Abstract
In filamentous fungi, RNA silencing is an attractive alternative to disruption experiments for the functional analysis of genes. We adapted the gene encoding the autofluorescent DsRed protein as a reporter to monitor the silencing process in fungal transformants. Using the cephalosporin C producer Acremonium chrysogenum, strains showing a high level of expression of the DsRed gene were constructed, resulting in red fungal colonies. Transfer of a hairpin-expressing vector carrying fragments of the DsRed gene allowed efficient silencing of DsRed expression. Monitoring of this process by Northern hybridization, real-time PCR quantification, and spectrofluorometric measurement of the DsRed protein confirmed that downregulation of gene expression can be observed at different expression levels. The usefulness of the DsRed silencing system was demonstrated by investigating cosilencing of DsRed together with pcbC, encoding the isopenicillin N synthase, an enzyme involved in cephalosporin C biosynthesis. Downregulation of pcbC can be detected easily by a bioassay measuring the antibiotic activity of individual strains. In addition, the presence of the isopenicillin N synthase was investigated by Western blot hybridization. All transformants having a colorless phenotype showed simultaneous downregulation of the pcbC gene, albeit at different levels. The RNA-silencing system presented here should be a powerful genetic tool for strain improvement and genome-wide analysis of this biotechnologically important filamentous fungus.
The study of gene functions in filamentous fungi has advanced substantially in recent years, and diverse genetic approaches have been used to disrupt target genes by homologous recombination (46). However, in most filamentous fungi, substitution of the target genes by homologous recombination occurs at very low frequencies due to ectopic integration of the transforming DNA. In wild-type strains of Aspergillus nidulans, Neurospora crassa, and Sordaria macrospora, for example, homologous recombination has been observed at frequencies of about 0.1% to 5% (4, 6, 30). One traditionally used approach is generation of multiple knockout strains; however, this approach is both time-consuming and labor-intensive. It also requires multiple resistance markers for selecting individual transformants showing homologous recombination (9). Therefore, discovery and sustained development of the RNA interference (RNAi) system are a reliable alternative for studying gene functions.
An advantage of the RNAi mechanism is its locus independence due to its mediation by a mobile trans-acting signal in the cytoplasm. Consequently, this mechanism can be used in fungi having multinuclear hyphae or a low targeting efficiency, and even heterokaryotic fungal strains can exhibit downregulation of target genes (8, 25). Generation of a knockout strain often requires a considerable length (>1 kb) of homologous sequences, whereas at least 132 bp of sequence homology is sufficient for double-stranded (dsRNA)-induced silencing of a target gene (5, 46). Typically, approximately 500 bp of coding sequence is used for RNAi approaches (19). This is advantageous when the information about genomic sequences is restricted. Beyond this, it is possible to silence different genes or gene families simultaneously (1, 9, 19).
Another advantage of knockdown strains is the possibility of controlling expression of the silencing construct. Inducible promoters, for example, can turn on gene expression at specific stages during development (10, 24). Even genes showing low-level expression can be downregulated efficiently with RNAi constructs expressing hairpin dsRNA (10). Furthermore, the RNAi system is useful when genes that are indispensable for fungal growth are investigated. In such cases, downregulation of gene expression close to zero results in survival of transformants.
Diverse RNAi systems have been developed for a broad range of filamentous fungi (25). Similar to the genomes of other eukaryotes, the genomes of filamentous fungi encode conserved components, such as RNA-dependent RNA polymerases that are known to be involved in the RNAi process. In fungi, downregulation of gene expression is variable in individual transformants, and gene functions can be studied only with preselected strains. To overcome these difficulties, several investigators have used vector systems in which a reporter gene is simultaneously downregulated together with an endogenous target gene. Such RNAi systems use either strain-specific marker genes that can be identified phenotypically (10, 19) or, alternatively, the generally applicable gfp reporter gene, encoding the green fluorescent protein (GFP) (8, 9). In the latter case, fluorescence microscopy identifies nonfluorescent strains and thus silenced transformants (8, 9, 14).
In this investigation, we used an alternative reporter system that is even simpler to identify silenced transformants. As the fungal organism, we used the major cephalosporin C producer Acremonium chrysogenum, which lacks an efficient host system, to study gene functions by homologous recombination. This biotechnologically relevant fungus propagates strictly asexually and therefore can be modified genetically only by molecular genetic techniques. Over the past few years, we and other workers have developed different selection and reporter gene systems to efficiently manipulate this important fungus (17, 18, 45). These systems include, for example, use of the gfp gene as a reporter of gene expression (31). Recently, an alternative application became feasible because of the discovery of the red fluorescent protein (DsRed) in Discosoma sp., which has an emission spectrum in the far-red zone (20). So far, the DsRed protein has been used as a highly effective marker in only a few filamentous fungi, including A. nidulans, Penicillium paxilli, and Trichoderma species (22, 43).
In this study, the DsRed gene was used to monitor gene expression in the cephalosporin C producer A. chrysogenum. Remarkably, colonies of strains carrying the DsRed reporter gene on solid media are red and thus can serve as recipients to monitor the RNA-silencing process. Consequently, a change in the color of the mycelium is an indicator of successful downregulation of gene expression. In addition, to quantify the RNA-silencing efficiencies, we developed a new spectrofluorometric approach to measure DsRed protein concentrations. We demonstrated the applicability of this RNA-silencing system by investigating cosilencing of the DsRed reporter gene together with a gene involved in secondary metabolism.
MATERIALS AND METHODS
Construction of the RNAi vectors.
Escherichia coli K-12 strain XL1-Blue (Stratagene) was used for general plasmid construction and maintenance (2). Plasmid pSilent-1 was generated previously as a vector for gene silencing in Magnaporthe oryzae and Colletotrichum lagenarium (26). For our approach, the hygromycin B resistance marker gene had to be replaced by the nat1 gene conferring nourseothricin resistance (16). Plasmid pNAT1 carrying the nat1 gene was hydrolyzed with SacI and NotI. The resulting 1,453-bp fragment was used for ligation with the SacI-NotI-restricted fragment of pSilent-1, and the resulting recombinant plasmid was designated pS-NAT1. In the next step, two 430-bp DsRed fragments, DsRedsense and DsRedanti, were amplified using primers DsRed-1 and DsRed-2 and primers DsRed-3 and DsRed-4, respectively, and plasmid pRHN1 as the template. pRHN1 carries the full-size DsRed gene (DsRed-Express; BD Biosciences Clontech, Heidelberg, Germany) under control of the gpd promoter and trpC terminator of A. nidulans (I. Godehardt and U. Kück, unpublished data). Then the DsRed fragments were inserted in inverse orientation into XhoI and StuI-ApaI restriction sites of plasmid pS-NAT1, generating plasmid pREDi.
In the case of plasmid pPCBCi, two inversely oriented pcbC fragments were PCR amplified with primers pcbC-1 and pcbC-2 and primers pcbC-3 and pcbC-4 using genomic DNA from A. chrysogenum A3/2 as the template. The PCR fragments were ligated into the vector pDrive (QIAGEN, Hilden, Germany) and subsequently isolated using SnaBI-HindIII (pcbCanti) and BglII-StuI (pcbCsense) restriction sites. These fragments were ligated into plasmid pREDi using the corresponding restriction sites. The sequences of all oligonucleotides and the recombinant plasmids are shown in Tables 1 and 2, respectively.
TABLE 1.
Sequences of oligonucleotides used in this work to generate PCR amplicons for construction of RNAi-inducing vectors and for real-time PCR
| Oligonucleotide | Sequence (5′-3′)a | Specificity |
|---|---|---|
| DsRed-1 | CTCGAGATGGCCTCCTCCGAGGACGTCATCAAGG | DsRed (positions 1 to 28) + XhoI |
| DsRed-2 | CTCGAGCCCAGCCCATAGTCTTCTTCTGC | DsRed (positions 430 to 408) + XhoI |
| DsRed-3 | GGGCCCATGGCCTCCTCCGAGGACGTCATCAAGG | DsRed (positions 1 to 28) + ApaI |
| DsRed-4 | AGGCCTCCCAGCCCATAGTCTTCTTCTGC | DsRed (positions 430 to 408) + StuI |
| pcbC-1 | AAGCTTATGGGTTCCGTTCCAGTTCC | pcbC (positions 1 to 20) + HindIIIb |
| pcbC-2 | TACGTATGAAGAAGTCCTCGTCGCGAC | pcbC (positions 520 to 500) + SnaBIb |
| pcbC-3 | AGATCTATGGGTTCCGTTCCAGTTCC | pcbC (positions 1 to 20) + BglIIb |
| pcbC-4 | AGGCCTTGAAGAAGTCCTCGTCGCGAC | pcbC (positions 520 to 500) + StuIb |
| DsRed_sense | GATCCACAAGGCCCTGAAGC | DsRed (positions 480 to 499) |
| DsRed_anti | GCTCCACGATGGTGTAGTCC | DsRed (positions 637 to 618) |
| SSU rRNA_for | ATCCAAGGAAGGCAGCAGGC | Small-subunit rRNA (N. crassa) |
| SSU rRNA_rev | TGGAGCTGGAATTACCGCG | Small-subunit rRNA (N. crassa) |
Restriction sites are underlined.
The nucleotide positions are the positions in the accession no. M33522 sequence.
TABLE 2.
Plasmids used for RNAi analysis
| Plasmid | Characteristics | Reference |
|---|---|---|
| pRHN1 | gpd promoter, DsRed gene (amino acids 1 to 226) and trpC terminator of A. nidulans | I. Godehardt and U. Kück, unpublished data |
| pNAT1 | gpd promoter of A. nidulans and nat1 gene of Streptomyces noursei | I. Godehardt, D. Janus, and U. Kück, unpublished data |
| pSilent-1 | trpC promoter and terminator of A. nidulans; hph resistance gene and cutinase intron 2 of M. oryzae | 26 |
| pS-NAT1 | gpd promoter of A. nidulans and nat1 gene in pSilent-1 | This study |
| pREDi | DsRed fragments (amino acids 1 to 143) in sense and antisense orientations in pS-NAT1 | This study |
| pPCBCi | pcbC fragments (amino acids 1 to 173) in sense and antisense orientations in pREDi | This study |
Fungal strains and culture conditions.
Transformation of A. chrysogenum strain ATCC 14553 and producer strain A3/2 was performed using conventional transformation procedures (32, 45). All strains used in this study are listed in Table 3, and their genotypes were verified by Southern blot analysis (data not shown) using gene-specific probes for hybridization of genomic DNA. To generate strains for RNA silencing of the DsRed or pcbC gene, ATCC:DsRed or A3/2:DsRed was transformed with plasmid pREDi or pPCBCi. The resulting transformants were selected on media containing hygromycin B and nourseothricin at concentrations described previously (17, 32, 45).
TABLE 3.
Recipient and transgenic strains used in this study
| Strain | Characteristic(s) | Source or reference |
|---|---|---|
| ATCC 14553 | Wild-type strain | American Type Culture Collection |
| A3/2 | Producer strain | 32 |
| ATCC:DsRed | gpd(p)::DsRed::trpC(t), trpC(p)::hph | This study |
| A3/2:DsRed | gpd(p)::DsRed::trpC(t), trpC(p)::hph | This study |
| ATCC:DsRedi T8, T20, T25, T43, T52, T53, T54, T123, T131, T144, T145, T179, and T180 | gpd(p)::DsRed::trpC(t), trpC(p)::hph, trpC(p)::DsReds::DsRedas::trpC(t), gpd(p)::nat1 | This study |
| A3/2:DsRedi T27 | gpd(p)::DsRed::trpC(t), trpC(p)::hph, trpC(p)::DsReds::DsRedas::trpC(t); gpd(p)::nat1 | This study |
| A3/2:pcbCi T4, T8, T9, and T10 | gpd(p)::DsRed::trpC(t), trpC(p)::hph, trpC(p)::DsReds::pcbCas::pcbCs::DsRedas::trpC(t), gpd(p)::nat1 | This study |
Liquid cultures of A. chrysogenum strains were grown at 27°C and 180 rpm for 3 days in CCM medium as described previously (23). Cultivation of strains for RNA and protein isolation was started with a 5% inoculum from a 2.5-day-old liquid culture.
Preparation and analysis of RNA.
RNA was isolated as described by Jekosch and Kück (12), and the integrity of all RNAs was verified by agarose gel electrophoresis and Northern blot analyses (35). The blots were hybridized with 32P-radiolabeled DNA probes as described in the Results.
Quantitative real-time PCR.
Quantitative real-time PCR was performed as described previously (29), with the following modifications: reverse transcription was performed with 400 U Superscript II (Invitrogen) and deoxynucleoside triphosphates at a concentration of 0.33 mM, and real-time PCR was carried out with a DNA Engine Opticon 2 (MJ Research).
Protein purification and Western blot analysis.
Protein purification was carried out as described previously (36), except that the mycelium was resuspended in 2 ml of buffer A (0.1 M MOPS [morpholinepropanesulfonic acid] [pH 7.5], 0.2 M KCl, 10 mM MgCl2, 1 mM EDTA, 10 mM dithiothreitol, 4.2 mM phenylmethylsulfonyl fluoride, 40% [wt/vol] glycerol). Ten micrograms of each protein extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane (44). Western blotting and immunodetection of the pcbC gene product were performed by using previously described methods (13).
Preparation of A. chrysogenum crude mycelial extract.
To isolate A. chrysogenum crude extracts, mycelium was first filtered and then resuspended in 40 ml of buffer A (see above). The mycelium was disrupted with a standard household blender (Siemens, Munich, Germany) for 3 min on ice. Spectrofluorometric measurement was performed with 20 μl of each crude extract that was diluted in 2 ml of storage buffer (BD Biosciences) (10 mM EDTA, 10% glycerol in phosphate-buffered saline [20 mM NaH2PO4·H2O, 80 mM Na2HPO4·2H2O, 100 mM NaCl]). The remaining portion was vacuum filtered and dried for 24 to 48 h at 65°C to determine the dry weight of the mycelium.
Spectrofluorometric measurement.
A calibration curve was produced using purified DsRed protein (BD Biosciences) as a prelude to determining DsRed levels in protein or crude mycelial extracts. The fluorescence of the DsRed protein at 576 nm was measured by using an excitation wavelength of 554 nm and a JASCO FP-6500 spectrofluorometer (JASCO, Tokyo, Japan). Both the emission and excitation bandwidths were set at 5 nm. The protein or mycelial extract measurements were obtained using a 1:100 or 1:1,000 dilution in storage buffer. All the results described below are means of three independent measurements. The amount of DsRed protein was expressed in ng DsRed/mg mycelium or ng DsRed/μg total protein.
Quantification of cephalosporin C synthesis.
Cephalosporin C synthesis was quantified with a bioassay as described previously (45). Alcaligenes faecalis was used as a gram-negative indicator bacterium for antibiotic production. Fungal transformants were grown for 3 days in liquid culture, and each supernatant was applied to wells in the test medium seeded with A. faecalis. Test plates were incubated for at least 12 h at 37°C. Simultaneously, the dry weight of the mycelium was estimated to calculate the ratio of the halo size to the mycelial biomass. On all plates, the halo of the A3/2:DsRed recipient was used as a control, and its size was defined as 100%. Accordingly, the areas of the halos were determined and grouped into four classes: large (100 to 70% of the control strain A3/2:DsRed halo size), medium (70 to 30%), small (30 to >0%), and no halos.
RESULTS
Use of the DsRed gene to monitor RNA silencing.
A. chrysogenum exhibits a high frequency of nonhomologous recombination, and therefore, gene substitution to generate knockout strains is inefficient (11, 36, 45). As an alternative, we developed a reliable RNA-silencing system using the DsRed gene from Discosoma sp. as a reporter. For our studies, two A. chrysogenum strains with different properties were used. ATCC 14553 is closely related to the wild-type strain and gives transformation frequencies of about 20 transformants per μg DNA (45). The cephalosporin C titer of this strain is relatively low compared to that of the producer strain A3/2, and antibiotic production can be measured with conventional bioassays only for the latter strain (32). In contrast, transformation of A3/2 resulted in a very low number of transformants (45), and therefore, our initial experiments to establish the RNAi system were performed with ATCC 14553.
As a first step to develop an RNAi system, we generated a transgenic recipient A. chrysogenum strain expressing the DsRed gene. After transformation of ATCC 14553 with pRHN1 (Table 2), about 70% of the strains obtained were pink or red on solid medium containing hygromycin B. To avoid heterokaryon formation, a single red transformant was selected and used for preparation of protoplasts. After regeneration of protoplasts, a single red colony was propagated on solid media. This strain had a stable phenotype even after at least five passages on solid medium and was used as a recipient (ATCC:DsRed) in further RNAi experiments. In a comparable experiment, strain A3/2:DsRed was generated using A3/2 as the recipient.
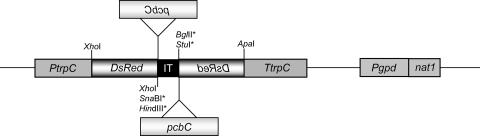
To silence the DsRed gene, plasmid pREDi was constructed as shown in Fig. 1. pSilent-1 carrying cutinase intron 2 from M. oryzae was used as the basic vector. This plasmid was reported previously to induce strong and relatively stable silencing in fungal transformants like M. oryzae and C. lagenarium (26). pSilent-1 also contained unique cloning sites for integration of two approximately 430-bp inversely oriented DsRed fragments. As an alternative selectable marker, the nourseothricin resistance gene from plasmid pNAT1 (Table 2) was inserted as described in Materials and Methods. Plasmid pREDi itself is not sufficient to generate a functional DsRed protein.
FIG. 1.
Schematic map of the pREDi vector and the derived recombinant plasmid pPCBCi. pREDi is a derivative of plasmid pSilent-1, carrying inversely oriented DsRed gene fragments. In the case of plasmid pPCBCi, the pcbC gene from A. chrysogenum was introduced into singular restriction sites of the pREDi vector (indicated by asterisks). Genes are not drawn to scale. For details concerning construction, see the text. Abbreviations: DsRed, DsRed gene from Discosoma sp.; IT, intron 2 of the cutinase gene from M. oryzae; nat1, nourseothricin acetyltransferase gene from Streptomyces noursei; pcbC, isopenicillin N synthase gene from A. chrysogenum; Pgpd, A. nidulans gpd promoter; PtrpC, A. nidulans trpC promoter; TtrpC, A. nidulans trpC terminator.
To establish the RNAi system in A. chrysogenum, the recipient strains and vectors described above were tested in three independent experiments. Transformation of pREDi into ATCC:DsRed resulted in generation of 478 nourseothricin- and hygromycin B-resistant transformants. These transformants exhibited at least three different types of coloring. As shown in Fig. 2 and summarized in Table 4, we found that about 20% of the colonies were dark red, while about 40% of the colonies were pink. More than 30% of the transformants showed no coloring at all and thus were most severely silenced.
FIG. 2.
Phenotypes of three representative colonies of A. chrysogenum transformants (ATCC:DsRedi) on solid medium, showing different colors due to silencing of the DsRed gene.
TABLE 4.
Summary of RNAi transformants having different colors of colonies
| Recipient strain | No. (%) of transformants
|
||||
|---|---|---|---|---|---|
| Red | Pink | Colorless | Heterokaryons | Total | |
| ATCC:DsRed(pREDi)a | 89 (18.6) | 194 (40.6) | 159 (33.3) | 36 (7.5) | 478 (100) |
| A3/2:DsRed(pREDi)a | 22 (14.3) | 64 (41.6) | 54 (35) | 14 (9.1) | 154 (100) |
| A3/2:DsRed(pPCBCi)b | 17 (25) | 29 (42.6) | 18 (26.5) | 4 (5.9) | 68 (100) |
Recipient strains ATCC:DsRed and A3/2:DsRed were transformed with vector pREDi to silence the DsRed gene.
Transformants were obtained when recipient strain A3/2:DsRed was transformed with pPCBCi in order to cosilence the DsRed and pcbC genes.
To exclude the possibility that the recipient strain, or alternatively the transformation procedure itself, was responsible for the phenotype observed, we conducted two different control experiments. In the first experiment, the DsRed-expressing producer strain A3/2:DsRed was transformed with plasmid pREDi. As shown in Table 4, the ratio of the three classes of colored transformants was very similar to the ratio obtained with strain ATCC:DsRed. In the second experiment, plasmid pS-NAT1, which was almost identical to pREDi but lacked the two 430-bp DsRed fragments, was transformed into ATCC:DsRed and A3/2:DsRed. In these cases, all 50 and 9 transformants, respectively, had a red phenotype.
In conclusion, the DsRed reporter gene provides a powerful tool for identification of silenced fungal transformants. These transformants can be identified easily by their pink or colorless phenotype compared to the red recipient.
Characterization of DsRed-silenced transformants.
For further analysis, Northern blot hybridization, quantitative real-time PCR, and spectrofluorometric assays were used to characterize the fungal transformants mentioned above. We randomly selected two red colonies, four pink colonies, and five colorless colonies and examined these colonies together with the wild-type strain (ATCC 14553) and the ATCC:DsRed recipient as controls. After RNA isolation, Northern blot hybridization was conducted with the DsRed and gpd genes as radiolabeled probes. To detect only the full-length DsRed transcript and not RNAs derived from the pREDi vector, a 240-bp fragment was used to detect the 3′ end of DsRed. This gene fragment was present in pRHN1 but not in pREDi. With the exception of the ATCC 14553 strain, all strains carried plasmid pRHN1, and thus the DsRed transcript could be detected in Northern hybridization blots (Fig. 3A).
FIG. 3.
Expression of the DsRed reporter gene in A. chrysogenum transformants (strain ATCC 14553) and recipient strains (ATCC 14553 and ATCC:DsRed). Open, shaded, and solid stars indicate white, pink, and red fungal colonies, respectively. (A) Northern blot hybridization using the RNAs indicated at the top. Radiolabeled DsRed- and gpd-specific probes were used for hybridization. (B) Quantitative real-time PCR of the DsRed transcript. The data are the ratios of the log2 values for the transformants (ATCC:DsRedi) to the log2 values for the recipient strain (ATCC:DsRed). (C) Spectrofluorometric quantification of the DsRed protein (ng DsRed/mg mycelium) in crude mycelial extracts.
However, the hybridizing bands for the DsRed transcript had different intensities. The strongest signal was obtained from red colonies, while in pink and colorless colonies there was a clear reduction in the DsRed transcript level. The most significant reduction was seen in transformants T180 and T131, both of which appeared to be white on solid media. The gpd probe was used as a reference and gave almost identical signals with RNA from all transformants tested. Comparable results were obtained when A3/2:DsRed was used as a recipient (data not shown).
More detailed quantification was achieved by real-time PCR using oligonucleotides that hybridized only to the 240-bp 3′ part of the DsRed gene, as described above. For normalization, small-subunit rRNA levels were determined as previously described (29). As a typical example of this analysis, the log2 ratios for individual transformants compared to the ATCC:DsRed recipient strain are shown in Fig. 3B. These data are in good accordance with the Northern blot hybridization data shown in Fig. 3A. The colorless transformant T180 showed the most drastic change, with a 16-fold reduction in the DsRed transcript level.
Finally, DsRed gene expression was measured on the protein level by using a simple and reproducible spectrofluorometric measurement for quantification. The results of spectrofluorometric DsRed protein quantification for the crude mycelial extracts are shown in Fig. 3C and demonstrate that the red transformants ATCC:DsRedi T25 and T43 had an increased amount of DsRed compared to the amount in the recipient strain. All pink transformants gave intermediate values for DsRed, while the white transformants clearly had the lowest levels of DsRed. Once again, the most remarkable examples are ATCC:DsRedi T54 and T180, which had a DsRed synthesis rate close to zero. The wild-type strain (ATCC 14553) served as a negative control which demonstrated that no background fluorescence was observed in this analysis.
Our results for the spectrofluorometric measurements are consistent with the results of the RNA analysis. To the best of our knowledge, this is the first study using spectrofluorometric measurement of the DsRed protein in microbial transformants, an approach that also has the advantage of being a fast and efficient procedure for protein quantification. Taken together, our data clearly show that the RNA-silencing approach described here for A. chrysogenum is effective and can be used for targeted inhibition of genes.
Cosilencing of DsRed and the endogenous pcbC gene.
To examine the effect of RNAi on the expression of an endogenous fungal gene product, an interference construct was generated to inhibit specifically secondary metabolism in A. chrysogenum. The pcbC gene encodes the isopenicillin N synthase and is strongly expressed on the transcriptional level compared with the expression of other cephalosporin C biosynthesis genes. Previously, inactivation of pcbC has resulted in strains having a reduced antibiotic titer (45) and thus can be used as a representative marker to demonstrate downregulation of the cephalosporin C biosynthesis pathway.
To achieve this, pcbC was silenced simultaneously with DsRed, which was used as a marker for selecting knockdown transformants. Starting with plasmid pREDi, inversely oriented pcbC fragments, which were about 520 bp long, were cloned using SnaBI/HindIII and BglII/StuI restriction sites for the pcbCanti and pcbCsense fragments, respectively. In relation to the DsRed gene, each fragment was inserted in the opposite orientation. A linear map of the resulting plasmid, pPCBCi, is shown in Fig. 1.
In the next step, plasmid pPCBCi was transformed into recipient strain A3/2:DsRed. As shown in Table 4, about 70% of the transformants had an altered phenotype compared to the phenotype of the recipient strain. From 68 transformants, we selected 4 that had different phenotypes for further analysis. A3/2:DsRed and A3/2:DsRedi T27 were used as appropriate controls. A3/2:DsRedi T27 was generated as described above and carried plasmid pREDi instead of pPCBCi. As shown in Fig. 4A, RNA from the six strains described above was used for Northern blot analysis. Similar to the previous hybridization analysis, we generated a probe that detected only the full-size DsRed transcript. Similarly, the mature pcbC transcript hybridized with a probe that was complementary to the residual 500 bp of the 3′ region of pcbC and did not exhibit any homology to plasmid pPCBCi. As shown in Fig. 4A, we found that there was a good correlation between the color of the colonies and the RNA transcript levels. In T10, which is a red strain, neither the level of pcbC nor the level of DsRed mRNA was reduced. The opposite was observed for T4, T8, and T9. In these cases, the DsRed expression levels, as well as the pcbC expression levels, were reduced. Remarkably, T8 showed the smallest reduction in the pcbC mRNA level, a finding that is consistent with data obtained later in protein quantification and bioassay analyses. On the other hand, the results for T27, which was used as a control, confirmed that downregulation of DsRed itself was not sufficient to reduce the amount of the pcbC mRNA transcript.
FIG. 4.
Analysis of A. chrysogenum transformants generated with strain A3/2:DsRed as the recipient and construct pPCBCi. (A) Northern blot hybridization analysis using radiolabeled probes for hybridization, as indicated on the right. (B) Spectrofluorometric quantification of the DsRed protein (ng DsRed/μg total protein). The error bars indicate the standard deviations of three independent measurements. (C) Western blot analysis using polyclonal antibody against the isopenicillin N synthase (IPNS). As a loading control, the Coomassie blue-stained protein gel is shown. The colors of fungal colonies are indicated as explained in the legend to Fig. 3.
Protein analysis of silenced transformants.
To further correlate the levels of mRNA and protein synthesis in silenced transformants, we performed spectrofluorometric measurement and Western blot analyses. For the spectrofluorometric analysis, we used crude protein extracts from all of the transformants described above. As shown in Fig. 4B, the amount of the DsRed protein correlated perfectly with the intensity of the signals obtained in the Northern blot analysis. As expected, the A3/2 producer strain exhibited no background fluorescence (data not shown), while for all silenced strains values less than 0.05 ng DsRed/μg protein were obtained.
In parallel, we detected isopenicillin N synthase using a polyclonal antibody that was described previously (13). As shown in Fig. 4C, all control strains, as well as the red knockdown strain T10, gave a strong signal in the Western blot analysis. Unexpectedly, the colorless strain T8 also showed a strong signal in the Western analysis. However, considering the results of the Northern hybridization experiment, this can be explained by the amount of the pcbC mRNA transcript, which was detectable in this transgenic strain. The most dramatic reduction in the level of the polypeptide encoded by pcbC was observed in T4, a result that correlates perfectly with the amount of pcbC mRNA transcript detected.
Bioassay to measure cephalosporin C titers.
The antibiotic cephalosporin C inhibits the growth of gram-positive as well as gram-negative bacteria (for a review, see reference 37). During biosynthesis, cephalosporin C is secreted into the culture broth, which can be used for bioassays with A. faecalis as the cephalosporin C-sensitive indicator (41).
Figure 5 shows the results of a cephalosporin C bioassay with the four pcbC-silenced transformants (A3/2:pcbCi T4, T8, T9, and T10) and the three control strains. As this figure shows, the three controls produced halos that were almost the same size, indicating that very similar amounts of cephalosporin C were secreted into the culture broth. As expected, the extracts from the red transformant T10 and from the three control strains exhibited the same inhibition of the bacterial lawn. The pink transformants and the two colorless transformants produced smaller halos, and the most extreme results were obtained when supernatants from transformant T4 were used. In this case, no halo was visible. The overall results of a bioassay performed with extracts from 40 different transformants are shown in Table 5, and these results demonstrate that the four transformants analyzed in detail were representative of all strains investigated. Generally, the red transformants produced large halos, but some halos were significantly smaller than those produced by the recipient strain. We found some pink transformants which produced small halos representing less than 30% of the activity observed in the control. The most drastic reduction was found in the colorless transformants. Six members of this group formed no halos, which indicates that complete knockdown of pcbC expression occurred. In conclusion, the chimeric hairpin construct described here induced silencing of both target genes in most but not all transformants investigated. Nevertheless, cosilencing should be a reliable alternative to construction of knockout strains for efficient functional genomic analysis of the filamentous fungus A. chrysogenum.
FIG. 5.
Bioassay to measure the antibiotic activity of cosilenced transformants and recipient strains (A3/2:DsRed and A3/2:DsRedi T27) using A. faecalis as an indicator bacterium. Large, medium, and small halos are shown. Some transformants, like A3/2:pcbCi T4, did not show any activity in this test. For further comparison, the halo of the nontransformed producer strain A3/2 is shown. For an explanation of the stars see the legend to Fig. 3.
TABLE 5.
Summary of bioassay to measure cephalosporin C titers for 40 different A. chrysogenum transformants (A3/2:pcbCi)a
| Color of transgenic strain | No. (%) of transformantsb
|
|||
|---|---|---|---|---|
| Large | Medium | Small | None | |
| Red | 4 | 9 | ||
| Pink | 1 | 7 | 5 | |
| Colorless | 2 | 6 | 6 | |
| Total | 5 (12.5) | 18 (45) | 11 (27.5) | 6 (15) |
For comparison, the activity of recipient strain A3/2:DsRed was defined as 100%. Transformants were grouped into four classes (large, medium, small, and none) according to the size of halos on solid medium. A. faecalis was used as the indicator organism in this analysis.
Large, 100 to 70% of the activity of strain A3/2:DsRed; medium, 70 to 30% of the activity of strain A3/2:DsRed; small, 30 to >0% of the activity of strain A3/2:DsRed; none, no activity.
DISCUSSION
DsRed gene is a favorable and reliable alternative for screening fungal transformants.
We recently used reverse genetic techniques to investigate genes controlling cephalosporin C biosynthesis in the biotechnologically important fungus A. chrysogenum (11, 36, 45). Several genes were replaced by a resistance marker gene in order to study the resulting knockout strains. However, the frequency of homologous recombination in A. chrysogenum is extremely low, and therefore screening for a knockout strain is both time-consuming and labor-intensive. Recently, other investigators have developed the double-marker enrichment technique for A. chrysogenum, which increases the frequency of homologous recombination by a factor of 10 (18). Nevertheless, the frequencies (about 5 to 30%) are still low compared to the frequencies of integration events occurring in strains deficient in nonhomologous end joining. For example, disruption of the ku70 or ku80 gene resulted in almost 100% homologous recombination in Aspergillus species, N. crassa, and S. macrospora (7, 15, 27, 28, 30). A comparable recipient strain for A. chrysogenum has not yet been constructed, and therefore we devised and developed an RNA-silencing system for A. chrysogenum that has the potential to become a powerful tool for functional genomic studies. For example, this system offers the possibility of generating double-knockdown strains in A. chrysogenum, allowing studies of the functions of genes involved in a common biosynthetic pathway.
Previously, fungal RNA-silencing systems were developed for different species using reporters like the GFP, and screening of the silenced strains was performed predominantly by fluorescent microscopy (14). However, the DsRed reporter gene used here is a more favorable alternative that allows effortless detection of silenced transformants by a change in the easily recognizable red phenotype. Recently, a similar observation was reported for P. paxilli carrying the DsRed gene (22). As a consequence, preselection of silenced transformants is possible by visual inspection without the need for any molecular characterization (e.g., Northern blot hybridization). In addition to phenotypic detection, use of the DsRed protein has the advantage that it is possible to quantify gene expression using simple-to-obtain spectrofluorometric measurements. Only crude mycelial extracts are necessary to determine the amount of DsRed protein, and we are currently developing procedures to quantify the DsRed protein in intact fungal cells. Another application of the spectrofluorometric measurements could be quantification of promoter strength in expression studies. Such studies could be particularly apt for investigating mutated promoter sequences conferring constitutive or induced expression properties. Similarly, high-expression constructs could be tested with the DsRed as a reporter. To the best of our knowledge, this is the first report to describe successful quantification of the DsRed protein in crude protein extracts from microbes using a spectrofluorometry approach. Comparable approaches to quantify an autofluorescent protein spectrofluorometrically were described recently for the GFP in transgenic yeasts, filamentous fungi, and plants (3, 26, 33). Beyond this, our RNAi system offers the possibility of developing a high-throughput screening procedure for functional genomics. With the DsRed fluorescent marker, clearly visible phenotypes were generated that should drastically reduce the screening workload, as well as increase our current understanding of fungal functional genomics. As an alternative to use of the DsRed protein, workers may consider using the monomeric red fluorescent protein having accelerated folding properties. This fluorescent protein has already been used successfully in filamentous fungi, but compared with the brightness of DsRed, the brightness of the fully mature protein is reduced (38). Further improvements might be obtained with second-generation fluorescent proteins, such as tdTomato (38, 39); however, so far there have been no successful applications of these proteins in filamentous fungi.
Simultaneous silencing of multiple targets occurs with variable efficiencies.
Recently, it has been demonstrated that vectors generating hairpin RNAs are highly effective in inducing silencing (10, 14, 21, 26, 40). For this purpose, plasmid pSilent-1, which was shown previously to provide stable silencing in different filamentous fungi (26), was used for simultaneous inhibition of two target genes in A. chrysogenum.
Our quantitative and qualitative data demonstrate that the DsRed and pcbC genes were successfully downregulated. Simultaneous silencing of multiple target genes has several advantages. First, only a single silencing construct is needed to achieve simultaneous silencing of multiple target genes, and this minimizes the number of selectable marker genes required. In contrast, multiple knockouts in a single strain require use of different selectable markers in order to monitor the integration process. Consequently, the number of genes that can be deleted is limited by the fact that only a few dominant resistance marker genes are available (46). The applicability of the RNA-silencing system can be extended further by using vectors with inducible promoters. In this case, gene expression can be controlled in specific stages during development, or alternatively, functional analysis can be performed for genes indispensable for fungal growth (24, 26).
Another important result of our study is the discovery that the two target genes (pcbC and DsRed) are not cosilenced to the same extent. Table 4 shows that the red phenotype was silenced with the same efficiency irrespective of whether it was used in cosilencing experiments. Using hairpin-expressing plasmid pPCBCi, we obtained different levels of repression of the two target genes. Some transformants with an almost completely downregulated DsRed gene, for example, showed a moderate reduction in cephalosporin C synthesis. Nevertheless, all colorless transformants in this experiment had a more or less reduced rate of pcbC expression. This effect has been reported previously for other RNAi systems suppressing two target genes simultaneously. For example, in Venturia inaequalis, all transformants showed a cosilencing effect on the two target genes encoding GFP and trihydroxynaphthalene reductase (thn); however, quantification by real-time PCR indicated that RNAi did not occur to the same extent (9). Comparable results were obtained when Cryptococcus neoformans was used to cosilence ADE2, which encodes the phosphoribosylaminoimidazole carboxylase, and CAP59, which is required for synthesis of the polysaccharide capsule. Although more than 80% of all transformants exhibited reduction of expression of both genes, some exhibited only a single silenced phenotype (19). An explanation for this irregular RNAi efficiency could be that incomplete formation of the hairpin RNA stem derived from one target gene might lead to a reduction in the length of the dsRNA. This would result in a decrease in the efficiency of knockdown effects. Recently, an increase in the stem length of the hairpin RNA was shown to enhance the number of transformants having a mutant phenotype (10).
RNAi system could unravel components regulating secondary metabolism.
The RNA-silencing system developed in this study may make it possible to conduct high-throughput loss-of-function screening to identify novel regulators of secondary metabolism. It was recently suggested that in order to generate a fungal RNAi hairpin vector library, the Gateway (Invitrogen) in vitro recombination technology could be used for high-throughput cloning (46). In a comparable approach mammalian cells were screened successfully with a lentiviral RNAi library for genome-scale loss-of-function screening (34). The use of RNA silencing with microbial systems provides a potential alternative to conventional strain improvement programs in which UV rays or chemical mutagens are used for construction of strains with novel properties. Preliminary success in this direction using the RNAi system has been reported previously for a few filamentous fungi. For example, in Aspergillus and Fusarium species it was possible to reduce mycotoxin yields in order to eliminate secondary product contamination of food (21). In another attempt, fatty acid production was recently improved in Mortierella alpina, the commercially used producer of arachidonic acid, using the RNAi system (42). The authors suggested that RNA silencing could enable biosynthesis of previously rare and economically valuable compounds.
We have shown previously that disruption of the pcbC gene results in transformants exhibiting no antibiotic activity (45). The same phenotype was found in this investigation in about 15% of cosilenced transformants. Thus, secondary metabolism can be studied by downregulation of genes that directly or indirectly control cephalosporin C biosynthesis. The RNAi approach also provides the possibility of unraveling signal cascades in secondary metabolism. Silencing of repressor proteins, for example, may result in an increase in cephalosporin C production and thus in construction of strains with an enhanced production rate.
In conclusion, here we describe a quantitative and qualitative investigation of cosilenced fungal transformants using hairpin vector technology. The successful coinhibition of the heterologous DsRed gene and the endogenous pcbC gene indicates that RNAi provides an effective tool for studying secondary metabolism in A. chrysogenum. Thus, dsRNA-based silencing is a reliable alternative to gene inactivation experiments, which currently have low rates of efficiency with A. chrysogenum. Our data indicate that this RNA-silencing system along with other technologies should accelerate the genome-wide analysis of A. chrysogenum and thus promote rational strain improvement in this industrial cephalosporin C producer.
Acknowledgments
We thank Kerstin Kalkreuter, Stefanie Mertens, and Ingeborg Godehardt for their excellent technical assistance, Minou Nowrousian for advice concerning the quantitative real-time PCR, Gabriele Frenßen-Schenkel for the artwork, and E. Friedlin and H. Kürnsteiner (Kundl, Austria) for their interest and support. We acknowledge receipt of the vector pSilent-1 from H. Nakayashiki (Kobe, Japan). Protein quantification by spectrofluorometric measurement was done in the collaborative research center (SFB 480, projects A1 and C6).
This work was supported by Sandoz GmbH (Austria).
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Allen, R. S., A. G. Millgate, J. A. Chitty, J. Thisleton, J. A. Miller, A. J. Fist, W. L. Gerlach, and P. J. Larkin. 2004. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 22:1559-1566. [DOI] [PubMed] [Google Scholar]
- 2.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 3.Cha, H. J., N. G. Dalal, and W. E. Bentley. 2004. In vivo monitoring of intracellular expression of human interleukin-2 using green fluorescent protein fusion partner in Pichia pastoris. Biotechnol. Lett. 26:1157-1162. [DOI] [PubMed] [Google Scholar]
- 4.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker, and G. Macino. 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15:3153-3163. [PMC free article] [PubMed] [Google Scholar]
- 6.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103:10352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silva Ferreira, M. E., M. R. Kress, M. Savoldi, M. H. Goldman, A. Härtl, T. Heinekamp, A. A. Brakhage, and G. H. Goldman. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong, J. F., H. J. Deelstra, H. A. Wösten, and L. G. Lugones. 2006. RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl. Environ. Microbiol. 72:1267-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, A., J. A. van Kan, and K. M. Plummer. 2004. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 41:963-971. [DOI] [PubMed] [Google Scholar]
- 10.Goldoni, M., G. Azzalin, G. Macino, and C. Cogoni. 2004. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016-1024. [DOI] [PubMed] [Google Scholar]
- 11.Hoff, B., E. K. Schmitt, and U. Kück. 2005. CPCR1, but not its interacting transcription factor AcFKH1, controls fungal arthrospore formation in Acremonium chrysogenum. Mol. Microbiol. 56:1220-1233. [DOI] [PubMed] [Google Scholar]
- 12.Jekosch, K., and U. Kück. 2000. Glucose dependent transcriptional expression of the cre1 gene in Acremonium chrysogenum strains showing different levels of cephalosporin C production. Curr. Genet. 37:388-395. [DOI] [PubMed] [Google Scholar]
- 13.Jekosch, K., and U. Kück. 2000. Loss of glucose repression in an Acremonium chrysogenum beta-lactam producer strain and its restoration by multiple copies of the cre1 gene. Appl. Microbiol. Biotechnol. 54:556-563. [DOI] [PubMed] [Google Scholar]
- 14.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2003. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 16:769-776. [DOI] [PubMed] [Google Scholar]
- 15.Krappmann, S., C. Sasse, and G. H. Braus. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krügel, H., G. Fiedler, C. Smith, and S. Baumberg. 1993. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 127:127-131. [DOI] [PubMed] [Google Scholar]
- 17.Kück, U., and B. Hoff. 2006. Application of the nourseothricin acetyltransferase gene (nat1) as a dominant marker for the transformation of filamentous fungi. Fungal Genet. Newsl. 53:9-11. [Google Scholar]
- 18.Liu, G., J. Casqueiro, O. Bañuelos, R. E. Cardoza, S. Gutiérrez, and J. F. Martín. 2001. Targeted inactivation of the mecB gene, encoding cystathione-gamma-lyase, shows that the reverse transsulfuration pathway is required for high-level cephalosporin biosynthesis in Acremonium chrysogenum C10 but not for methionine induction of the cephalosporin genes. J. Bacteriol. 183:1765-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matz, M. V., A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969-973. [DOI] [PubMed] [Google Scholar]
- 21.McDonald, T., D. Brown, N. P. Keller, and T. M. Hammond. 2005. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant-Microbe Interact. 18:539-545. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen, L., S. Sarrocco, M. Lübeck, and D. F. Jensen. 2003. Expression of the red fluorescent protein DsRed-Express in filamentous ascomycete fungi. FEMS Microbiol. Lett. 223:135-139. [DOI] [PubMed] [Google Scholar]
- 23.Minuth, W., P. Tudzynski, and K. Esser. 1982. Extrachromosomal genetics of Cephalosporium acremonium. Curr. Genet. 5:227-231. [DOI] [PubMed] [Google Scholar]
- 24.Mouyna, I., C. Henry, T. L. Doering, and J. P. Latgé. 2004. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 237:317-324. [DOI] [PubMed] [Google Scholar]
- 25.Nakayashiki, H. 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579:5950-5957. [DOI] [PubMed] [Google Scholar]
- 26.Nakayashiki, H., S. Hanada, B. Q. Nguyen, N. Kadotani, Y. Tosa, and S. Mayama. 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42:275-283. [DOI] [PubMed] [Google Scholar]
- 27.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101:12248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowrousian, M., C. Ringelberg, J. C. Dunlap, J. J. Loros, and U. Kück. 2005. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genomics 273:137-149. [DOI] [PubMed] [Google Scholar]
- 30.Pöggeler, S., and U. Kück. 2006. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Pöggeler, S., S. Masloff, B. Hoff, S. Mayrhofer, and U. Kück. 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54-61. [DOI] [PubMed] [Google Scholar]
- 32.Radzio, R., and U. Kück. 1997. Efficient synthesis of the blood-coagulation inhibitor hirudin in the filamentous fungus Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 48:58-65. [DOI] [PubMed] [Google Scholar]
- 33.Richards, H. A., M. D. Halfhill, R. J. Millwood, and C. N. Stewart, Jr. 2003. Quantitative GFP fluorescence as an indicator of recombinant protein synthesis in transgenic plants. Plant Cell Rep. 22:117-121. [DOI] [PubMed] [Google Scholar]
- 34.Root, D. E., N. Hacohen, W. C. Hahn, E. S. Lander, and D. M. Sabatini. 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3:715-719. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Habor, NY.
- 36.Schmitt, E. K., A. Bunse, D. Janus, B. Hoff, E. Friedlin, H. Kürnsteiner, and U. Kück. 2004. Winged helix transcription factor CPCR1 is involved in regulation of beta-lactam biosynthesis in the fungus Acremonium chrysogenum. Eukaryot. Cell 3:121-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, E. K., B. Hoff, and U. Kück. 2004. Regulation of cephalosporin biosynthesis. Adv. Biochem. Eng. Biotechnol. 88:1-43. [DOI] [PubMed] [Google Scholar]
- 38.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 39.Shu, X., N. C. Shaner, C. A. Yarbrough, R. Y. Tsien, and S. J. Remington. 2006. Novel chromophores and buried charges control color in mFruits. Biochemistry 45:9639-9647. [DOI] [PubMed] [Google Scholar]
- 40.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 41.Stauffer, J. F., L. J. Schwartz, and C. W. Brady. 1966. Problems and progression in a strain selection program with cephalosporin-producing fungi. Dev. Ind. Microbiol. 7:104-113. [Google Scholar]
- 42.Takeno, S., E. Sakuradani, A. Tomi, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2005. Improvement of the fatty acid composition of an oil-producing filamentous fungus, Mortierella alpina 1S-4, through RNA interference with Δ12-desaturase gene expression. Appl. Environ. Microbiol. 71:5124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toews, M. W., J. Warmbold, S. Konzack, P. Rischitor, D. Veith, K. Vienken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 44.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walz, M., and U. Kück. 1993. Targeted integration into the Acremonium chrysogenum genome: disruption of the pcbC gene. Curr. Genet. 24:421-427. [DOI] [PubMed] [Google Scholar]
- 46.Weld, R. J., K. M. Plummer, M. A. Carpenter, and H. J. Ridgway. 2006. Approaches to functional genomics in filamentous fungi. Cell Res. 16:31-44. [DOI] [PubMed] [Google Scholar]