactobacillales (Alkalibacterium, Marinilactibacillus-related clone BX005, and Lactobacillus) undetectable by both SSCP-RFLP and culture. Other genera, namely Facklamia, Weissella, and Yania, were identified by molecular profiling but not by culture. This can be ascribed to special growth requirements of these bacteria, as in the case of Alkalibacterium, which grows at pH 9 to 12 (38), and Yania, which requires a high salt concentration for growth (32). Conversely, few bacterial isolates that were sporadically recovered from tobacco (e.g., Brevundimonas sp., P. pentosaceus-related isolate, and S. cohnii) did not match any of the taxa resolved by DGGE or SSCP. The actual population density and the biological role in tobacco fermentation of such sporadic isolates cannot be assessed on available data.
A crucial point is that bacteria referable to Bacillus species, A. viridans, and C. ammoniagenes, were not only detected by DGGE and SSCP community profiling but also were cultured from the tobacco bulk (Fig. 2 and 3 and Tables 3 and 4). Since a correlation exists between culture-independent (qualitative) detection, culturability, and activity of bacteria in natural environments (reference 12 and references therein), it can reasonably be proposed that these species represent the major bacterial components of the tobacco ecosystem during fermentation.
The Toscano cigar tobacco fermentation process implies changes in composition and activity of the microbial community concomitant with periodic shuffling of the fermenting bulk. In fact, time course analysis of the prokaryotic community by DGGE and, to a lesser extent, SSCP fingerprinting shows remarkable variation in correspondence of shuffling times (days 5, 10, and 15), consistent with the semicontinuous nature of the process. Three phases can be distinguished in this process.
Early phase.
Both culture-independent and culture-based approaches suggest a major role for yeasts, Staphylococcaceae, and Lactobacillales during the early stages of tobacco fermentation. The slightly acidic pH and the mesophilic temperature (RT) promote growth of these groups of microorganisms which are supposed to rapidly metabolize reducing sugars and available malic and citric acids. Organic acids are important constituents of tobacco because they directly influence the pH during fermentation and, consequently, smoke aroma and taste (34). The pH shift from acidic to alkaline values is a hallmark of Toscano cigar tobacco fermentation; it defines the final cigar basic characteristics and distinguishes cigars from cigarettes which are made with acidic nonfermented tobacco. Oxalic acid is generated during tobacco fermentation (Table 2) and represents an undesirable end product. In yeasts and filamentous fungi, biosynthesis of oxalic acid occurs as a by-product of malic and citric acid metabolism via hydrolysis of oxaloacetic acid and glyoxylate oxidation, respectively (16, 44). Oxalic acid production in TB medium was maximal with C. rugosa and D. hansenii, which preferentially catabolized malic acid, while it was barely detectable with C. parapsilosis, which preferentially utilized citric acid (see Fig. S3 in the supplemental material).
Among Staphylococcaceae, S. cohnii and a strain belonging to the genus Jeotgalicoccus were isolated. Notably, DGGE analysis detected the presence of Salinicoccus, a Jeotgalicoccus-related taxon, during the first days of fermentation (Fig. 2). This stage is largely dominated by Lactobacillales belonging to four distinct families. DNA signals corresponding to A. viridans (Aerococcaceae) and Weissella spp. (Leuconostocaceae) characterized both DGGE and SSCP patterns of the early fermentation stages, though they were also detectable at later time points (Fig. 2 and 3 and Table 3). The DGGE profile also demonstrated the presence of DNA belonging to a microorganism taxonomically close to A. iburiense (Carnobacteriaceae) (38) which persisted during the entire process. Signals corresponding to three Lactobacillus spp. (Lactobacillaceae) and a Marinilactibacillus-related species (Carnobacteriaceae) appeared at the end of the early phase.
The SSCP and culture analyses demonstrated that D. hansenii was the predominant yeast species (Fig. 2). Noteworthy is the specificity of the association between D. hansenii and several lactic acid and coryneform bacteria, including those we isolated from fermenting tobacco. These microbial consortia are typical of certain fermenting substrates that are characterized by a low C/N ratio, such as ripening cheeses (2, 4, 10, 31, 36, 43, 61), fresh sausages (7, 47), vegetables (35), and algae (57). An association between D. hansenii and P. pentosaceus, revealed for tobacco samples (Tables 4 and 5), has previously been described for sorghum fermentation (35), and the same yeast has been isolated from the surface microflora of many brick cheeses in association with several coryneform bacteria, including C. ammoniagenes (58). In these consortia, D. hansenii stimulates the metabolism of the lactic acid bacteria, providing them with a number of growth factors, including vitamins and amino acids. In semisoft cheeses with surface films and in mold-ripened cheeses, D. hansenii utilizes lactic acid as a main carbon source and produces ammonia by proteolysis, leading to an increase of pH that stimulates growth of other microorganisms during the second stage of maturation (2, 4). The predominance of D. hansenii in a number of fermented dairy products prevents spoilage by clostridial species through the production of antibacterial metabolites (14). In addition, D. hansenii produces a killer toxin that is active against various yeast species and molds (19, 27). By analogy, the presence of D. hansenii during the early stages of tobacco fermentation might facilitate the growth of lactic acid bacteria and prevent outgrowth of molds.
Middle phase.
Probably as a consequence of the activity of yeasts, Staphylococcaceae, and Lactobacillales, the temperature of fermenting tobacco bulk increases. Production of ammonia via proteolysis and utilization of the organic acids by both yeasts and bacteria might be responsible for the pH shift from acidic to alkaline values. Probably as a consequence of these changes, the population of the above taxa declines (Fig. 1 and 2). In contrast, endospore-forming low-G+C content gram-positive bacilli (several of which are also able to grow at slightly acidic pH [Table 4]) grow (Fig. 1 to 3). DNA signals corresponding to a number of Bacillus species characterized both DGGE and SSCP patterns of the middle fermentation phase, although they were detectable even at later time points (Fig. 2 and 3 and Table 3). Strains referable to B. subtilis, B. pumilus, B. licheniformis, B. mojavensis, and B. thuringiensis were frequently isolated (Table 4). Although mesophilic, most B. licheniformis, B. subtilis, and B. pumilus tobacco isolates were also able to grow at 60°C (Table 4). This finding, common to many other Bacillus spp. strains isolated from self-heating material (51), extends the maximum temperature for growth of these species previously reported (50).
Most of tobacco Bacillus spp. isolates hydrolyzed carbon sources less readily available than reducing sugars and organic acids and showed proteolytic activity (Table 4). In addition to proteolysis, they are expected to produce a large amount of ammonia by other mechanisms. Most of them, except B. pumilus and one of the two B. thuringiensis-related strains, are able to reduce nitrate to nitrite (Table 4). While nitrate reduction by B. licheniformis led to production of a considerable amount of gas (N2O and N2), in B. subtilis nitrite evolves to ammonia by assimilative and dissimilative (nitrite ammonification) pathways (21, 40).
Late phase.
As a consequence of the metabolic activities of these microorganisms, pH further increases promoting growth of moderately halotolerant and alkaliphilic Actinomycetales. Two DNA signals referable to Yania (best match, Y. flava, Micrococcineae, Yaniaceae) and one to Corynebacterium (best match, C. ammoniagenes, Corynebacterineae, Corynebacteriaceae) dominated the DGGE profile of the middle-late stages (Fig. 2 and Table 3). A sequence related to C. ammoniagenes was also detectable by SSCP during the entire process (Fig. 3). This species was frequently isolated from tobacco samples at later stages, in addition to an unspeciable urease-negative coryneform belonging to the same genus (TOB-B32) (Table 4). It is reasonable that C. ammoniagenes further increases the pH by means of urease activity and accumulates nitrite that does not evolve to gas or ammonia, likely contributing to TSNA generation. In addition to the above-mentioned Actinomycetales, a microorganism related to F. tabacinasalis (Lactobacillales, Aerococcaceae) was detected in late-phase samples by DGGE (Fig. 2 and Table 3). It is noteworthy that this bacterium has been isolated from powdered snuff tobacco, which is a typical alkaline pH tobacco product (8). However, because this microorganism was biochemically unreactive in common metabolic tests (8), little can be inferred from its presence in the process.
In conclusion, this study predicts a plausible and unprecedented model for Toscano cigar tobacco fermentation. Although the model does not take into due account some microorganisms that could not be cultivated or that are poorly characterized, it could provide a basis for future development of strategies aimed at minimizing the production of undesirable fermentation by-products, such as oxalic acid, nitrite, and TSNA.
Supplementary Material
Acknowledgments
This work was supported as part of the Progetto MIUR 593 (Rif. prot. 976, 06-02-2002) by Ministero per l'Università e la Ricerca (MIUR), Rome, Italy, “Applicazione delle nuove tecnologie-nanomateriali e biotecnologie-ai prodotti del tabacco” coordinated by British American Tobacco Italia.
P.A., F.I., G.V., and P.V. declare no conflict of interest in relation to the publication of results presented in this article.
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addis, E., G. H. Fleet, J. M. Cox, D. Kolak, and T. Leung. 2001. The growth, properties and interactions of yeasts and bacteria associated with the maturation of Camembert and blue-veined cheeses. Int. J. Food Microbiol. 69:25-36. [DOI] [PubMed] [Google Scholar]
- 3.Berber, I., and E. Yenidünya. 2005. Identification of alkaliphilic Bacillus species isolated from Lake Van and its surroundings by computerized analysis of extracellular protein profiles. Turk. J. Biol. 29:181-188. [Google Scholar]
- 4.Besancon, X., C. Smet, C. Chabalier, M. Rivemale, J. P. Reverbel, R. Ratomahenina, and P. Galzy. 1992. Study of surface yeast flora of Roquefort cheese. Int. J. Food Microbiol. 17:9-18. [DOI] [PubMed] [Google Scholar]
- 5.Brunnemann, K. D., and D. Hoffmann. 1991. Analytical studies on tobacco-specific N-nitrosamines in tobacco and tobacco smoke. Crit. Rev. Toxicol. 21:235-240. [DOI] [PubMed] [Google Scholar]
- 6.Cappuccino, J. G., and N. Sherman. 2001. Microbiology: a laboratory manual, 6th ed. Benjamin Cummings Publishing, San Francisco, Calif.
- 7.Cocolin, L., K. Rantsiou, L. Iacumin, R. Urso, C. Cantoni, and G. Comi. 2004. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 70:1883-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, M. D., R. A. Hutson, E. Falsen, and B. Sjödén. 1999. Facklamia tabacinasalis sp. nov., from powdered tobacco. Int. J. Syst. Bacteriol. 49:1247-1250. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. F., W. W. Lawrence, and J. F. Williams. 1972. An automated procedure for the determination of ammonia in tobacco. Beitr. Tabakforsch. Int. 6:167-172. [Google Scholar]
- 10.Cosentino, S., M. E. Fadda, M. Deplano, A. F. Mulargia, and F. Palmas. 2001. Yeasts associated with Sardinian ewe's dairy products. Int. J. Food Microbiol. 69:53-58. [DOI] [PubMed] [Google Scholar]
- 11.Dees, P. M., and W. C. Ghiorse. 2001. Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol. Ecol. 35:207-216. [DOI] [PubMed] [Google Scholar]
- 12.Ellis, R. J., P. Morgan, A. J. Weightman, and J. C. Fry. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ Microbiol. 69:3223-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.English, C. F., E. J. Bell, and A. J. Berger. 1967. Isolation of thermophiles from broadleaf tobacco and effect of pure culture inoculation on cigar aroma and mildness. Appl. Microbiol. 15:117-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatichenti, F., J. L. Bergere, P. Deiana, and G. A. Farris. 1983. Antagonistic activity of Debaryomyces hansenii towards Clostridium tyrobutyricum and Cl. butyricum. J. Dairy Res. 50:449-457. [DOI] [PubMed] [Google Scholar]
- 15.Finstein, M. S., and M. L. Morris. 1975. Microbiology of municipal solid waste composting. Adv. Appl. Microbiol. 19:113-151. [DOI] [PubMed] [Google Scholar]
- 16.Gadd, G. M. 1999. Fungal production of citric and oxalic acid: importance in metal speciation, physiology and biogeochemical processes. Adv. Microb. Physiol. 41:47-92. [DOI] [PubMed] [Google Scholar]
- 17.Giovannozzi, M. 1948. La fermentazione dei tabacchi. Società Anonima Editrice Dante Alighieri, Roma, Italia.
- 18.Granger, D. L., R. R. Taintor, K. S. Boockvar, and J. B. Hibbs, Jr. 1995. Determination of nitrate and nitrite in biological samples using bacterial nitrate reductase coupled with the Griess reaction. Methods 7:78-83. [DOI] [PubMed] [Google Scholar]
- 19.Gunge, N., K. Fukuda, S. Morikawa, K. Murakami, M. Takeda, and A. Miwa. 1993. Osmophilic linear plasmids from the salt-tolerant yeast Debaryomyces hansenii. Curr. Genet. 23:443-449. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann, D., K. D. Brunnemann, B. Prokopczyk, and M. V. Djordevic. 1994. Tobacco-specific N-nitrosamines and Areca-derived N-nitrosamines: chemistry, biochemistry, carcinogenicity, and relevance to humans. J. Toxicol. Environ. Health 41:1-52. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, T., N. Frankenberg, M. Marino, and D. Jahn. 1998. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J. Bacteriol. 180:186-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori, T., S. Haruta, Y. Ueno, M. Ishii, and Y. Igarashi. 2006. Direct comparison of single-strand conformation polymorphism (SSCP) and denaturing gradient gel electrophoresis (DGGE) to characterize a microbial community on the basis of 16S rRNA gene fragments. J. Microbiol. Methods 66:165-169. [DOI] [PubMed] [Google Scholar]
- 23.Ishii, K., M. Fukui, and S. Takii. 2000. Microbial succession during a composting process as evaluated by denaturing gradient gel electrophoresis analysis. J. Appl. Microbiol. 89:768-777. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, C. O., and H. B. Parmele. 1950. Fermentation of cigar-type tobacco. Ind. Eng. Chem. 42:519-522. [Google Scholar]
- 25.Jones, D., and M. D. Collins. 1986. Irregular, nonsporing gram-positive rods, p. 1261-1434. In P. H. A. Sneath (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 26.Kadivar, H., and A. E. Stapleton. 2003. Ultraviolet radiation alters maize phyllosphere bacterial diversity. Microb. Ecol. 45:353-361. [DOI] [PubMed] [Google Scholar]
- 27.Klassen, R., S. Teichert, and F. Meinhardt. 2004. Novel yeast killer toxins provoke S-phase arrest and DNA damage checkpoint activation. Mol. Microbiol. 53:263-273. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 30.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leclercq-Perlat, M. N., A. Oumer, J. L. Bergere, H. E. Spinnler, and G. Corrieu. 2000. Behavior of Brevibacterium linens and Debaryomyces hansenii as ripening flora in controlled production of smear soft cheese from reconstituted milk: growth and substrate consumption. J. Dairy Sci. 83:1665-1673. [DOI] [PubMed] [Google Scholar]
- 32.Li, W. J., P. Schumann, Y. Q. Zhang, P. Xu, G. Z. Chen, L. H. Xu, E. Stackebrandt, and C. L. Jiang. 2005. Proposal of Yaniaceae fam. nov. and Yania flava sp. nov. and emended description of the genus Yania. Int. J. Syst. Evol. Microbiol. 55:1933-1938. [DOI] [PubMed] [Google Scholar]
- 33.Lord, N. S., C. W. Kaplan, P. Shank, C. L. Kitts, and S. L. Elrod. 2002. Assessment of fungal diversity using terminal restriction fragment (TRF) pattern analysis: comparison of 18S and ITS ribosomal regions. FEMS Microbiol. Ecol. 42:327-337. [DOI] [PubMed] [Google Scholar]
- 34.Mendell, S., E. C. Bourlas, and M. Z. DeBardeleben. 1984. Factors influencing tobacco leaf quality: an investigation of the literature. Beitr. Tabakforsch. Int. 12:153-167. [Google Scholar]
- 35.Mohammed, S. I., L. R. Steenson, and A. W. Kirleis. 1991. Isolation and characterization of microorganisms associated with the traditional sorghum fermentation for production of Sudanese Kisra. Appl. Environ. Microbiol. 57:2529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mounier, J., R. Gelsomino, S. Goerges, M. Vancanneyt, K. Vandemeulebroecke, B. Hoste, S. Scherer, J. Swings, G. F. Fitzgerald, and T. M. Cogan. 2005. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 71:6489-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima, K., K. Hirota, Y. Nodasaka, and I. Yumoto. 2005. Alkalibacterium iburiense sp. nov., an obligate alkaliphile that reduces an indigo dye. Int. J. Syst. Evol. Microbiol. 55:1525-1530. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura, K., S. Haruta, H. L. Nguyen, M. Ishii, and Y. Igarashi. 2004. Enzyme production-based approach for determining the functions of microorganisms within a community. Appl. Environ. Microbiol. 70:3329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano, M. M., T. Hoffmann, Y. Zhu, and D. Jahn. 1998. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 180:5344-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters, S., S. Koschinsky, F. Schwieger, and C. C. Tebbe. 2000. Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes. Appl. Environ. Microbiol. 66:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen, K. M., S. Westall, and L. Jespersen. 2002. Microbial succession of Debaryomyces hansenii strains during the production of Danish surfaced-ripened cheeses. J. Dairy Sci. 85:478-486. [DOI] [PubMed] [Google Scholar]
- 44.Pronk, J. T., H. Y. de Steensma, and J. P. Van Dijken. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607-1633. [DOI] [PubMed] [Google Scholar]
- 45.Pucher, G. W., and H. B. Vickery. 1933. The katabolism of the non-volatile organic acids of tobacco leaves during curing. Proc. Natl. Acad. Sci. USA 19:623-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu, J. 1999. Statistics aided optimization for high-performance liquid chromatographic analysis of organic acids in tobacco. J. Chromatogr. 859:153-158. [DOI] [PubMed] [Google Scholar]
- 47.Rantsiou, K., R. Urso, L. Iacumin, C. Cantoni, P. Cattaneo, G. Comi, and L. Cocolin. 2005. Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 71:1977-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sneath, P. H. A. 1986. Endospore-forming gram-positive rods and cocci, p. 1104-1207. In P. H. A. Sneath, Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 51.Strom, P. F. 1985. Effect of temperature on bacterial species diversity in thermophilic solid-waste composting. Appl. Environ. Microbiol. 50:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strom, P. F. 1985. Identification of thermophilic bacteria in solid-waste composting. Appl. Environ. Microbiol. 50:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamburini, E., A. G. León, B. Perito, and G. Mastromei. 2003. Characterization of bacterial pectinolytic strains involved in the water retting process. Environ. Microbiol. 5:730-736. [DOI] [PubMed] [Google Scholar]
- 54.Testa, F., and A. Marconi. 2000. Il Toscano, guida completa al sigaro italiano. Giunti, Firenze, Italia.
- 55.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tso, T. C. 1990. Production, physiology and biochemistry of tobacco plant. IDEALS, Inc., Beltsville, Md.
- 57.Uchida, M., and M. Murata. 2004. Isolation of a lactic acid bacterium and yeast consortium from a fermented material of Ulva spp. (Chlorophyta). J. Appl. Microbiol. 97:1297-1310. [DOI] [PubMed] [Google Scholar]
- 58.Valdés-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 59.Van der Walt, J. P., and D. Yarrow. 1984. Methods for the isolation, maintenance, classification and identification of yeasts, p. 45-104. In N. J. W. Kreger-van Rij (ed.), The yeasts, a taxonomic study. Elsevier Science Publishers BV, Amsterdam, Holland.
- 60.Wahlberg, I., and T. Ringberger. 1999. Smokeless tobacco p. 452-460. In D. L. Davis and M. T. Nielsen (ed.), Tobacco production, chemistry and technology. Blackwell Science, Oxford, United Kingdom.
- 61.Welthagen, J. J., and B. C. Viljoen. 1998. Yeast profile in Gouda cheese during processing and ripening. Int. J. Food Microbiol. 41:185-194. [DOI] [PubMed] [Google Scholar]
- 62.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 63.Yu, Z., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.