Abstract
It is well documented that microbial contamination of coastal waters poses a significant risk to human health through recreational exposure and consumption of shellfish. Identifying the source of microbial contamination (microbial source tracking) plays a dominant role in enabling effective management and remediation strategies. One method used to determine the source of the contamination is quantification of the ratio of the four subgroups of F+-specific RNA coliphages (family Leviviridae) in impacted water samples. Because of typically low concentrations in the environment, enrichment assays are performed prior to detection, even though differential replication rates have been reported. These assays are also compromised by differential loss of phage infectivity among subgroups after release into the environment, thus obscuring the initial ratio. Here, a culture-independent multiplex real-time reverse transcriptase-PCR (RT-PCR) protocol for the simultaneous quantification of all four subgroups of F+-specific RNA coliphages using novel primer sets and molecular beacons is presented. This assay is extremely sensitive, achieving detection with as few as 10 copies of isolated coliphage RNA, and is linear for a minimum of six orders of magnitude. During survival experiments, the real-time RT-PCR technique was able to quantify coliphages in seawater when culture-based double agar layer assay failed. While infectivity was lost at different rates at the subgroup level, decay constants in seawater, calculated using the real-time RT-PCR estimates, did not vary among subgroups. The accurate determination of the in situ concentration of F+-specific RNA coliphages using this method will facilitate more effective remediation strategies for impacted environments.
Microbial contamination of lakes, rivers, estuarine, and coastal waters poses a major risk to human health through consumption of organisms that inhabit these environments as well as through recreational exposure. Identifying the source of microbial contamination (microbial source tracking) plays a dominant role in the types of actual risk and enabling effective management and remediation strategies. In their current form, monitoring programs and identification protocols for determining the magnitude of fecal contamination are unable to differentiate between human and nonhuman sources of fecal contamination. Therefore, there is a critical need for a method that allows reliable discrimination of human and nonhuman microbial contamination.
Over the last three decades, several methods have been proposed to discriminate between human and nonhuman fecal contamination in surface waters using different microorganisms (e.g., Bifidobacterium spp., Bacterioides fragilis phages, Clostridium perfringens, Escherichia coli, F+-specific RNA coliphages, fecal streptococci, Rhodococcus coprophilus, and bovine enterovirus) as well as various chemical compounds (e.g., caffeine, laundry detergents, fecal sterols, stanols, etc.) (30, 32). As an indicator organism, Escherichia coli has been most extensively studied. Several phenotypic and molecular methods have been applied for microbial source tracking using this organism, including multiple antibiotic resistance patterns (18), immunological assays (24), denaturing gradient gel electrophoresis (2, 8), repetitive element PCR (3, 5), amplified fragment length polymorphism (15), and ribotyping, among others (25).
While some of these methods have been relatively successful in field studies, many concerns have been expressed lately, especially with E. coli-based techniques. These criticisms include reference database issues (limited and location specific), changes in subpopulations during transition from primary (host) to secondary (outside the host) habitat, variations in methodology, reproducibility concerns, requirement for cultivation, and cost and time demands (15, 31). Based on extensive genetic diversity studies, Gordon (12) stated that any program attempting to identify the sources of fecal contamination focusing on commensal E. coli appears to have very limited value. A similar conclusion was reached more recently based on discrepancies between cluster and discriminant function analyses results (19). Therefore, it has been suggested (7) that tracking bacteria other than E. coli may be a more valid approach, and the quest for an ideal indicator organism continues.
F+-specific RNA coliphages constitute the taxonomically distinct family Leviviridae (21). This family contains two genera, Allolevivirus and Levivirus (21), which both contain two distinct subgroups based upon serological cross-reactivity (9). Levivirus contains Leviviridae subgroups I and II, while Allolevivirus contains subgroups III and IV. The key trait on which the F+-specific RNA coliphage approach of source tracking is based is that subgroups I and IV are predominantly isolated from nonhuman feces, while subgroups II and III are predominantly isolated from human feces and sewage (9, 23). This segregation appears to hold worldwide, which is a great advantage over the other techniques in use.
One of the current limitations of the F+-specific RNA coliphage approach is their low concentrations in the environment. Because the concentrations of these phages in water samples is low, enrichment assays are recommended (30) and deployed (14) for their detection, but this method of coliphage amplification has an inherent bias as a result of differential burst size and infection efficiencies among the subgroups (10). In addition, loss of phage infectivity in the environment reduces the sensitivity of culture-based assays. Therefore, a sensitive, culture-independent method is needed in order to make the coliphage approach a useful tool for microbial source tracking. We developed a novel molecular beacon-based multiplex real-time reverse transcriptase-PCR (RT-PCR) assay for quantification and identification of F+-specific RNA coliphages from the environmental samples for this purpose.
MATERIALS AND METHODS
Bacterial and coliphage stocks.
E. coli HS(pFamp)R was propagated on tryptone broth and used as a host for all coliphage cultures. Seven F+-specific RNA coliphage stocks, MS2 (2), GA (2), Q-β (2), and FI (1), were used to test the real-time RT-PCR protocol as well as to evaluate its specificity and efficiency.
Genome sequences.
All sequences available for F-specific RNA coliphages were downloaded from GenBank and used for primer and beacon design. In addition, the genome of FI was sequenced (Beckman Coulter CEQ 8000 Genetic Analysis System; Fullerton, CA) (see the supplemental material).
Primers and molecular beacons.
The alignment of sequences revealed high levels of variability. The primers and beacons were positioned in the replicase gene for subgroups I, II, and IV and in the coat protein gene for subgroup III because of the relatively small amount of sequence variation encountered in those areas. All oligonucleotides were designed using primer3 and mfold 3.11 (26, 29, 40). The primer and beacon sequences were verified to be subgroup-specific using GenBank's BLAST (1). The specifications of primers are given in Table 1, and molecular beacons are given in Table 2.
TABLE 1.
Primer sequences and characteristics
| Primer | Sequence | Start position | Length (bp) | Target | Tma (°C) | Product length (bp) |
|---|---|---|---|---|---|---|
| Subgroup I | ||||||
| Forward | 5′-CGTGGTTCCATACTGGAGGT-3′ | 3265 | 20 | RNA replicase | 62.6 | |
| Reverse | 5′-CTTTCGAGCACACCCACC-3′ | 3441 | 18 | Beta chain | 61.1 | 177 |
| Subgroup II | ||||||
| Forward | 5′-GGTTCAAGTTGCGGGATG-3′ | 2155 | 18 | RNA replicase | 60.2 | |
| Reverse | 5′-GAAAACAAACCGTTGCCG-3′ | 2338 | 18 | Beta chain | 59.0 | 184 |
| Subgroup III | ||||||
| Forward | 5′-CCGCGTGGGGTAAATCC-3′ | 1373 | 17 | Coat protein | 62.2 | |
| Reverse | 5′-TTACGATTGCGAGAAGGCTG-3′ | 1485 | 20 | Coat protein | 61.3 | 113 |
| Subgroup IV | ||||||
| Forward | 5′-CGTGGAAGCATGCCTGT-3′ | 2944 | 17 | RNA replicase | 61.9 | |
| Reverse | 5′-TTCCAGCCRGGCTCGAT-3′ | 3127 | 18 | Beta chain | 64.1 | 182 |
Tm, predicted melting temperature.
TABLE 2.
Molecular beacon sequences and characteristics
| Beacon | Sequencea | Fluorophore | Quencher | dGb (kcal/mol) | Tmc (°C) |
|---|---|---|---|---|---|
| Subgroup I | |||||
| Beacon 1 | CTGGGGCCAGCGAGCTCTCCTCCCCAG | FAMd | Dabcyl | −1.26 | 65.1 |
| Subgroup II | |||||
| Beacon 2 | CTCGCGCAGGAAACGTATGGCGATGAGACGCGAG | HEXe | Dabcyl | −1.55 | 65.2 |
| Subgroup III | |||||
| Beacon 3 | ACGGCAAGCGGGTGCAGTTCCTGCCGT | CY-5f | BHQ2g | −1.7 | 68.2 |
| Subgroup IV | |||||
| Beacon 4 | CCCAGGCAAATAAAGCAGTCACTGTTCCTGGG | Texas-Red | Dabcyl | −0.83 | 61.1 |
Subgroup-specific sequences are in boldface.
dG, predicted minimum free energy.
Tm, predicted melting temperature.
FAM, 6-carboxyfluorescein.
HEX, hexachloro-6-carboxyfluorescein.
CY-5, indodicarbocyanine.
BHQ-2, black hole quencher 2.
Initial primer and molecular beacon specificity tests.
RNA was extracted from phage stocks using TriReagent LS (MRC Inc., Cincinnati, OH) according to the manufacturer's protocol, with modifications (see the supplemental material). The RT-PCRs were performed using a ProStar Ultra HF RT-PCR System (Stratagene, La Jolla, CA) on Programmable Thermal Block II (Lab-Line, Melrose Park, IL). The RT-PCR products were sized on 3% agarose gels stained with ethidium bromide (2 μg/liter) and visualized with UV excitation. The functionality and specificity of molecular beacons was verified using melting curve analyses. All reactions were conducted in 1× Core PCR Buffer (4 mM magnesium chloride) using equal amounts (200 nM) of beacons and matching or mismatching synthesized oligonucleotides. Synthesized DNA targets of the three other subgroups were used as mismatching oligonucleotide controls. Melting curve analyses were performed on a Stratagene MX4000 (95°C for 3 min, followed by stepwise [1°C/min] decrease of temperature from 85°C to 30°C).
Standards. (i) RNA standards.
RNA was extracted from filtered (0.22 μm) coliphage stocks and quantified spectrophotometrically (SmartSpec 3000; Bio-Rad, Hercules, CA).
(ii) DNA standards.
RNA extracted from coliphage stocks was amplified using the RT-PCR protocol specified above. PCR products were cleaned using a QIAquick PCR purification kit (QIAGEN, Valencia, CA) and quantified using a Quant-IT kit (Invitrogen, Carlsbad, CA) on the Wallac Victor2 1420 Multilabel Counter (PerkinElmer Life Sciences, Wellesley, MA) according to the manufacturer's protocols.
Optimized real-time RT-PCR protocol.
A 2-Step Brilliant QRT-PCR Core Reagent kit (Stratagene, La Jolla, CA) was used for all real-time RT-PCRs. The optimal (see the supplemental material) multiplexed RT reactions (10 μl) contained 100 nM of subgroup I reverse primer, 100 nM of subgroup II reverse primer, 100 nM of subgroup III reverse primer, 200 nM of subgroup IV reverse primer, 4 mM deoxynucleoside triphosphate, and 10 U of StrataScript reverse transcriptase in 1× Core RT Buffer. The optimized multiplexed real-time PCRs (50 μl) contained 100 nM of subgroup II forward and reverse primers, 100 nM of subgroup III forward and reverse primers, 100 nM of subgroup IV forward primer, 200 nM of subgroup IV reverse primer, 200 nM of each molecular beacon, 4 mM deoxynucleoside triphosphate, and 5 U of SureStart Taq DNA polymerase in 1× Core PCR Buffer (4 mM MgCl2). RT reactions were performed on Programmable Thermal Block II (Lab-Line, Melrose Park, IL) and were incubated at 48°C for 1 h, followed by the reverse transcriptase inactivation at 95°C for 3 min. Real-time PCRs were performed on an MX4000 (Stratagene, La Jolla, CA). Thermal cycling consisted of an initial cycle of 95°C for 10 min, followed by 40 cycles of 1 min at 95°C, 30 s at 60°C, and 1 min at 72°C.
Sensitivity tests.
Quantified DNA was serially diluted at concentrations from 107 to 101 copies of each subgroup. Simultaneously, additional sets of tubes were seeded at 50 copies of DNA of one subgroup and 107 copies of each of the three other subgroups. Three replicate experiments were conducted.
Standards.
Because RNA is more labile and contamination from host RNA contamination can bias quantification, DNA standards were evaluated as an alternative by comparing amplification efficiencies of multiplexed real-time PCRs with RNA standards. This comparison was carried out in three replicate experiments.
Coliphage survival.
The performance of the real-time PCR technique compared to the culture-based double layer agar technique (DAL) was tested using survival experiments by spiking polypropylene tubes containing 10 ml of filtered (0.45 μm) seawater with a coliphage stock (∼106 PFU/ml). Triplicate experiments were conducted with each subgroup. The samples were incubated away from direct sunlight at ambient room temperature (21 to 23°C). Samples were assayed for infective phages using the DAL technique and for total phage concentrations using the real-time RT-PCR technique at 24-h intervals. Volumes tested by the DAL technique varied; initially, 1:100 dilutions were necessary. The sample volume analyzed using real-time RT-PCR was 0.1 ml. The sample was considered to be negative based on the DAL when no phages were present in 1 ml of sample. An additional direct real-time RT-PCR assay was performed after 20 days. The decay constant was estimated for each replicate as a slope of a first-order linear regression line fitted to log-linear plots (ln). The real-time PCR data did not fit to the linear regression model, and the decay constant (k) was calculated as k = ln[(C0/Ct)/tx], where C0 is initial coliphage concentration and Ct is coliphage concentration at a given time (tx) during the experiment (22).
Field samples.
The culture-based, DAL method was used to confirm high concentrations of F+-specific RNA phages in a raw sewage sample from the Narragansett Wastewater Facility (Rhode Island) and a chicken stool sample (Casey Farm, Saunderstown, Rhode Island) diluted in 5 ml of filtered (0.2 μm) and autoclaved seawater. Twenty phage isolates, which did not form plaques in the presence of RNase, were typed from each sample using the real-time RT-PCR protocol. In addition, F+-specific RNA coliphages were quantified directly from the mixed samples using the RT-PCR assay. The slurries were vortexed and centrifuged at 3,000 × g for 10 min to remove larger solids. Supernatant of both samples was mixed at ratios of 1:0, 0.75:0.25, 0.5:0.5, 0.25:0.75, and 0:1 (vol:vol), followed by RNA extraction (as described above) and real-time RT-PCR quantification. Initial phage concentrations were determined by using a serial dilution of 1 ml for DAL and 0.1 ml for RT-PCR, and the results were normalized to phage per milliliter.
PCR inhibition.
Samples of different origin (commercial drinking water, freshwater pond, seawater, and cat and cow feces [0.5 g wet weight/ml]) were verified to contain no detectable levels of F+-specific RNA coliphages using DAL and real-time PCR techniques and subsequently seeded with a cocktail of coliphage stocks, i.e., 171.7 × 104, 114 × 104, 6.7 × 104, and 3.5 × 104 copy/ml final of subgroups I, II, III, and IV, respectively. Phage stocks were mixed and diluted in SM buffer (28) parallel to other treatments and used as controls of initial concentrations. RNA was extracted from each treatment using a TriReagent LS extraction protocol and quantified using the real-time RT-PCR protocol specified above. Quadruplicate experiments were conducted for each treatment.
Statistical analyzes.
Real-time PCR data were analyzed using MX4000 v.4.20 software (Stratagene, La Jolla, CA). Briefly, the baseline was corrected using the default adaptive baseline algorithm, and the threshold was manually adjusted by visually analyzing log-scale amplification plots. Linear regression analyses were applied to calculate the standard curve and estimate the r2 and slope values. Assuming 100% efficiency if DNA template is doubled in each cycle, the PCR efficiency was calculated as E = 10(−1/slope) − 1, where E is PCR efficiency.
One-way analysis of variance was used to calculate the F statistic and corresponding P value in survival and inhibition experiments. Pair-wise multiple comparisons were performed using a “Student” t test, and Bonferroni correction (α = 0.05) (39) was applied to identify which treatment was different. Differences were concluded to be significant when P ≤ 0.05 after adjusting for multiple tests; adjusted P values are reported. The number of pair-wise comparisons made (n) was 6 for survival studies and 15 for inhibition studies. SigmaStat v.9.0 (Systat Software Inc, Point Richmond, CA) was used for the analyses.
Nucleotide sequence accession number.
Final alignments and the sequence of FI were constructed in BioEdit (16) and submitted to the GenBank under accession number EF068134.
RESULTS
Initial primer and molecular beacon specificity tests.
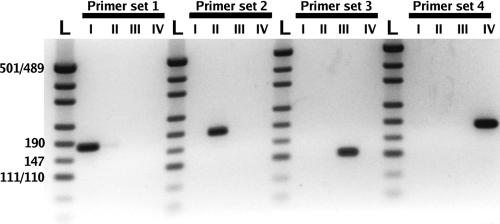
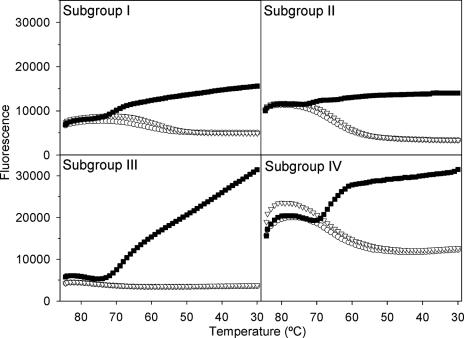
Each primer set is subgroup specific, yielding a single RT-PCR product of the predicted size. No amplification occurred using RNA of subgroups not matched to the primer set as a potential template (Fig. 1). RT-PCRs containing all primer sets and RNA of only one subgroup at a time also resulted in a single amplicon per reaction. There was a strong signal for each molecular beacon in the presence of matching template, while the cocktail of nonspecific templates did not result in hybridization when the temperature was decreased during the melting curve analyses (Fig. 2). Therefore, this set of molecular beacons is functional, template specific, and lacks cross-hybridization.
FIG. 1.
Negative image of ethidium bromide-stained agarose gel (3%) indicating the specificity of each primer set. I, subgroup I; II, subgroup II; III, subgroup III; IV, subgroup IV. L, pUC 19/HpaII digest double-stranded DNA marker (Ambion).
FIG. 2.
Melting curves demonstrating the specificity of each molecular beacon (▪, matching template; ▿, mismatching template cocktail; ○, no template control). At high temperature the stem hybrid structure of a beacon is dissociated. As the temperature decreases the beacon hybridizes with the specific template (▪), forcing the stem hybrid to remain dissociated and, therefore, producing a fluorescent signal. Treatments with templates of other subgroups (▿) do not exhibit an increase in fluorescence, as the beacon forms closed stem-loop structure.
Simplex versus multiplex real-time RT-PCR.
The correlation coefficients of the simplex reactions were 0.998 to 1.000, and efficiencies were in the range of 95.1 to 103.5%. The correlation coefficients for multiplexed (fourplex) reactions were 0.997 to 1.000, and efficiencies were in the range of 95.4 to 101.9%. As both parameters did not vary more than 5% for each subgroup between the reactions, multiplexing was considered a valid option.
RNA versus DNA standard.
The RNA- and DNA-based standard curves had similar amplification efficiencies, indicating that both types of standard are suitable for this assay. The correlation coefficients were >0.995, and the reaction efficiencies range from 97.2 to 103.3%.
Sensitivity of the real-time RT-PCR protocol.
As few as 10 copies of DNA were detectable, and the standard curves of the multiplexed reactions remained linear over six orders of magnitude. Fifty copies of each subgroup remained detectable when 107 copies of the three other subgroups were added to the RT-PCR.
Phage survival.
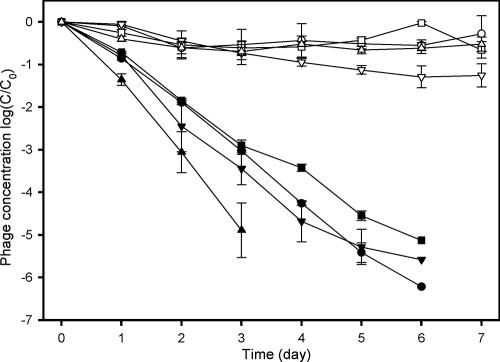
F+-specific RNA coliphages of all four subgroups were inactivated in less than 7 days in filtered seawater (Fig. 3). The inactivation of coliphage subgroups was more rapid than in a previous freshwater-based study (20), where subgroup I remained detectable for more than 80 days by single agar layer (SAL). While the phages were inactivated rapidly, there was little decrease in real-time PCR signal (Fig. 3), and the real-time RT-PCR assay was able to detect all four subgroups after 20 days. The decay constant for coliphage infectivity was in the range of 2.09 to 3.97 day−1, whereas the mean decay constant of the intact coliphage genome in seawater was in the range of 0.81 to 1.51 day−1 (Table 3). Subgroup IV exhibited a significantly higher loss of infectivity (P values ranged from <0.001 to 0.001 depending on the test; n = 6) compared to other subgroups based on the DAL technique. While subgroup IV is known to exhibit rapid inactivation (20), the subgroup III decay rates were not significantly different from subgroups I and II in any of the tests conducted. The real-time PCR-based decay constant was not statistically different among the four subgroups at the end of the experiment.
FIG. 3.
Inactivation of F-specific RNA coliphages in seawater (•, subgroup I infective; ▪, subgroup II; ▾, subgroup III; ▴, subgroup IV) and the quantity of the phage particles detected using the real-time RT-PCR protocol (○, subgroup I; □, subgroup II; ▿, subgroup III; Δ, subgroup IV RT-PCR). Symbols indicate the means, and bars indicate the standard errors. The detection limit of the DAL assay was 1 PFU/ml, which corresponds to −6 on the y axis.
TABLE 3.
Decay constant (k) based on the infective fraction of phages (PFU) and the real-time RT-PCR estimatesa
| Subgroup | Decay constant
|
|
|---|---|---|
| Plaque (ln/day) | Real-time RT-PCR (ln/day) | |
| I | 2.53 (±0.09) | 0.81 (±0.36) |
| II | 2.09 (±0.06) | 1.12 (±0.06) |
| III | 2.54 (±0.21) | 1.51 (±0.09) |
| IV | 3.97 (±0.22)* | 1.07 (±0.01) |
Values in parentheses represent the standard errors (an asterisk denotes statistical difference at P < 0.05).
Phage discrimination.
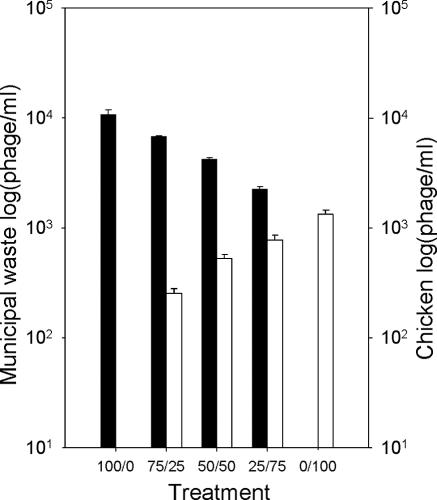
The real-time RT-PCR assay was successful in detection and quantification of F+-specific RNA coliphage groups originating from different sources in mixed samples (Fig. 4). The proportion of each subgroup of coliphage correlated with the initial dosage of sewage and stool slurry mixed. Raw municipal sewage contained high concentrations (1.1 × 104 phage/ml) of subgroup III phages (conventionally assigned as human source), and chicken stool samples contained lower concentrations (1.3 × 103 phage/ml) of subgroup I phages (conventionally assigned as nonhuman source). These findings were supported by the isolates typed using this real-time RT-PCR technique from unmixed samples; 20 isolates from the raw sewage all belonged to subgroup III, and 20 isolates from the chicken stool belonged to subgroup I. The concentrations based on the culture technique (6.4 × 103 PFU/ml and 5.7 × 102 PFU/ml, respectively) were approximately half the concentrations tested directly by the RT-PCR (sensitivity, 0.5 PFU/reaction).
FIG. 4.
Simultaneous quantification of two different contamination sources (▪, raw municipal sewage [subgroup III]; □, chicken stool [subgroup I]) mixed at different ratios. Columns indicate the means, and bars indicate the standard errors.
PCR inhibition.
There were significant differences between the treatments for each subgroup. Compared to the baseline estimates (SM buffer), coliphage concentration estimates of all subgroups were significantly different in saltwater and in both fecal slurry samples studied (subgroup I, P < 0.001; subgroup II, P < 0.001; subgroup III, P < 0.001; and subgroup IV, P = 0.015 [saltwater], P = 0.014 [cow], and P = 0.016 [cat] [n = 15 samples for each subgroup). The freshwater sample matrix revealed significant deviation from the baseline estimates for subgroups I and II (P = 0.031 and 0.005; n = 15). The estimates of coliphage concentrations were reduced 29.9 to 46.7% for drinking water, 45.8 to 62.9% for freshwater, 90.5 to 94.3% for saltwater samples, and 92.5 to 97.0% for the fecal slurry samples compared to the baseline (Table 4). While there were significant differences between the treatments, the initial ratio of the subgroups was maintained in each treatment (Table 4).
TABLE 4.
Inhibition of the real-time RT-PCR estimates in different matrices determined by dosing known quantities of each subgroup to the samplesa
| Sample | Concn (104 phages/ml) in subgroup:
|
Ratio of subgroup I:II:III:IV | |||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| SM Buffer | 171.7 (±21) | 114.6 (±20.2) | 6.7 (±1.8) | 3.5 (±1.2) | 1.4:1.3:1.1:1.0 |
| Drinking water | 117.6 (±7.2) | 78.4 (±9.3) | 4.7 (±0.5) | 1.9 (±0.7) | 1.4:1.4:1.1:1.0 |
| Freshwater | 110.3 (±19) | 53.1 (±8.6) | 3.2 (±0.4) | 1.3 (±0.3) | 1.5:1.4:1.1:1.0 |
| Saltwater | 16.3 (±2.0) | 10.6 (±4.2) | 0.4 (±0.2) | 0.2 (±0.1) | 1.6:1.5:1.1:1.0 |
| Cow feces | 9.3 (±1.9) | 4.1 (±1.0) | 0.2 (±0.1) | 0.2 (±0.1) | 1.5:1.4:1.0:1.0 |
| Cat feces | 10.2 (±2.1) | 6.2 (±1.5) | 0.5 (±0.2) | 0.2 (±0.1) | 1.5:1.4:1.1:1.0 |
Values in parentheses represent the standard errors.
DISCUSSION
Many approaches to microbial source tracking have been pursued over several decades. The use of F+-specific coliphages, while promising, has been hampered by the relatively low abundance of these viruses in the environment. Culture-based enrichment assays designed to overcome the low abundance introduce a bias that renders this approach ineffective. PCR-based techniques can be more sensitive and rapid than culture-based techniques and have been used successfully to detect a variety of microorganisms in environmental samples. RT-PCR protocols for detection of F+-specific RNA coliphages on group and subgroup levels exist (36). Also, quantitative most probable number-PCR has been described for F-specific RNA coliphages (27). More recently, real-time PCR has become a method of choice to quantify nucleic acids as a result of its simplicity and reproducibility (37). The real-time RT-PCR technique described here allows for rapid quantification of all four subgroups of F+-specific RNA coliphages in a single tube. This ability allows for streamlining sample preparation and a reduction in cost relative to performing four separate real-time RT PCRs, as reagents common to all reactions are conserved.
Culture-based assays using the F+-specific RNA coliphages suffer from the fact that these phages are rapidly inactivated when released into the environment. Using the real-time RT-PCR protocol, it was possible to detect and quantify the phages throughout the survival experiment, even after no infective phages were detected. This greatly increases the sensitivity of the real-time PCR-based method compared to most culture-based methods.
Inactivation rates of different groups of viruses are functions of temperature (38) and the presence of microorganisms (11) and sunlight (33). In saltwater, F+-specific RNA coliphages were inactivated faster than in lake water (20). It is plausible that ions (Na+ and Mg2+) have negative impacts on survival rates of F+-specific RNA coliphages. It has been shown that the presence of calcium ions increases MS2 decay rates, although the actual calcium concentration had no significant correlation with the decay rate (38).
Inactivation rates differ significantly among the subgroups, thus altering the initial ratio of coliphages (20). Findings in this study agree with this conclusion. Contrary to the culture-based estimates, the real-time RT-PCR data showed no significant differences among the decay constants of the subgroups, thus preserving the initial ratio throughout the duration of the experiment. Therefore, in saltwater samples, the real-time RT-PCR eliminates environment-based biases associated with the survival characteristics of F+-specific RNA coliphages in the environment as the ratio of subgroups is maintained.
In addition to microbial source tracking, F+-specific RNA coliphages have been suggested as alternative indicators for enteric viruses (4, 6, 17, 34). Though this function is vastly debatable (13), the total F+-specific RNA coliphage abundance can be obtained for environmental samples by summing the signal from all four subgroups from the multiplexed real-time RT-PCR assay presented here. The increased sensitivity this method provides could result in detecting the coliphages after the health risk has dissipated as a result of the inactivation of the enteric viruses. This study examined a small number of strains during a relatively short timeframe. Therefore, more strains on longer time scales need to be evaluated by the real-time RT-PCR method in parallel with survival experiments with enteric viruses.
The sewage-dosing and the coliphage survival experiments demonstrate that this technique can be used to estimate the relative contributions of F-specific coliphages by various animals as well as detect and quantify phages that may have become noninfective after release to the outside environment. One significant concern is the coelution of PCR inhibitors during the RNA isolation step. This requires a more rigorous RNA cleanup and makes coliphage quantification directly from environmental samples more complicated. In pilot studies, when extracted RNA was cleaned and concentrated from the environmental samples using Croma Spin-1000 (Clontech Laboratories Inc., Mountain View, CA) and Microcon YM-100 (Millipore Corporations, Billerica, MA) spin columns, up to 10 copies RNA/100 ml was detectable, while SAL failed. In this study, the real-time PCR was inhibited in all environmental samples. Therefore, additional cleanup techniques and utilization of extraction and inhibition controls should be considered. Consequently, the total phage particle numbers for raw sewage and chicken stool presented in this paper are estimates. Without the use of internal controls, this technique would underestimate coliphage concentrations in environmental samples (Table 4), but it is still orders of magnitude more sensitive than the cultivation-based DAL technique (Fig. 3). This enables the detection of 0.1 PFU/reaction in seawater and 0.5 PFU/reaction in stool samples. Overall the sensitivities observed were similar to those of traditional RT-PCR protocols developed for the F+-specific RNA coliphages (27) and separately for Q-β (subgroup III) (19a). It is possible that, because of the increased sensitivity, the assay will detect the presence of these coliphages in more animals, thus alleviating the infrequency with which they are isolated. Further studies comparing this technique to SAL assay are warranted.
One-step RT-PCR protocols allow for the analysis of the entire concentrated sample. This method was tested for this application but resulted in low efficiency in multiplexed reactions. Because a two-step RT-PCR protocol was used instead, the volume of the RT product that can be transferred to the PCR step without causing inhibition of the amplification is limited (≤40%), though the entire sample could be analyzed by performing three separate PCRs of the RT products if so desired.
Although it may turn out that a combination of methods will be required to conclusively answer the difficult problem of microbial source tracking, the use of F+-specific RNA coliphages has great potential among the many alternatives. The real-time RT-PCR protocol for F+-specific RNA coliphages presented here can provide water quality managers worldwide with a rapid and effective tool to help protect the public health.
Supplementary Material
Acknowledgments
We are very grateful to D. Griffin for providing us with E. coli HS(pFamp)R stock and advice on initial stages of this study. We are also very grateful to A. Dishlers (Biomedical Research Study Center, Latvia), L. Liljas (Uppsala University, Sweden), and M. Sobsey (University of North Carolina, Chapel Hill) for providing coliphage stocks. Our sincere thanks also go to P. Johnson (URI Genomics and Sequencing Services) for assistance with sequencing and real-time PCR and to M. Zuker (Rensselaer Polytechnic Institute) for advice and access to full mfold services.
This research was supported by NA03N03S4190195 from the Cooperative Institute for Coastal and Estuarine Environmental Control (CICEET) and the URI Proposal Development Fund DCS.
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchan, A., M. Alber, and R. E. Hodson. 2001. Strain-specific differentiation of environmental Escherichia coli isolates via denaturing gradient gel electrophoresis (DGGE) analysis of the 16S-23S intergenic spacer region. FEMS Microbiol. Ecol. 35:313-321. [DOI] [PubMed] [Google Scholar]
- 3.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, H., and M. D. Sobsey. 1993. Comparative survival of indicator viruses and enteric viruses in seawater and sediment. Water Sci. Technol. 27:425-428. [Google Scholar]
- 5.Dombek, P. E., L. K. Johnson, S. T. Zimmerly, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doré, W. J., K. Henshilwood, and D. N. Lees. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl. Environ. Microbiol. 66:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykes, G. A. 2002. Tracing contamination and Escherichia coli diversity. Appl. Environ. Microbiol. 68:4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farnleitner, A. H., N. Kreuzinger, G. G. Kavka, S. Grillenberger, J. Rath, and R. L. Mach. 2000. Simultaneous detection and differentiation of Escherichia coli populations from environmental freshwaters by means of sequence variations in a fragment of the beta-d-glucuronidase gene. Appl. Environ. Microbiol. 66:1340-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuse, K. 1987. Distribution of coliphages in environment: general considerations. John Wiley & Sons, New York, NY.
- 10.Furuse, K. 1982. Phylogenetic studies on RNA coliphages. J. Keio Med. Soc. 59:265-274. [Google Scholar]
- 11.Gordon, C., and S. Toze. 2003. Influence of groundwater characteristics on the survival of enteric viruses. J. Appl. Microbiol. 95:536-544. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 13.Grabow, W. O. K. 2001. Bacteriophages: update on application as models for viruses in water. Water SA 27:251-268. [Google Scholar]
- 14.Griffin, D. W., R. Stokes, J. B. Rose, and J. H. Paul III. 2000. Bacterial indicator occurrence and the use of an F+-specific RNA coliphage assay to identify fecal sources in Homosassa Springs, Florida. Microb. Ecol. 39:56-64. [DOI] [PubMed] [Google Scholar]
- 15.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Havelaar, A. H., M. Van Olphen, and Y. C. Drost. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl. Environ. Microbiol. 59:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasalde, C., R. Rodriguez, and G. A. Toranzo. 2005. Statistical analyses: possible reasons for unreliability of source tracking efforts. Appl. Environ. Microbiol. 71:4690-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Limsawat, S., N. Kamiko, K. Yamamoto, and S. Ohgaki. 1995. Application of polymerase chain reaction to detect RNA coliphage Q beta in environmental water samples. Water Sci. Technol. 31:383-390. [Google Scholar]
- 20.Long, S. C., and M. D. Sobsey. 2004. A comparison of the survival of F+ RNA and F+ DNA coliphages in lake water microcosms. J. Water Health 2:15-22. [PubMed] [Google Scholar]
- 21.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Arch. Virol. Suppl. 10:51-54. [Google Scholar]
- 22.Noble, R. T., and J. A. Fuhrman. 2000. Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl. Environ. Microbiol. 66:3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osawa, S., K. Furuse, and I. Watanabe. 1981. Distribution of ribonucleic acid coliphages in animals. Appl. Environ. Microbiol. 41:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Phenotypic and genotypic characterization of human and non-human Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 25.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peyret, N. 2000. Prediction of nucleic acid hybridization: parameters and algorithms. Wayne State University, Detroit, MI.
- 27.Rose, J. B., X. Zhou, D. W. Griffin, and J. H. Paul. 1997. Comparison of PCR and plaque assay for detection and enumeration of coliphage in polluted marine waters. Appl. Environ. Microbiol. 63:4564-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1 to 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.SantaLucia, J., Jr. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 32.Sinton, L. W., R. K. Finlay, and D. J. Hannah. 1998. Distinguishing human from animal faecal contamination in water: a review. N. Z. J. Mar. Freshw. Res. 32:323-348. [Google Scholar]
- 33.Sinton, L. W., R. K. Finlay, and P. Lynch. 1999. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 65:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Springthorpe, S. V., C. L. Loh, W. J. Robertson, and S. A. Sattar. 1993. In situ survival of indicator bacteria, MS-2 phage and human pathogenic viruses in river water. Water Sci. Technol. 27:413-420. [Google Scholar]
- 35.Reference deleted.
- 36.Vinje, J., S. J. Oudejans, J. R. Stewart, M. D. Sobsey, and S. C. Long. 2004. Molecular detection and genotyping of male-specific coliphages by reverse transcription-PCR and reverse line blot hybridization. Appl. Environ. Microbiol. 70:5996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, N. J. 2002. Tech. Sight. A technique whose time has come. Science 296:557-559. [DOI] [PubMed] [Google Scholar]
- 38.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zar, J. H. 1999. Biostatistical analyses, 4th ed. Prentice Hall, Inc., Upper Saddle River, NJ.
- 40.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.