Abstract
Microbially induced concrete corrosion (MICC) in sewer systems has been a serious problem for a long time. A better understanding of the succession of microbial community members responsible for the production of sulfuric acid is essential for the efficient control of MICC. In this study, the succession of sulfur-oxidizing bacteria (SOB) in the bacterial community on corroding concrete in a sewer system in situ was investigated over 1 year by culture-independent 16S rRNA gene-based molecular techniques. Results revealed that at least six phylotypes of SOB species were involved in the MICC process, and the predominant SOB species shifted in the following order: Thiothrix sp., Thiobacillus plumbophilus, Thiomonas intermedia, Halothiobacillus neapolitanus, Acidiphilium acidophilum, and Acidithiobacillus thiooxidans. A. thiooxidans, a hyperacidophilic SOB, was the most dominant (accounting for 70% of EUB338-mixed probe-hybridized cells) in the heavily corroded concrete after 1 year. This succession of SOB species could be dependent on the pH of the concrete surface as well as on trophic properties (e.g., autotrophic or mixotrophic) and on the ability of the SOB to utilize different sulfur compounds (e.g., H2S, S0, and S2O32−). In addition, diverse heterotrophic bacterial species (e.g., halo-tolerant, neutrophilic, and acidophilic bacteria) were associated with these SOB. The microbial succession of these microorganisms was involved in the colonization of the concrete and the production of sulfuric acid. Furthermore, the vertical distribution of microbial community members revealed that A. thiooxidans was the most dominant throughout the heavily corroded concrete (gypsum) layer and that A. thiooxidans was most abundant at the highest surface (1.5-mm) layer and decreased logarithmically with depth because of oxygen and H2S transport limitations. This suggested that the production of sulfuric acid by A. thiooxidans occurred mainly on the concrete surface and the sulfuric acid produced penetrated through the corroded concrete layer and reacted with the sound concrete below.
Concrete corrosion has an enormous economic impact worldwide, especially when the replacement or repair of municipal sewer systems is required. Concrete corrosion is the result of several causes such as carbonation, chloride erosion, and other chemical reactions. In sewer systems and wastewater treatment facilities where high concentrations of hydrogen sulfide, moisture, and oxygen are present in the atmosphere, the deterioration of concrete is caused mainly by biogenic acid (i.e., sulfuric acid) and is known as microbially induced concrete corrosion (MICC). The biogenic acid is generated by various microbial species and complex mechanisms. The general mechanism for the sulfuric acid-caused corrosion of sewer systems has been described in the literature (17, 36). In the first step, hydrogen sulfide (H2S) is produced by sulfate-reducing bacteria under anaerobic conditions in sewer pipes. This hydrogen sulfide enters the sewer atmosphere by volatilization and dissolves in the condensate on the sewer crown. Finally, sulfur-oxidizing bacteria (SOB) oxidize the dissolved H2S and other sulfur compounds (e.g., S2O3− and S0) to sulfuric acid, which corrodes the concrete. Current theory suggests that because the pH of uncorroded concrete surface is approximately 12, a pH at which SOB cannot grow, a succession of microorganisms is required to enable colonization of the concrete surface by SOB and their production of sulfuric acid (17). However, the initial colonization and the microbial succession involved in the MICC process are very poorly understood because most of the previous studies were focused on corroded concrete and conducted with conventional culture-dependent techniques that could detect only a limited range of microorganism in such sewer environments (10, 14, 17). To efficiently control MICC, it is necessary to understand the microbial succession of SOB and other bacteria that are responsible for the initial colonization, the production of sulfuric acid, and the subsequent deterioration of concrete under real sewer conditions. Although molecular-based techniques have proven to be useful in more accurately describing the microbial ecology of acidic environments, only a few studies have been carried out with these techniques (15, 44).
In most cases, only the rate of weight loss of concrete coupons was evaluated in experimental chambers on a bench scale in which fully grown pure cultures of SOB in acidic artificial culture media were used to inoculate the concrete coupons (28, 29, 36, 40). The rates derived from these experiments do not necessarily reflect the rates that would be found in real sewer systems. To date, the MICC has not been quantitatively investigated, especially the initial stages of microbial colonization and succession of SOB populations in relation to the pH change and loss of weight of concrete in real sewer systems.
The objective of this study was, therefore, to acquire a quantitative understanding of the population dynamics of microbial communities inducing concrete corrosion by using several 16S rRNA gene-based molecular techniques: 16S rRNA gene-cloning analysis and fluorescent in situ hybridization (FISH). We placed concrete coupons in a severely corroding manhole of a real sewer system and quantitatively analyzed time-dependent changes in microbial communities, especially SOB species, growing on these corroding coupons. We also determined the vertical distribution of dominant SOB species in heavily corroded concrete (wet gypsum layer) to obtain a better understanding of the mechanism of MICC.
MATERIALS AND METHODS
Concrete coupons.
Concrete coupons were prepared by mixing cement, sand, and water at a weight ratio of 2.5:5:1, according to the Japanese JIS R 5201 Standard. The resulting paste was put into a frame (4 by 4 by 16 cm) and allowed to solidify. The specimens were cured in moist air for 24 h and then cured in water at 20°C for 14 days. After curing, the specimens were cut into small pieces (concrete coupons, 4 by 1.8 by 0.8 cm). A total of 60 concrete coupons were placed in stainless steel frames and then installed in a manhole connecting to sewer pipes from a sludge-thickening tank in Hachinohe, Japan, which has exhibited severe corrosion of concrete sewer pipes. Although returned water flows into this manhole for 45 min of each hour, these coupons were placed 30 cm above the water level. The water qualities of the returned water were as follows: pH, 6.9 ± 0.2; oxidation reduction potential, 34 ± 92 mV; chemical oxygen demand concentration, 226 ± 76 mg liter−1; and dissolved oxygen concentration, 2.5 ± 0.6 mg liter−1. The concentration of H2S gas in the manhole atmosphere was 30 ± 20 ppm. Oxygen and carbon dioxide were at atmospheric concentrations.
Sampling.
After 1 month of exposure in the sewer, two coupons were taken from the stainless steel frames at approximately 2-week intervals, up to day 174. After that time, we were unable to access the experimental site (the manhole) for about 6 months because of unexpected construction in the sewer system. Thus, we were able to obtain only 1-year-old samples after the construction was completed. The corroded products were collected from the entire concrete surface of each coupon by scraping with a clean metal spatula. Immediately following their collection, we transferred each sample scraped from an individual coupon to a separate, sterile 50-ml centrifuge tube containing sterilized, distilled water and shook the tubes vigorously for 3 min for subsequent treatment (each of these samples is defined hereafter as a master sample). Most of the corrosion products (mainly gypsum) easily dissolved into the water. For 16S rRNA gene-cloning analysis and chemical analysis, the master samples were directly used after large particles were allowed to settle for 1 minute. For FISH analysis and total cell counts, subsamples (various volumes) were taken from the master sample after large particles were allowed to settle for 1 minute, mixed with the equal amounts of fresh 8% paraformaldehyde solution (the final concentration was 4%), and stored for 4 h at 4°C. All analyses were performed in duplicate.
DNA extraction and PCR amplification.
Total DNA was extracted directly from each master sample (an approximately 0.2-ml subsample) with a FastDNA spin kit for soil (Bio 101; Qbiogene, Inc., Carlsbad, CA) as described in the manufacturer's instructions. 16S rRNA gene fragments were amplified from the extracted total DNA with Taq DNA polymerase (TaKaRa Bio, Inc., Ohtsu, Japan) by using bacterial primer sets 11F (46) and 1492R (46). The PCR products were electrophoresed on a 1% (wt/vol) agarose gel and purified with a WIZARD PCR Preps DNA purification system (Promega). To reduce the possible PCR bias, the rRNA gene was amplified in duplicate tubes for each sample, and all four tubes were combined for the next cloning step.
Cloning and sequencing of the 16S rRNA gene and phylogenetic analysis.
The purified PCR products were ligated into a qCR-XL-TOPO vector with a TOPO XL PCR cloning kit (Invitrogen, Carlsbad, CA). The ligated products were transformed into TOP10-competent Escherichia coli cells (Invitrogen). Plasmids were extracted from the cloned cells and purified with a Wizard Plus Minipreps DNA purification system (Promega). Nucleotide sequencing was performed with an automatic sequencer (3100 Avant genetic analyzer; Applied Biosystems). All sequences were checked for chimeric artifacts by using a CHECK_CHIMERA program from the Ribosomal Database Project (23). Partial sequences (approximately 600 bp) were compared with similar sequences of the reference organisms by a BLAST search (2). Sequences with 97% or greater similarity were grouped into operational taxonomic units (OTUs) by a SIMILARITY_MATRIX program from the Ribosomal Database Project (23). Nearly complete sequencing of the 16S rRNA gene of each representative OTU was performed, and the sequences were aligned with a CLUSTAL W package (43). A phylogenetic tree was constructed by the neighbor-joining method (39). Bootstrap resampling analysis of 500 replicates was performed to estimate the degree of confidence in tree topologies.
Sample fixation and washing.
After fixation with a 4% paraformaldehyde solution for 4 h at 4°C, samples were washed three times with phosphate-buffered saline (10 mM sodium phosphate buffer, 130 mM sodium chloride [pH 7.2]) using sequential centrifugation (10 min at 10,000 × g) and resuspension (34). After the washing steps, samples were spotted onto a gelatin-coated glass slide with six glass surface windows separated by a hydrophobic coating (31).
Fluorescent in situ hybridization.
The 16S and 23S rRNA-targeted oligonucleotide probes used in this study are listed in Table 1. The probes were labeled with fluorescein isothiocyanate (FITC) or tetramethyl rhodamine 5-isothiocyanate (TRITC). Three probes were newly designed in this study by using a PROBE_DESIGN tool of ARB software (22), according to the current version of the 16S rRNA sequence database. The specificity of the probes was checked against the ARB database, and the optimal hybridization conditions were experimentally determined (Table 1). We used the following bacterial strains to evaluate the specificities of the newly designed probes: Halothiobacillus neapolitanus (DSM 15147), Acidithiobacillus thiooxidans (DSM 14887), Thiomonas intermedia (NBRC 14564), and Thiobacillus plumbophilus (DSM 6690). We also used the following bacterial strains as negative controls: Halothiobacillus halophilus (DSM 6132), Thiobacillus plumbophilus (DSM 6690), Conchiformibius steedae (DSM 2580), and Nitrosomonas europaea (ATCC 25978). All probes designed in this study were named in accordance with the standard introduced by Alm et al. (1), while the names of previously described probes were not changed, to avoid confusion. Dehydration and FISH analysis were performed by using the procedure described by Amann et al. (3) and by Okabe et al. (31). The hybridization conditions used for FISH are shown in Table 1.
TABLE 1.
A list of sequences and hybridization conditions of oligonucleotide probes used in this study
| Probe | Sequence (5′ to 3′) | Specificity | FAa (%) | Reference(s) |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | Most bacteria | 4 | |
| EUB338 II | GCAGCCACCCGTAGGTGT | Planctomycetales | 8 | |
| EUB338 III | GCTGCCACCCGTAGGTGT | Verrucomicrobiales | 8 | |
| ARC915 | GTGCTCCCCCGCCAATTCCT | Archaea | 35 | 41 |
| ALF1b | CGTTCG(C/T)TCTGAGCCAG | Alphaproteobacteria, some Deltaproteobacteria, Spirochaetes | 20 | 24 |
| BET42a | GCCTTCCCACTTCGTTT | Betaproteobacteria | 35 | 24 |
| GAM42a | GCCTTCCCACATCGTTT | Gammaproteobacteria | 35 | 24 |
| G123T | CCTTCCGATCTCTATGCA | Thiothrix eikelboomii, T. nivea, T. unzii, T. fructosivorans, T. defluvii, Eikelboom type 021N groups I, II, and III | 40 | 19 |
| 21N | TCCCTCTCCCAAATTCTA | Eikelboom type 021N strain II-26 | 35 | 45 |
| Thio820 | ACCAAACATCTAGTATTCATCG | Acidithiobacillus thiooxidans, Acidithiobacillus ferrooxidans | 30 | 15, 34 |
| S-S-T.int-0442-a-A-18 | TCGATATTTCGCCCCCGC | Thiomonas intermedia | 10 | This study |
| S-S-T.plum-450-a-A-18 | ATTAGCCTCAACTGTTTC | Thiobacillus plumbophilus | 20 | This study |
| S-S-H.neap-635-a-A-19 | TAGAATCCCAGTATCCAAT | Halothiobacillus neapolitanus | 35 | This study |
| Ntspa712 | CGCCTTCGCCACCGGCCTTCC | Most members of the Nitrospirae phylum | 50 | 9 |
| LF655 | CGCTTCCCTCTCCCAGCCT | Leptospirillum groups I, II, and III | 35 | 5 |
| ACD840 | CGACACTGAAGTGCTAAGC | Acidiphilium genus | 10 | 5 |
| HGC69A | TATAGTTACCACCGCCGT | Actinobacteria (high G+C gram-positive bacteria) | 20 | 37 |
| LGC354a | TGGAAGATTCCCTACTGC | Firmicutes (gram-positive bacteria with low G+C content) | 35 | 25 |
| LGC354b | CGGAAGATTCCCTACTGC | Firmicutes (gram-positive bacteria with low G+C content) | 35 | 25 |
| LGC354c | CCGAAGATTCCCTACTGC | Firmicutes (gram-positive bacteria with low G+C content) | 35 | 25 |
FA, formamide concentration.
Total cell and probe-hybridized cell count.
The samples, fixed in 4% paraformaldehyde solution, were subjected to 4′,6′-diamidino-2-phenylindole (DAPI) staining and FISH. After DAPI staining, the samples (various volumes) were filtered with 25-mm, 0.2-μm-pore-sized black nucleopore polycarbonate filters (Costar Scientific). Total DAPI-stained cells were enumerated by direct counting according to the method described by Hobbie et al. (16). For the determination of probe-hybridized cell numbers, the ratios of total probe-hybridized cells to total DAPI-stained cells were determined on glass slides with six glass surface windows. To determine the ratios, we randomly chose three windows for each sample and directly counted probe-hybridized and DAPI-stained cells for at least 20 randomly chosen microscopic fields for each sample, which corresponded to 700 to 1,500 DAPI-stained cells. Subsequently, we determined the number of probe-hybridized cells (cells cm−2) by multiplying the total DAPI-stained cells and the ratios of total probe-hybridized cells to total DAPI-stained cells. This measurement was performed in duplicate, and then the average probe-hybridized cell numbers were reported in this study.
Vertical distribution of bacteria in heavily corroded concrete.
The vertical distributions of microbial populations in heavily corroded concrete samples (wet gypsum layer) were determined in duplicate. Since the gypsum layer had the consistency of “cottage cheese,” the gypsum layer was sliced into 1,000- to 1,500-μm sections by means of a Microslicer (model DTK 1000; D.S.K., Osaka, Japan), without any pretreatment, as described previously (32). The sliced samples were immediately transferred to sterile 50-ml centrifuge tubes containing sterilized, distilled water and shaken for 3 min. The samples were fixed and spotted onto gelatin-coated glass slides as described above. We determined the numbers of probe-hybridized cells and the total DAPI-stained cells as described above. The counts were then converted to cell densities (where a unit is cells per cm3 of corroded concrete).
Microelectrode measurement.
The in situ pH profile and the O2 penetration depth for the heavily corroded concrete (1-year-old samples) were determined in the laboratory using microelectrodes. Clark-type microelectrodes for O2 (35), LIX-type microelectrodes for pH (11), and amperometric microelectrodes for H2S (18) were prepared, calibrated, and used as described previously. The pH reference microelectrode was also constructed with a microcapillary tube. For all measurements, the concrete coupons were placed in an atmosphere-controlled chamber. A tray containing synthetic medium in the bottom of the chamber supplied sufficient moisture and H2S. The synthetic medium consisted of MgCl2·6H2O (450 μM), CaCl2 (400 μM), NH4Cl (1,000 μM), NaCl (1,000 μM), KH2PO4 (100 μM), NaHCO3 (100 μM), EDTA ([ethylenediaminetetraacetic acid] 270 μM), and Na2S·9H2O (various amounts). The pH of the medium was adjusted to pH 3 to 4. The chamber was kept at room temperature (20 to 23°C) and at 95 to 99% humidity (condensing conditions). The H2S concentration in the chamber atmosphere was periodically measured with a membrane H2S sensor and manually controlled to within 20 to 50 ppm by adding fresh Na2S·9H2O crystals and blowing air. The O2, pH, and reference microelectrodes were inserted directly into the corroded gypsum layer from the top through a small window in the chamber. The pH and reference microelectrodes were mounted on a micromanipulator at the same horizontal level. The distance between the tips of the microelectrodes was less than 0.5 cm. All electrode assemblies were placed in a chemical chamber. Each measurement was performed three to five times.
Sulfuric acid production rate.
The potential aerobic sulfuric acid production rate was determined in standard batch experiments by measuring the initial production of sulfate (33). The corrosion products were taken from the surface of 1-year-old concrete coupons, homogenized, and inoculated into duplicate serum vials (130 ml) containing 100 ml of a synthetic medium (21, 33). Na2S2O3·5H2O (2.5 mM) and Na2S·9H2O (1.6 mM) were used as the sole electron donors. The pH was adjusted to 4.0. The serum vials inoculated with corrosion products and uninoculated controls were incubated aerobically on a rotary shaker at 100 rpm at 20°C in the dark. At regular time intervals, subsamples were withdrawn for SO42− measurement. The SO42− concentrations during the initial 72-h incubation were used to calculate the SO42− production rate (micromolar concentration of SO42− per cubic centimeter of corroded concrete per hour) (33).
Chemical analyses.
The pH at the surface of the concrete coupon was measured using a flat surface electrode (HORIBA, Japan). A small droplet of deionized water was placed on the coupon surface and allowed to equilibrate (36). The pH of this droplet was then measured directly. At least two locations from duplicate coupons were measured. For the determination of SO42−, S2O32−, and S0 concentrations on the concrete coupons, the concentrations of SO42−, S2O32−, and S0 in the supernatant of the master samples were measured. The SO42− and S2O32− concentrations were measured with an ion chromatograph (model DX-100 with an AS4A column; Nippon DIONEX, Osaka, Japan) after filtration with 0.2-μm-pore-sized membrane filters (DISMIC-13CP; Advantec Co., Ltd.). Elemental sulfur (S0) was extracted in 99.5% ethanol and analyzed by high-performance liquid chromatography using a UV detector as described previously (12).
Weight loss.
After careful removal of surface precipitates (e.g., gypsum) with a toothbrush (after the sampling as described above), the coupons were dried in an oven at 80°C for 3 days, transferred to a chamber kept at a humidity of 100% for 24 h, and then weighed.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of the clones used for the phylogenetic analysis are AB255052 to AB255122.
RESULTS
pH, sulfate, and elemental sulfur on concrete surfaces.
Figure 1 shows a change in the appearance of concrete coupons exposed to the sewer atmosphere, demonstrating the occurrence of concrete corrosion. White powder-like depositions (most likely gypsum, CaSO4·2H2O) appeared on the surface of concrete coupons after day 102. After 1 year, a thick wetted gypsum layer was formed on the concrete surface. The gypsum layer had the consistency of “cottage cheese” and provided no structural strength.
FIG. 1.
Concrete coupons exposed to the sewer atmosphere [H2S(g), ca. 30 ppm] for 42 days (A), 102 days (B), and 1 year (C and D), showing the progression of concrete corrosion.
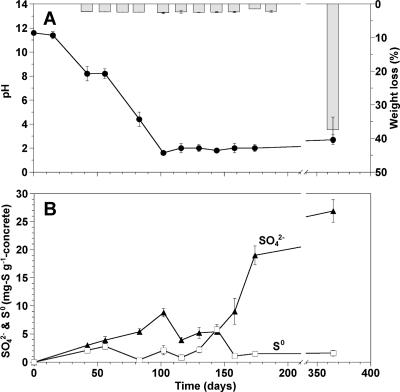
The initial pH of the new concrete surface was about 12 and gradually decreased to 8.2 within the initial 56 days (Fig. 2A). Thereafter, the pH decreased further to about 1.6 by day 102 and remained in this range. There were no significant weight losses of the concrete coupons (less than 3%) for the initial 174 days. A significant weight loss (37%) was observed after 1 year (Fig. 2A). A significant increase in SO42− concentration at the concrete surface, up to 20 mg S g−1 concrete, was observed on day 174 (Fig. 2B), while the level of pH remained unchanged. The concentration of S2O32− was below the detection limit throughout the entire monitoring period. Elemental sulfur gradually accumulated, up to 5.8 mg S g−1 concrete by day 144, and thereafter decreased with increasing SO42− concentrations.
FIG. 2.
Time-dependent changes in surface pH and weight loss of the concrete coupons exposed to the sewer atmosphere (A) and SO42− and S0 concentrations on the surface of concrete coupons placed in the sewer system (B). In panel A, the line graph refers to pH measurements and the bar graph to weight loss measurements. Error bars represent the standard errors of duplicate measurements.
Total and bacterial cell counts.
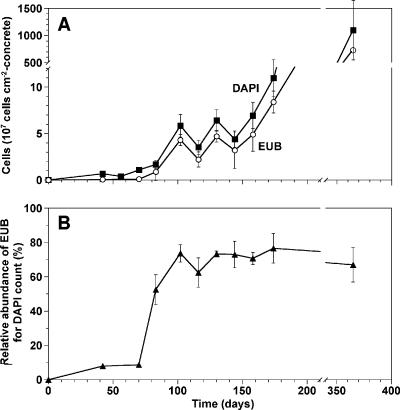
Figure 3 shows time course monitoring of numbers of DAPI-stained total cells and bacteria hybridized with EUB338-mixed probes on the concrete surface over 1 year. Significant bacterial growth occurred twice. The first growth was observed between day 73 and day 102 when the pH on the concrete surface became neutral. The second growth occurred after day 130, which was accompanied by the rapid increase in SO42− concentration. The abundance of the EUB338-mixed probe-hybridized cells relative to that of the total DAPI-stained cells exhibited a sharp increase, from 8% on day 73 to 80% on day 102, and thereafter remained unchanged (Fig. 3B). After 1 year, the total number of DAPI-stained cells reached 1.2 × 1010 cells cm−2. The abundance of Archaea hybridized with the ARC915 probe was below the detection limit of FISH during the entire experimental period.
FIG. 3.
(A) Time-dependent changes of total DAPI-stained (DAPI) cell numbers and EUB338-mixed probe-hybridized (EUB) cell counts on the concrete surface and (B) the relative abundance of EUB338-mixed probe-hybridized cells in relation to total DAPI-stained cells. Error bars represent the standard errors of duplicate measurements.
Phylogenetic analysis of microbial community on concrete surfaces.
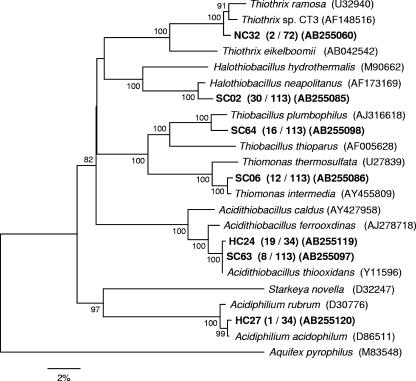
Microbial communities at different stages of the MICC process were analyzed by 16S rRNA gene-cloning analysis. Three clone libraries (uncorroded concrete taken at day 42 [72 clones], slightly corroded concrete taken at day 102 [115 clones], and heavily corroded concrete taken after 1 year [34 clones]) were constructed (Table 2 and Fig. 4). Only two clones (represented by NC32) were related to an axenic SOB, Thiothrix sp. strain CT3, in the uncorroded sample, comprising only 3% of total clones (2/72) (Fig. 4). In contrast, the clones belonging to four phylotypes of axenic SOB, Halothiobacillus neapolitanus, Thiomonas intermedia, Thiobacillus plumbophilus, and Acidithiobacillus thiooxidans, were detected in the slightly corroded sample. In the heavily corroded sample, the SOB microbial community was different from that in the uncorroded and slightly corroded samples. About 60% of clones (20/34) were closely related to Acidithiobacillus thiooxidans and Acidiphilium acidophilum, with 99% similarity, both of which were acidophilic SOB and able to oxidize S0 and S2O32− at a low pH (down to pH 0.5). One clone was closely related to an iron-oxidizing bacterium, Leptospirillum ferrooxidans (98% similarity).
TABLE 2.
Phylogenetic relatives and detection frequency of clones obtained from different stages of concrete corrosion
| Closest isolate (accession no.a) | No. of clones obtained
|
Similarity (%) | Sequence length (bp) | ||
|---|---|---|---|---|---|
| 42 days | 102 days | 1 yr | |||
| Nitrospira | |||||
| Leptospirillum ferrooxidans (X86776) | 1 | 98 | 1,480 | ||
| Alphaproteobacteria | |||||
| Rubritepida flocculans (AF465832) | 1 | 96 | 1,244 | ||
| Rhodobacter apigmentum (AF035433) | 1 | 98 | 1,387 | ||
| Sinorhizobium sp. (AF357225) | 1 | 97 | 1,440 | ||
| Devosia sp. (AJ786801) | 1 | 98 | 1,427 | ||
| Xanthobacter agilis (X94198) | 1 | 97 | 1,319 | ||
| Kaistia adipata (AY039817) | 1 | 2 | 98 | 1,337 | |
| Brevundimonas subvibrioides (AJ227784) | 1 | 6 | 98 | 1,401 | |
| Paracoccus aminophilus (AY014176) | 2 | 97 | 1,330 | ||
| Paracoccus alcaliphilus (AJ294415) | 2 | 95 | 1,413 | ||
| Caulobacter henricii (AJ227758) | 1 | 96 | 1,422 | ||
| Caulobacter intermedius (AB023784) | 1 | 99 | 1,397 | ||
| Ochrobactrum tritici (AY429607) | 2 | 98 | 1,438 | ||
| Acidiphilium acidophilum (D8651) | 1 | 99 | 1,419 | ||
| Betaproteobacteria | |||||
| Acidovorax delafieldii (AF078764) | 1 | 99 | 1,490 | ||
| Aquaspirillum psychrophilum (AF078755) | 1 | 98 | 1,489 | ||
| Rhodoferax ferrireducens (AF435948) | 1 | 99 | 1,467 | ||
| Polaromonas naphthalenivorans (AY166684) | 2 | 98 | 1,396 | ||
| Variovorax paradoxus (AJ420329) | 1 | 98 | 1,462 | ||
| Thiobacillus plumbophilus (AJ316618) | 16 | 97 | 1,469 | ||
| Thiomonas intermedia (AY455809) | 12 | 99 | 1,390 | ||
| Achromobacter xylosoxidans (AF411020) | 1 | 5 | 96 | 1,491 | |
| Azonexus caeni (AB166882) | 1 | 93 | 1,464 | ||
| Unidentified Betaproteobacteria | 1 | ||||
| Gammaproteobacteria | |||||
| Thiothrix sp. strain CT3 (AF148516) | 2 | 98 | 1,444 | ||
| Legionella lytica (X66835) | 2 | 96 | 1,484 | ||
| Psychrobacter fozii (AJ430827) | 18 | 98 | 1,477 | ||
| Frateuria sp. (AY495957) | 11 | 99 | 1,428 | ||
| Stenotrophomonas maltophilia (AB008509) | 6 | 99 | 1,467 | ||
| Halothiobacillus neapolitanus (AF173169) | 30 | 97 | 1,413 | ||
| Acidithiobacillus thiooxidans (Y11596) | 8 | 19 | 99 | 1,469 | |
| Acinetobacter junii (X81664) | 1 | 98 | 1,460 | ||
| Unidentified Gammaproteobacteria | 1 | ||||
| Firmicutes | |||||
| Carnobacterium alterfunditum (L08623) | 1 | 98 | 1,520 | ||
| Clostridium lentocellum (X71851) | 1 | 90 | 1,313 | ||
| Clostridium bartlettii (AY438672) | 1 | 2 | 97 | 1,263 | |
| Planococcus antarcticus (AJ314745) | 18 | 6 | 98 | 1,485 | |
| Turicibacter sanguinis (AF349724) | 1 | 98 | 1,473 | ||
| Actinobacteria | |||||
| Leucobacter komagatae (AJ746337) | 1 | 96 | 1,080 | ||
| Frankia sp. (AF063642) | 1 | 92 | 1,251 | ||
| Microbacterium oxydans (Y17227) | 1 | 99 | 1,453 | ||
| Arthrobacter bergeri (AJ609631) | 1 | 97 | 1,477 | ||
| Mycobacterium farcinogenes (AY457084) | 1 | 95 | 1,473 | ||
| Mycobacterium florentinum (AJ616230) | 1 | 98 | 1,470 | ||
| Bacteroidetes | |||||
| Flavobacterium psychrophilum (AB078060) | 1 | 95 | 1,452 | ||
| Subsaxibacter broadyi (AY693999Z) | 1 | 96 | 1,351 | ||
| Pedobacter piscium (AJ438174) | 1 | 1 | 95 | 1,390 | |
| Pedobacter heparinus (AJ438172) | 1 | 97 | 1,451 | ||
| Algoriphagus antarcticus (AJ577142) | 1 | 96 | 1,195 | ||
| Dyadobacter fermentens (AF137029) | 1 | 95 | 1,397 | ||
| Sphingobacterium thalpophilum (AJ438177) | 1 | 92 | 1,286 | ||
| Unidentified Bacteroidetes | 6 | ||||
| Unidentified bacteria | 4 | ||||
| Total | 72 | 113 | 34 | ||
Accession numbers are from the EMBL/GenBank/DDBJ databases.
FIG. 4.
Phylogenetic tree showing the distributions of the OTUs related to sulfur-oxidizing bacteria, which were obtained from 42-day-old (uncorroded, NC), 102-day-old (slightly corroded, SC), and 1-year-old (heavily corroded, HC) samples. The tree was generated by using approximately 1,400 bp of the 16S rRNA genes and the neighbor-joining method. The scale bar represents 2% sequence divergence. The values at the nodes are bootstrap values (500 resampling analysis). The Aquifex pyrophilus sequence served as the outgroup for rooting the tree. The numbers in parentheses indicate the frequencies of appearance of identical clones in the clones analyzed.
Succession of SOB.
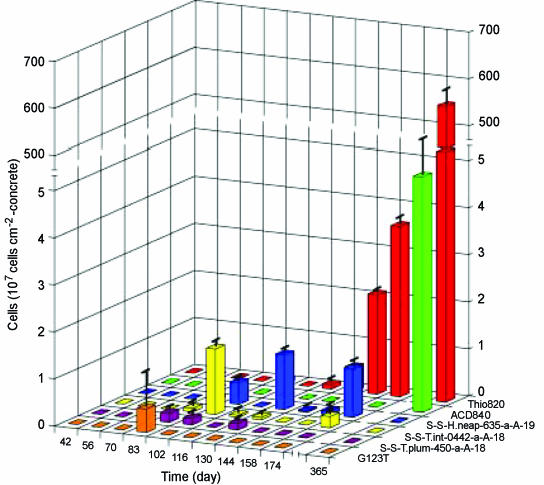
The results of phylogenic analysis suggested that at least six phylotypes of putative SOB (Thiothrix, Thiomonas, Thiobacillus, Halothiobacillus, Acidiphilium and Acidithiobacillus) species were involved in the MICC process in this study. Therefore, the population dynamics of these SOB species were determined by FISH. Figure 5 shows the time-dependent changes in abundances of these SOB species on the concrete surface. We could not detect any SOB species with the probes used in this study for the initial 70 days. On day 83, we detected for the first time filamentous bacteria that hybridized with the G123T probe specific for Thiothrix spp. but not with probe 21N. These filamentous bacteria accounted for 23% of the cells hybridized with the EUB338-mixed probe (defined as total bacteria hereafter). On day 102, probe G123T-hybridized Thiothrix spp. could no longer be detected. Instead, we noted hybridization with probe S-S-T.int-0442-a-A-18 for Thiomonas intermedia, accounting for 32% of the total bacteria. In addition, we found a moderate abundance of Halothiobacillus neapolitanus, identified by hybridization with probe H.neap-634. In the 130-day sample, H. neapolitanus, identified with the H.neap-634 probe, replaced the previously dominant SOB species. After day 144, we observed a significant increase in cells detected with the Thio820 probe. After 1 year, H. neapolitanus dominated the microbial community on the concrete surface, reaching an abundance of 5.5 × 109 ± 3.8 × 108 cells cm−2, corresponding to ca. 70% of the total bacteria. The abundance of T-plumbo-449-hybridized cells was at most 0.2 × 107 cells cm−2 between day 83 and day 130. Although none of cells was detected with the ACD840 and LF655 probes, specific for the Acidiphilium genus and Leptospirillum groups, until day 174, their numbers increased to 5 × 107 and 1.5 × 108 cells cm−2 after 1 year, respectively.
FIG. 5.
Time-dependent changes in numerically important SOB cell numbers detected by FISH analysis with SOB species- or genus-specific probes (Table 1). Error bars represent the standard errors of duplicate measurements.
Identification of Thio820-hybridized cells.
The Thio820-hybridized cells dominated the microbial community after 1 year. The Thio820 probe, however, hybridizes with both Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans (34). Terminal restriction fragment length polymorphism analysis was therefore conducted on 144-, 158-, and 174-day-old and 1-year-old samples to determine which species dominated in these communities (the detailed method of terminal restriction fragment length polymorphism is described in the supplemental material). The terminal restriction fragment peaks representing A. thiooxidans were detected in all samples, whereas the peaks representing A. ferrooxidans were found only in 174-day-old and 1-year-old samples (data not shown). The ratios of the peak heights of A. thiooxidans to those of A. ferrooxidans were about 15 to 1 in the 174-day-old sample and 30 to 1 in the 1-year-old sample, respectively. This result indicated that A. thiooxidans was the most dominant member of the community, as detected with the Thio820 probe.
Vertical distribution of microbial populations in the heavily corroded concrete.
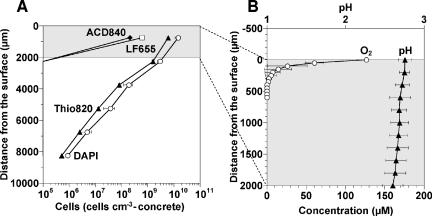
Figure 6A presents the vertical distributions of probe Thio820-, LF655-, and ACD840-hybridized cells in the 1-year-old heavily corroded concrete sample. Probe Thio820-hybridized Acidithiobacillus cells were the most dominant group (60% to 79% of the EUB338-mixed probe-hybridized cells) throughout the depth. Their number was highest (6.1 × 109 ± 1 × 109 cells cm−3 concrete) in the 1.5-mm layer near the surface and decreased logarithmically with the depth. The ALF1b-hybridized cells (ca. 8% of total DAPI-stained cells), the LF655-hybridized cells (ca. 3%), and the ACD840-hybridized cells (ca. 1%) were also detected only in the 1.5-mm layer near the surface. No hybridization signals were observed when probes Ntspa 712, HGC69a, LGC354a, LGC354b, and LGC354c were used.
FIG. 6.
(A) Vertical distribution of DAPI-stained total cells, Thio820 probe-hybridized Acidithiobacillus cells, ACD840 probe-hybridized Acidiphilium cells, and LF655 probe-hybridized Leptospirillum cells in the heavily corroded gypsum layer after a 1-year exposure to the sewer atmosphere. (B) Concentration profiles of O2 and pH in the top 2,000 μm of the heavily corroded gypsum layer that was exposed to the H2S atmosphere for 1 year. Stable total H2S concentration profiles could not be determined in this study. The surface of corroded concrete is at a depth of 0 μm. Error bars represent the standard errors of duplicate measurements (A) and of triplicate measurements (B).
The in situ pH profile of the heavily corroded gypsum layer that was exposed to the H2S atmosphere was determined with microelectrodes (Fig. 6B). The result indicated that the pH was within the range 2.6 to 2.7 throughout the depth (down to 2 mm). Oxygen concentration drastically decreased within the uppermost 200 μm, indicating a high O2 consumption rate probably due to high microbial sulfide-oxidizing activity. We could not determine reliable and stable total H2S concentration profiles, probably due to low H2S concentrations under such low pH conditions.
Potential sulfuric acid production rate.
In batch experiments, the SO42− production rates of a 1-year-old heavily corroded sample were determined to be 53 ± 4 μmol SO42− cm−3 of corroded concrete h−1 and 19 ± 5 μmol SO42− cm−3 of corroded concrete h−1 in the incubation with S2O32− and H2S as the sole electron donors, respectively (data not shown).
DISCUSSION
Succession of concrete corrosion and microbial community.
The results from our study of MICC indicate that the process can be divided into three stages: (i) the initial moderate lowering of pH, (ii) the continued decrease in pH with the appearance of a succession of neutrophilic SOB, and (iii) the emergence of acidophilic SOB and the development of severe concrete corrosion. The SOB species described in this paper are putative SOB species because we speculated only from the rRNA gene sequence similarity, without functional identification.
The pH decrease rate at the concrete surface during the first 56 days (the first stage) was 0.06 pH unit day−1 in this study. This rate is more than four times higher than the pure chemical pH decrease rate (0.013 pH unit day−1) that was calculated from the previously reported rate (4.38 × 10−5 pH unit day−1 ppm of H2S−1) (36) and the average H2S concentration (ca. 30 ppm) in the sewage manhole we studied. We suggest that, based on the rate of pH decrease we observed in our experiment, microbial activity was probably involved in addition to abiotic neutralization with CO2 and H2S. In fact, we found that some bacteria (approximately 1 × 107 cells cm−2) were already present on the concrete surface on day 42 (Fig. 3A), among which diverse bacterial species (mainly heterotrophic, halo-tolerant, and neutrophilic bacteria) were present (Table 2). Thus, the attachment and colonization of these pioneer microorganisms on the concrete surface could have a great impact on the initial pH decrease and establishment of suitable growth environments for the subsequent emergence of SOB species.
Continuing acidification of the concrete surface.
The second stage of pH decrease (pH 8.2 to 1.6) occurred between days 56 and 102 at a rate of 0.143 pH units day−1. During this period, microbial populations increased significantly (Fig. 3A) and a succession of neutrophilic SOB began to be detected by FISH (Fig. 5). However, significant weight loss by the concrete coupons was not yet detected (Fig. 2A). The dominant SOB species changed in the following order: Thiothrix spp., Thiobacillus plumbophilus, Thiomonas sp., and Halothiobacillus neapolitanus. These SOB species were probably responsible for the production of sulfuric acid that further reduced the pH of the concrete. General phenotypic characteristics of the SOB closely related to the clones obtained at the different stages of the MICC process are summarized (see Table S1 in the supplemental material). Although the involvement of Thiothrix spp. in MICC process was not reported in previous studies, Thiothrix spp. appeared first probably because Thiothrix spp. have an ability to grow heterotrophically or chemolithotrophically with H2S and S2O32− as electron donors at a neutral pH (38). Next, Thiomonas spp. emerged as the dominant organisms. Thiomonas spp. also can grow mixotrophically and use S0 and S2O32− as electron donors at a slightly lower pH range (pH 5.0 to 7.5) (20, 27). The third most numerous SOB in the second corrosion stage was Halothiobacillus neapolitanus. Unlike Thiothrix and Thiomonas, Halothiobacillus does not have an ability to grow mixotrophically but can grow in or adapt to a wider pH range (pH 4.5 to 8.5) (20). This is probably the reason Halothiobacillus appeared later than Thiothrix and Thiomonas and stayed for a long period, from day 83 to day 174.
In the third stage, at which the concrete pH remained around 2, we observed a weight loss by the concrete of 40% after 1 year, corresponding to a corrosion rate of ca. 3 to 4 mm year−1. This corrosion rate was within a range of previously reported corrosion rates for real sewer systems (28, 36). Coincident with this increased weight loss, the acidophilic SOB Acidithiobacillus thiooxidans appeared and became the most dominant microbial species. This is because A. thiooxidans can grow better at pH 2 with S0 and S2O32− as electron donors (20). It was previously reported that the Thio820 probe-hybridized Acidithiobacillus cells dominated in corroded sewerage collection systems (15). Thus, the optimum pH, trophic property (e.g., autotrophic or mixotrophic), and ability to utilize different sulfur compounds (e.g., H2S, S0, and S2O32−) of SOB probably determine the order of appearance of SOB species on corroding concrete surfaces in sewer systems.
Sulfide reacts with oxygen instantaneously, particularly when the pH is above 6 (6). This reaction may result in a number of products, including thiosulfate (S2O32−), sulfite (SO3−), and elemental sulfur (S0), which are readily utilized by the majority of SOB detected in this study (see Table S1 in the supplemental material) and are probably the true energy sources in the MICC process. The dominant Acidithiobacillus can preferentially utilize S2O32− over H2S.
In this study, we observed an accumulation of S0 around day 144, when the pH of the concrete surface was near 2. A previous report indicated that under such acidic conditions, abiotic conversion of S2O32− to S0 was rapid (17). S0 can also be produced by H. neapolitanus under oxygen-limited conditions and/or a higher load of H2S (20, 42). A decrease in S0 concentration accompanied by an increase in SO42− concentration after day 144 can be explained partly by the vigorous growth of A. thiooxidans, which has the ability to oxidize S0 (20).
Previous reports have mentioned that five Thiobacillus spp. (T. thioparus, T. novellus, T. neapolitanus, T. intermedius, and T. thiooxidans) were found to play important roles in concrete corrosion (17, 26, 36), but we could not find T. thioparus and T. novellus in this study. Instead, we found an involvement of Thiothrix spp. and Thiobacillus plumbophilus in the early stages of the MICC process. A. ferrooxidans was also found in the heavily corroded samples, which agrees with a previous report (47). However, the contribution of A. ferrooxidans to the corrosion process in our experiment must have been small because we found it to be much less abundant than A. thiooxidans. It was recently suggested that fungi also play an important role in the MICC process (7, 13, 30). We found no microscopic evidence at any time during the monitoring period of a significant presence of fungus-like microorganism after staining samples with DAPI.
Coexistence with heterotrophs.
Bacteria other than SOB coexisted with SOB during the bacterial succession in the corroding concrete (see Fig. S1 in the supplemental material). FISH analysis showed that more than 95% of total DAPI-stained microorganisms were bacteria other than SOB at day 42. Even after 1 year when the pH was around 2, bacteria other than SOB still accounted for ca. 50% of total DAPI-stained cells. Cloning analysis of 16S rRNA genes revealed that most of the bacteria other than SOB were related to heterotrophic bacteria and their community structure also changed with time (Table 2 and see Fig. S1 in the supplemental material). The most frequently detected clones were related to heterotrophic bacteria, which share the same physiological characteristics of the abilities to utilize various carbon sources, to produce acids, and to grow at low pH and high salt concentrations (see Table S2 in the supplemental material). Volatile organic compounds present in the sewer atmosphere could support the growth of heterotrophic bacteria on corroding concrete. In addition, since Acidithiobacillus excretes self-inhibitory organic compounds, it requires a mutualistic relationship with heterotrophs that can degrade such inhibitory organic compounds (7, 14, 34, 44). It is very likely that these heterotrophic bacteria scavenged organic compounds excreted by Acidithiobacillus. Therefore, microbial succession of both SOB and other heterotrophic bacteria was indeed required for the initial colonization of concrete and the production of sulfuric acid.
Mechanism of concrete deterioration.
To our knowledge, this is the first report of the vertical distributions of SOB populations, O2, and pH in the corroded concrete. FISH analysis and microelectrode measurements revealed that the highest abundance and activity of A. thiooxidans were found only in the surface layer of corroded concrete, where both H2S and O2 concentrations were high. Since the corroded concrete (gypsum layer) was wet, mushy, and about 10 mm thick after 1 year, H2S and O2 could penetrate only a few hundred micrometers as demonstrated by microelectrode measurement. Therefore, we speculate that sulfuric acid production by A. thiooxidans occurs mainly in the surface of the corroded concrete. The sulfuric acid produced must penetrate through the corroded concrete layer (gypsum layer) and eventually react with the sound (intact) concrete surface. The results of in situ analyses would provide us with valuable information to efficiently control, prevent, and/or predict MICC in sewer systems.
Supplementary Material
Acknowledgments
We thank the persons concerned with the wastewater treatment facility in Hachinohe, Japan, for allowing us to install the concrete coupons.
This work was partially supported by a grant-in-aid (13650593) for developmental scientific research from the Ministry of Education, Science and Culture of Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I. 1995. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, P. L., and J. F. Banfield. 2001. Designed and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. [DOI] [PubMed] [Google Scholar]
- 6.Chen, K. Y., and J. C. Morris. 1972. Kinetics of oxidation of aqueous sulfide by O2. Environ. Sci. Technol. 6:529-537. [Google Scholar]
- 7.Cho, K.-S., and T. Mori. 1995. A newly isolated fungus participates in the corrosion of concrete sewer pipes. Water Sci. Technol. 31:263-271. [Google Scholar]
- 8.Daims, H., A. Bruhl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Daims, H., J. L. Nielsen, P. H. Nielsen, K.-H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, J. L., D. Nica, K. Shields, and D. J. Roberts. 1998. Analysis of concrete from corroded sewer pipe. Int. Biodeterior. Biodegrad. 42:75-84. [Google Scholar]
- 11.de Beer, D., A. Schramm, C. M. Santegoeds, and M. Kühl. 1997. A nitrite microsensor for profiling environmental biofilms. Appl. Environ. Microbiol. 63:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 13.Gu, J.-D., T. E. Ford, N. S. Berke, and R. Mitchell. 1998. Biodeterioration of concrete by the fungus Fusarium. Int. Biodeterior. Biodegrad. 41:101-109. [Google Scholar]
- 14.Harrison, A. P., Jr. 1984. The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38:265-292. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez, M., E. A. Marchand, D. Roberts, and J. Peccia. 2002. In situ assessment of active Thiobacillus species in corroding concrete sewers using fluorescent RNA probes. Int. Biodeterior. Biodegrad. 49:271-276. [Google Scholar]
- 16.Hobbie, J. E., R. H. Daley, and S. Jasper. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Islander, R. L., J. S. Devinny, F. Mansfeld, A. Postyn, and H. Shih. 1991. Microbial ecology of crown corrosion in sewers. J. Environ. Eng. 117:751-770. [Google Scholar]
- 18.Jeroschewski, P., C. Steuckart, and M. Kühl. 1996. An amperometric microsensor for the determination of H2S in aquatic environments. Anal. Chem. 68:4351-4357. [Google Scholar]
- 19.Kanagawa, T., Y. Kamagata, S. Aruga, T. Kohno, M. Horn, and M. Wagner. 2000. Phylogenetic analysis of and oligonucleotide probe development for Eikelboom type 021N filamentous bacteria isolated from bulking activated sludge. Appl. Environ. Microbiol. 66:5043-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, D. P., and A. H. Harrison. 1989. Genus Thiobacillus, p. 1842-1858. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed), Bergey's manual of systematic bacteriology, vol. 3. Lippincott Williams & Wilkins, Baltimore, MD.
- 21.Kuenen, J. G., L. A. Robertson, and O. H. Tuovinen. 1991. The genera Thiobacillus, Thiomicrospira, and Thiosphaera, p. 2638-2657. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, NY. [Google Scholar]
- 22.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak, B. L., G. L. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 25.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 26.Milde, K., W. Sand, W. Wolff, and E. Bock. 1983. Thiobacilli of the corroded concrete walls of the Hamburg sewer system. J. Gen. Microbiol. 129:1327-1333. [Google Scholar]
- 27.Moreira, D., and R. Amils. 1997. Phylogeny of Thiobacillus cuprinus and other mixotrophic thiobacilli: proposal for thiomonas gen. nov. Int. J. Syst. Bacteriol. 47:522-528. [DOI] [PubMed] [Google Scholar]
- 28.Mori, T., T. Nonaka, K. Tazaki, M. Koga, Y. Hikosaka, and S. Noda. 1992. Interactions of nutrients, moisture and pH on microbial corrosion of concrete sewer pipes. Water Res. 26:29-37. [Google Scholar]
- 29.Mori, T., M. Koga, Y. Hikosaka, T. Nonaka, F. Mishina, Y. Sakai, and J. Koizumi. 1991. Microbial corrosion of concrete sewer pipes, H2S production from sediments and determination of corrosion rate. Water Sci. Technol. 23:1275-1282. [Google Scholar]
- 30.Nica, D., J. L. Davis, L. Kirby, G. Zuo, and D. J. Roberts. 2000. Isolation and characterization of microorganisms involved in the biodeterioration of concrete in sewers. Int. Biodeterior. Biodegrad. 46:61-68. [Google Scholar]
- 31.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okabe, S., T. Ito, K. Sugita, and H. Satoh. 2005. Succession of internal sulfur cycles and sulfide-oxidizing bacterial communities in microaerophilic wastewater biofilms. Appl. Environ. Microbiol. 71:2520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peccia, J., E. A. Marchand, J. Silverstein, and M. Hernandez. 2000. Development and application of small-subunit rRNA probes for assessment of selected Thiobacillus species and member of the genus Acidiphilium. Appl. Environ. Microbiol. 66:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 36.Roberts, D. J., D. Nica, G. Zuo, and J. L. Davis. 2002. Quantifying microbially induced deterioration of concrete: initial studies. Int. Biodeterior. Biodegrad. 49:227-234. [Google Scholar]
- 37.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 38.Rossetti, S., L. L. Blackall, C. Levantesi, D. Uccelletti, and V. Tandoi. 2003. Phylogenetic and physiological characterization of a heterotrophic, chemolithoautotrophic Thiothrix strain isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53:1271-1276. [DOI] [PubMed] [Google Scholar]
- 39.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Sand, W. 1987. Importance of hydrogen sulfide, thiosulfate, and methylmercaptan for growth of thiobacilli during simulation of concrete corrosion. Appl. Environ. Microbiol. 53:1645-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 42.Stefess, G. C., R. A. M. Torremans, R. de Schrijver, L. A. Robertson, and J. G. Kuenen. 1996. Quantitative measurement of sulphur formation by steady-state and transient-state continuous cultures of autotrophic Thiobacillus species. Appl. Microbiol. Biotechnol. 45:169-175. [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincke, E., N. Boon, and W. Verstraete. 2001. Analysis of the microbial communities on corroded concrete sewer pipes—a case study. Appl. Microbiol. Biotechnol. 57:776-785. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, M., R. Amann, P. Kämpfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K.-H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 46.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanaka, T., I. Aso, S. Togashi, M. Tanigawa, K. Shoji, T. Watanabe, N. Watanabe, K. Maki, and H. Suzuki. 2002. Corrosion by bacteria of concrete in sewerage systems and inhibitory effects of formats on their growth. Water Res. 36:2636-2642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.