Abstract
The m-AAA protease, an ATP-dependent proteolytic complex in the mitochondrial inner membrane, controls protein quality and regulates ribosome assembly, thus exerting essential housekeeping functions within mitochondria. Mutations in the m-AAA protease subunit paraplegin cause axonal degeneration in hereditary spastic paraplegia (HSP), but the basis for the unexpected tissue specificity is not understood. Paraplegin assembles with homologous Afg3l2 subunits into hetero-oligomeric complexes which can substitute for yeast m-AAA proteases, demonstrating functional conservation. The function of a third paralogue, Afg3l1 expressed in mouse, is unknown. Here, we analyze the assembly of paraplegin into m-AAA complexes and monitor consequences of paraplegin deficiency in HSP fibroblasts and in a mouse model for HSP. Our findings reveal variability in the assembly of m-AAA proteases in mitochondria in different tissues. Homo-oligomeric Afg3l1 and Afg3l2 complexes and hetero-oligomeric assemblies of both proteins with paraplegin can be formed. Yeast complementation studies demonstrate the proteolytic activity of these assemblies. Paraplegin deficiency in HSP does not result in the loss of m-AAA protease activity in brain mitochondria. Rather, homo-oligomeric Afg3l2 complexes accumulate, and these complexes can substitute for housekeeping functions of paraplegin-containing m-AAA complexes. We therefore propose that the formation of m-AAA proteases with altered substrate specificities leads to axonal degeneration in HSP.
Mitochondria are essential and versatile organelles. They house energy-producing respiratory complexes and represent the major site of cellular reactive oxygen production, but they also carry out crucial anabolic reactions and important functions during apoptotic processes and cellular signaling. Proteomic surveys of mitochondria isolated from different mammalian tissues revealed large versatility in the protein composition of the organelles, which may reflect the various metabolic and anabolic needs in different cells (14, 25). Given the importance of these organelles for cellular physiology, it is not surprising that mitochondrial dysfunctions have been linked to various prevalent human diseases, including neurodegenerative disorders (9, 21, 34, 35). Disease-causing mutations have been identified in both mitochondrial-encoded and nuclear-encoded mitochondrial proteins. Common features of mitochondrial disorders are an enormous variety in disease severity and striking tissue specificities. While this could be at least partly explained by heteroplasmy in the case of mitochondrial mutations, tissue-specific consequences of defective nuclear-encoded genes with housekeeping functions are currently poorly understood.
This is exemplified by an autosomal recessive form of hereditary spastic paraplegia (HSP), which is caused by mutations in paraplegin, a subunit of the conserved ATP-dependent m-AAA protease in the inner membrane of mitochondria (29). The loss of paraplegin results in the axonal degeneration of cortical motor neurons, leading to rigidity and weakness in the lower limbs (8). Studies with the yeast Saccharomyces cerevisiae assigned a dual activity to the m-AAA protease for protein degradation and activation (27): it conducts protein quality surveillance in the inner membrane and degrades nonassembled membrane proteins to peptides (4, 22); on the other hand, it mediates protein processing and thereby activates certain mitochondrial proteins. Substrates which are cleaved rather than degraded by the m-AAA protease include cytochrome c peroxidase (12) and the ribosomal subunit MrpL32 (26). The cleavage of MrpL32 by the m-AAA protease ensures ribosomal assembly and protein synthesis within mitochondria and thereby is essential for respiration (26).
Like all known subunits of AAA proteases, paraplegin belongs to the AAA superfamily of ATPases and is characterized by the presence of a conserved AAA ATPase module (15). Despite being involved in diverse cellular activities, a common feature of AAA proteins is that they are active upon only homo- or hetero-oligomeric assembly. This is explained by the activation of the ATPase activity by interdomain communication between neighboring AAA domains (18, 28). Accordingly, the yeast m-AAA protease forms a hetero-oligomeric complex built of homologous Yta10 (Afg3) and Yta12 (Rca1) subunits; the complex is inactivated in the absence of one subunit (4). Proteins homologous to Yta10 and Yta12 are ubiquitously present in multicellular eukaryotic cells (17), but the function of mammalian m-AAA proteases is only poorly understood. Complementation studies revealed the functional conservation of m-AAA proteases in yeast and mammals and identified a complex of mammalian paraplegin and Afg3l2 (for AFG3-like gene 2) as the functional orthologue of the yeast m-AAA protease (5, 26). A mouse model for HSP lacking paraplegin replicates important features of the human disease and reveals progressive and cell-specific axonal degeneration in long descending and ascending spinal tracts, in the optic nerves, and in the sciatic nerves (13). Neuropathological features and experimental evidence indicate that axonal degeneration is ultimately due to impaired axonal transport; however, one of the first signs of pathology is the appearance of giant and structurally abnormal mitochondria in distal regions of axons, suggesting that a neuronal specific mitochondrial dysfunction triggers the pathological cascade (13). The reproducibility of the human phenotype in the mouse model is remarkable considering that an additional homologue of paraplegin, Afg3l1 (for AFG3-like gene 1), is present in mouse but encoded by a pseudogene in humans (20).
Tissue-specific consequences of a loss of paraplegin are puzzling, as crucial housekeeping functions of the m-AAA protease, like the processing of the ribosomal subunit MrpL32, are conserved from yeast to mammals (27). The impaired maturation of MrpL32 in mammalian mitochondria is expected to result in defects in oxidative phosphorylation. However, while deficiencies in the assembly of complex I have been reported for HSP patient fibroblasts (5), only mild respiratory defects were observed in paraplegin-deficient mouse tissues (13). Recently, paraplegin has been linked to the proteolytic processing of OPA1 (16), a conserved dynamin-like GTPase in the inner membrane regulating mitochondrial dynamics (10). Mutations in OPA1 cause autosomal dominant optic atrophy characterized by the loss of retinal ganglion cells (1, 11). As enlarged and abnormal mitochondria accumulate at early stages in axons in Spg7−/− mice (13) and as optic atrophy is sometimes observed in complicated forms of HSP and in the paraplegin mouse model (8, 13), impaired OPA1 processing might be of pathogenic relevance.
To obtain further insights into the consequences of a loss of paraplegin, in the present study, we have analyzed the assembly of putative m-AAA protease subunits in murine and human mitochondria. We demonstrate tissue-specific differences in the relative abundance of m-AAA protease subunits in mouse and an unexpected variability in their assembly. Both homo-oligomeric Afg3l1 and Afg3l2 complexes as well as hetero-oligomeric assemblies containing paraplegin are formed. Our studies suggest that an altered assembly of homologous subunits rather than the complete loss of m-AAA protease activity is of pathogenic relevance in HSP.
MATERIALS AND METHODS
Cloning procedures.
For expression in yeast, the mature form of murine Afg3l1 (amino acids 25 to 789) was fused to the mitochondrial targeting sequence of Yta10 (amino acids 1 to 61). A HindIII/BamHI DNA fragment containing the YTA10 promoter (480 bp) and bp 1 to 183 of YTA10 and a BamHI/KpnI DNA fragment containing bp 73 to 2367 of Afg3l1 were cloned into the multicopy plasmid YEplac195. A c-Myc epitope was attached to the C terminus of Afg3l1 to allow the immunodetection of the hybrid protein Yta10(1-61)-Afg3l1(25-789)-Myc. Yeast plasmids allowing heterologous expression of human AFG3L2 (hAFG3L2)-Myc (in YEplac112ADH1), murine Afg3l2-hemagglutinin (in YEplac112YTA10), and paraplegin (in YEplac181YTA10) and the plasmids pGEM4-Atp7 and pGEM4-Yme2ΔC for SP6-polymerase-driven expression in vitro were described previously (5, 19, 26).
m-AAA protease subunits were inactivated by the substitution of catalytically active glutamate residues within the proteolytic centers with glutamine residues by using the QuikChange site-directed mutagenesis kit (Stratagene). The respective codons were mutated as follows: GAA to CAA for hAFG3L2 (amino acid 575) and murine Afg3l2 (amino acid 574) and GAG to CAG for murine paraplegin (amino acid 575) and Afg3l1 (amino acid 567). The following oligonucleotides were used: 5′-GGT CGC CTT CCA TCA GTC TGG CCA TGC C-3′ (forward primer) and 5′-GGC ATG GCC AGA CTG ATG GAA GGC GAC C-3′ (reverse primer) for murine paraplegin; 5′-CTG TAG CCT ACC ACC AGG CTG GGC ATG CAG-3′ (forward primer) and 5′-CTG CAT GCC CAG CCT GGT GGT AGG CTA CAG-3′ (reverse primer) for murine Afg3l1; and 5′-CGG TGG CTT ACC ACC AAG CAG GCC ATG CGG-3′ (forward primer) and 5′-CCG CAT GGC CTG CTT GGT GGT AAG CCA CCG-3′ (reverse primer) for murine Afg3l2. Mutagenesis was verified by DNA sequencing.
Yeast strains and growth conditions.
The Saccharomyces cerevisiae strains used in this study are derivatives of W303. The yta10Δ yta12Δ (YKO200) strain was obtained by the deletion of YTA12 in the yta10Δ (YGS101 [3]) strain by PCR-targeted homologous recombination using the kanMX6 deletion cassette (23). yta10Δ yta12Δ strains expressing human or murine m-AAA proteases (see Table S1 in the supplemental material) were generated by the transformation of YKO200 cells with the plasmids YEplac112ADH1-Yta10(1-61)-hAFG3L2(36-798)-Myc, YEplac181YTA10-Yta10(1-61)-paraplegin(44-781), YEplac195YTA10-Yta10(1-61)-Afg3l1(25-789)-Myc, and YEplac112YTA10-Yta10(1-61)-Afg3l2(36-802)-HA or plasmids encoding the respective proteolytic site variants (5, 26).
Yeast cells were grown according to standard procedures at 30°C in either yeast-peptone medium or minimal medium supplemented with the required auxotrophs. Two percent (wt/vol) glucose or, for isolation of mitochondria, 2% (wt/vol) galactose and 0.5% (wt/vol) lactate or, for examining respiratory growth, 3% (wt/vol) glycerol were added as carbon sources.
BN-PAGE.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed essentially as described previously (30). Mitochondrial proteins (150 μg) isolated from yeast or human primary fibroblasts were solubilized at a concentration of 5 mg/ml in 1% (wt/vol) digitonin, 30 mM Tris-HCl, pH 7.4, 4 mM Mg-acetate, 5 mM ɛ-amino-n-caproic acid, 50 mM NaCl, 1 mM ATP. After a clarifying spin for 30 min at 125,000 × g, extracts were loaded onto a 3 to 13% (wt/vol) polyacrylamide gel. Individual subunits of m-AAA protease complexes were detected by immunoblotting.
Gel filtration analysis of mitochondrial extracts.
Isolated yeast mitochondria containing hAFG3L2 or hAFG3L2E575Q (1 mg mitochondrial protein) were solubilized at a concentration of 5 mg/ml in 1% (wt/vol) digitonin, 30 mM Tris-HCl, pH 7.4, 4 mM Mg-acetate, 150 mM K-acetate, pH 7.4, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM ATP. Liver mitochondria from Spg7−/− mice (1 mg mitochondrial protein) were lysed at a concentration of 5 mg/ml in 2% (wt/vol) digitonin, 50 mM NaCl, 50 mM K-phosphate buffer, pH 7.0, 4 mM Mg-acetate, 10% (wt/vol) glycerol, 1 mM EDTA, 1 mM PMSF. Nonsolubilized material was removed by centrifugation for 30 min at 125,000 × g, and the supernatant was subjected to size exclusion chromatography using a Superose 6 column. Eluted proteins were collected in 500-μl fractions, trichloracetic acid (TCA) precipitated, and analyzed by sodium dodecyl sulfate (SDS)-PAGE and immunoblotting. Proteins present in eluate fractions were quantified by laser densitometry.
Determination of the relative abundance of m-AAA protease subunits.
To compare the relative amounts of paraplegin, Afg3l1, and Afg3l2 in liver and brain mitochondria, mitochondrial extracts were analyzed by SDS-PAGE and immunoblotting. Extracts containing similar amounts of Afg3l1 were fractionated by SDS-PAGE and analyzed by immunoblotting using Afg3l1-, paraplegin-, and Afg3l2-specific antibodies and, for control, antibodies directed against the 70-kDa subunit of succinate dehydrogenase (Sdha). Protein amounts were quantified using a fluorescence-based imaging system (Odyssey; LI-COR Biosciences). The signals for Afg3l2 and paraplegin were normalized using Afg3l1 as a loading control. The relative amount of paraplegin and Afg3l2 in liver and brain mitochondria was calculated as the ratio of their signals in both mitochondrial preparations.
Coimmunoprecipitation and immunodepletion.
For immunoprecipitation experiments, murine mitochondria (400 μg mitochondrial protein) were lysed at a concentration of 1 mg/ml in 2% (wt/vol) digitonin, 50 mM K-phosphate buffer, pH 7.0, 50 mM NaCl, 4 mM Mg-acetate, 10% (wt/vol) glycerol, 1 mM PMSF supplemented with protease inhibitor cocktail (Roche). After a clarifying spin for 15 min at 125,000 × g, extracts were loaded onto protein A-Sepharose beads (2 mg) coupled with affinity-purified antibodies or preimmune serum and incubated for 12 h at 4°C under gentle shaking. Beads were washed subsequently with 0.5% (wt/vol) digitonin, 50 mM K-phosphate buffer, pH 7.0, 50 mM NaCl, 4 mM Mg-acetate, 1 mM PMSF, 10 mM Tris-HCl, pH 7.4. Antibody-antigen complexes were eluted from the beads and fractionated based on their hydrophobicity using 1% (wt/vol) Triton-X114 and 1% (vol/vol) acetic acid. Eluted proteins were recovered from the detergent fraction by TCA precipitation and analyzed by SDS-PAGE and immunoblotting.
Immunodepletion experiments were performed using saturating amounts of Afg3l2 or Afg3l1C antibodies coupled to protein A-Sepharose beads (10 mg). Mitochondria (150 μg mitochondrial protein) were lysed at a concentration of 0.5 mg/ml in 2% (wt/vol) digitonin, 50 mM K-phosphate buffer, pH 7.0, 50 mM NaCl, 50 mM K-phosphate buffer, pH 7.0, 4 mM Mg acetate, 10% (wt/vol) glycerol, 1 mM PMSF supplemented with protease inhibitor cocktail (Roche) and applied onto antibody-coupled protein A-Sepharose beads. Precipitations were carried out as described above. Fifty percent of the supernatant fraction of the precipitation (corresponding to 75 μg mitochondrial protein) was subjected to TCA precipitation, SDS-PAGE, and immunoblotting.
Antibodies.
Polyclonal antibodies directed against m-AAA protease subunits were raised in rabbits (BioGenes, Germany) using peptides corresponding to amino acids 121 to 139 of murine paraplegin (C-PEDDEEEKRRKEREDQMYR), to amino acids 90 to 103 of Afg3l2 (C-KEAVGEKKEPQPSG) and to amino acids 104 to 118 (C-NAGPGGDGGNRGGKG) or amino acids 771 to 785 (C-WNKGREEGGTERGLQ) of Afg3l1 (Afg3l1 and Afg3l1C, respectively) Specific immunoglobulin Gs (IgGs) were purified from sera by immunoabsorption on a peptide-conjugated SulfoLink matrix (Pierce) and tested by immunoblotting and immunoprecipitation from yta10Δ yta12Δ yeast mitochondria containing either paraplegin, Afg3l1 or Afg3l2. The specificity of Afg3l1 and Afg3l2 antisera was also verified using protein fragments containing the corresponding antigenic regions expressed in Escherichia coli.
Commercially available monoclonal antibodies were used for the immunodetection of the 39-kDa subunit of complex I (Ndufa9; Molecular Probes), the 70-kDa subunit of succinate dehydrogenase (Sdha; Molecular Probes), the c-Myc (9B11; Cell Signaling Technology), and the hemagglutinin epitopes (3F10; Roche).
Miscellaneous.
The following procedures were performed essentially as described previously: isolation of mitochondria from yeast, human fibroblasts, and murine tissues (6, 24, 33); synthesis of radiolabeled mitochondrial preproteins; posttranslational protein import into isolated mitochondria; and proteolysis of newly imported proteins (33).
RESULTS
A homo-oligomeric hAFG3L2 complex in human mitochondria.
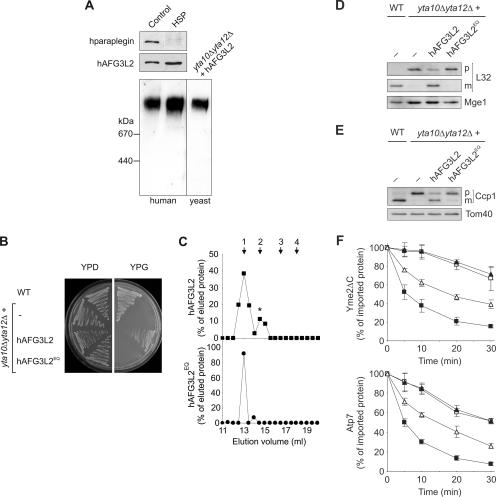
Paraplegin and hAFG3L2 subunits assemble in the inner membrane of human mitochondria into a functionally active m-AAA protease, whose formation is impaired in HSP patient cells lacking paraplegin (5). Surprisingly, a high-molecular-mass complex containing hAFG3L2 was detected when mitochondria isolated from paraplegin-deficient HSP fibroblasts were solubilized in digitonin and analyzed by BN-PAGE. The molecular mass of this complex was ∼900 kDa and similar to that of the hetero-oligomeric m-AAA protease containing hAFG3L2 and paraplegin present in human wild-type mitochondria (Fig. 1A).
FIG. 1.
hAFG3L2 forms a homo-oligomeric m-AAA protease complex with proteolytic activity. (A) A homo-oligomeric hAFG3L2 complex in mitochondria isolated from HSP fibroblasts. Mitochondria were solubilized in digitonin and analyzed by SDS-PAGE (50 μg mitochondrial protein; upper panel) or BN-PAGE (150 μg mitochondrial protein; lower panel), followed by immunoblotting using paraplegin-specific (upper panel) and hAFG3L2-specific (upper and lower panel) antisera. For the control, yeast yta10Δ yta12Δ mitochondria containing hAFG3L2 were fractionated by BN-PAGE in parallel. Thyroglobulin (669 kDa) and apoferritin (443 kDa) were used for calibration. (B) Maintenance of respiratory growth of yta10Δ yta12Δ yeast cells by hAFG3L2. Wild-type (WT) cells, yta10Δ yta12Δ cells, and yta10Δ yta12Δ cells expressing hAFG3L2 or the proteolytically inactive variant hAFG3L2E575Q (hAFG3L2EQ), were grown on fermentable (yeast extract-peptone-dextrose [YPD]) and nonfermentable (glycerol-containing media [YPG]) carbon sources at 30°C. (C) Homo-oligomeric hAFG3L2 complexes in yta10Δ yta12Δ mitochondria. hAFG3L2 or hAFG3L2E575Q (hAFG3L2EQ) were expressed in yta10Δ yta12Δ cells, and mitochondria were isolated. After solubilization in digitonin-containing buffer, mitochondrial extracts (1 mg mitochondrial protein) were fractionated by Superose 6 sizing chromatography. Eluate fractions were TCA precipitated and analyzed by SDS-PAGE and immunoblotting using hAFG3L2-specific antibodies. hAFG3L2 (filled squares) or hAFG3L2E575Q (filled circles) present in eluate fractions was quantified by laser densitometry, and results are given as a percent of the respective protein in the total eluate. A smaller hAFG3L2-containing complex, marked with an asterisk, most likely results from partial dissociation of the large complex. The following marker proteins were used for calibration: 1, Hsp60 (840 kDa); 2, apoferritin (443 kDa); 3, alcohol dehydrogenase (150 kDa); 4, bovine serum albumine (66 kDa). (D and E) Processing of yeast MrpL32 and Ccp1 by hAFG3L2. Protein processing was analyzed in wild-type (WT) cells, yta10Δ yta12Δ cells, and yta10Δ yta12Δ cells expressing either hAFG3L2 or hAFG3L2E575Q (hAFG3L2EQ) by SDS-PAGE. (D) Maturation of MrpL32 (L32) was monitored in isolated mitochondria (30 μg mitochondrial protein) by immunoblotting using polyclonal antisera directed against the mature and the precursor forms of MrpL32 and, as a loading control, against matrix-localized Mge1. (E) Ccp1 processing was examined in cell extracts using antisera directed against Ccp1 and, for control, the outer membrane protein Tom40. p, precursor; m, mature form. (F) Degradation of nonassembled membrane proteins by hAFG3L2 expressed in yeast. Radiolabeled Yme2ΔC (upper panel) or Atp7 (lower panel) was imported for 10 min at 25°C into mitochondria isolated from the yeast strains used for panels D and E. The stability of newly imported proteins at 37°C was determined by SDS-PAGE and autoradiography. The average of three independent experiments (± standard error of the mean) is shown. Filled squares, wild type; filled triangles, yta10Δ yta12Δ; open triangles, yta10Δ yta12Δ+hAFG3L2; open squares, yta10Δ yta12Δ+hAFG3L2E575Q.
To examine whether hAFG3L2 assembles with another AAA protease subunit or forms a homo-oligomeric complex in the inner membrane, we expressed the protein in yta10Δ yta12Δ yeast cells lacking any m-AAA protease subunit. Mitochondria were isolated from these cells and solubilized in digitonin, and the native molecular mass of hAFG3L2 was determined by BN-PAGE (Fig. 1A). hAFG3L2 was detected as part of a complex of ∼900 kDa and thus forms a complex similar in size to that in human paraplegin-deficient mitochondria. These findings suggest that hAFG3L2 can build a homo-oligomeric complex in the inner membrane of mitochondria.
The expression of hAFG3L2 alone restored the respiratory competence of yta10Δ yta12Δ cells (Fig. 1B). This observation is in contrast to our previous findings using another m-AAA protease-deficient yeast strain and most likely reflects strain-specific differences (5). To assess the dependence of respiratory growth on proteolysis by hAFG3L2, we replaced the catalytically active glutamate residue 575 by glutamine and expressed the mutant protein in yta10Δ yta12Δ cells. The mutation did not impair the assembly of hAFG3L2 into a high-molecular-mass complex as demonstrated by gel filtration analysis of digitonin-solubilized mitochondria (Fig. 1C). However, cell growth on glycerol-containing medium was completely abolished, indicating the dependence of respiration on the proteolytic activity of hAFG3L2 (Fig. 1B).
Proteolysis by homo-oligomeric hAFG3L2 complexes.
To directly monitor proteolysis by hAFG3L2, we examined the processing of two nuclear-encoded mitochondrial preproteins, namely the intermembrane space protein cytochrome c peroxidase (Ccp1) and the ribosomal subunit MrpL32, both of which are known to be cleaved by the yeast m-AAA protease (12, 26). The maturation of MrpL32 is required for ribosome assembly and thereby protein synthesis within mitochondria, a prerequisite for the assembly of respiratory complexes in the inner membrane (26). In agreement with the observed growth phenotypes, the expression of hAfg3l2 but not hAfg3l2E575Q in yta10Δ yta12Δ cells largely restored the processing of MrpL32, as revealed by Western blot analysis of mitochondria isolated from these cells (Fig. 1D). Similarly, mature Ccp1 accumulated in yta10Δ yta12Δ cells harboring hAfg3l2 but not in the presence of hAfg3l2E575Q (Fig. 1E).
In addition to its role for protein processing, the yeast m-AAA protease controls protein quality in the inner membrane and mediates the complete degradation of nonnative membrane proteins (4, 22). To examine whether the hAFG3L2 complex can also substitute for this activity, two known substrate proteins of the yeast m-AAA protease were synthesized in a cell-free system in the presence of [35S]methionine and imported into isolated yta10Δ yta12Δ mitochondria harboring hAFG3L2 or hAfg3l2E575Q. One of these substrates, Yme2ΔC, is localized in the inner membrane and contains a domain exposed to the matrix (22). The deletion of a carboxy-terminal intermembrane space domain, present in wild-type Yme2, results in the destabilization of Yme2ΔC and the subsequent degradation by m-AAA protease (Fig. 1F) (19). While accumulating in yta10Δ yta12Δ mitochondria, newly imported Yme2ΔC was degraded in the presence of hAFG3L2 (Fig. 1F). In contrast, Yme2ΔC remained stable in yta10Δ yta12Δ mitochondria harboring hAfg3l2E575Q. Similar effects were observed by analyzing the stability of newly imported Atp7, a peripheral inner membrane protein degraded by the m-AAA protease and another, as-yet-unidentified mitochondrial protease (Fig. 1F) (19). Whereas hAFG3L2 accelerates the degradation of Atp7 in yta10Δ yta12Δ mitochondria, a point mutation in the proteolytic center of hAFG3L2 (hAfg3l2E575Q) completely abolished this activity (Fig. 1F).
We therefore conclude that a homo-oligomeric hAFG3L2 complex exerts proteolytic activity in mitochondria and can substitute for the function of the yeast m-AAA protease in protein processing and the degradation of nonassembled membrane proteins. Thus, hAFG3L2 is proteolytically active both upon assembly with paraplegin into a hetero-oligomeric complex and upon homo-oligomerization.
Hetero-oligomeric m-AAA proteases in murine mitochondria.
These experiments revealed variability in the assembly of the human m-AAA protease subunit hAFG3L2. Another putative m-AAA protease subunit, Afg3l1, is encoded by a pseudogene in human but expressed in mouse pointing to an even higher variability in the latter (20). Murine Afg3l1 and Afg3l2 share 68% sequence identity, suggesting similar activities of both proteins; however, the function or assembly of Afg3l1 has not been analyzed.
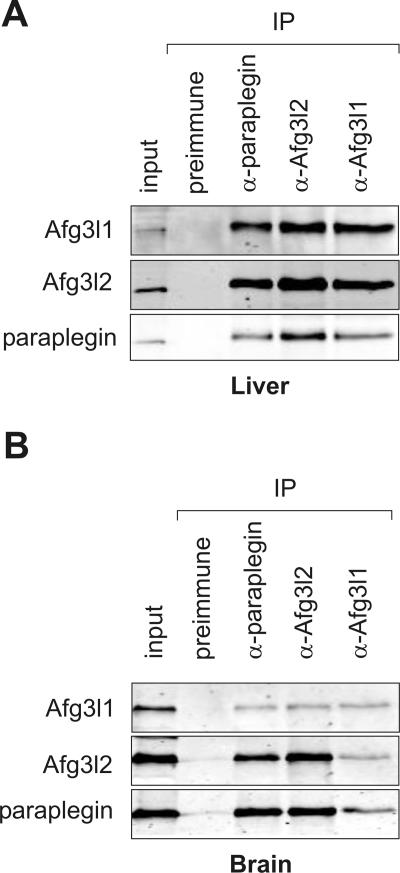
In order to examine the complex composition of m-AAA proteases in murine mitochondria, coimmunoprecipitation experiments were carried out. We raised peptide-specific antibodies directed against the putative m-AAA protease subunits Afg3l1, Afg3l2, and paraplegin. Mitochondria were isolated from murine liver and, considering the neuron-specific phenotype of HSP, also from brain. After the solubilization of mitochondria in digitonin, extracts were incubated with affinity-purified antisera directed against the three putative m-AAA protease subunits. Precipitates were subsequently analyzed by immunoblotting (Fig. 2). Paraplegin, Afg3l1, and Afg3l2 were detected in the immunoprecipitates irrespective of the antibody used for precipitation (Fig. 2A). In contrast, none of these proteins was precipitated with preimmune serum, demonstrating the specificity of the observed interaction. Similar results were obtained using liver or brain mitochondria (Fig. 2B). We therefore conclude that Afg3l1 directly interacts with paraplegin and Afg3l2 in both liver and brain mitochondria. In agreement with these findings, the proteins were detected in a high-molecular-mass complex and coeluted from the column upon fractionation of mitochondrial extracts by sizing chromatography (data not shown).
FIG. 2.
Assembly of paraplegin, Afg3l1, and Afg3l2 in murine mitochondria. Mitochondria (400 μg mitochondrial protein) were isolated from (A) liver and (B) brain and solubilized with digitonin. Ten (liver) or 20% (brain) of the sample was removed for control (input). Coimmunoprecipitations were carried out using affinity-purified paraplegin-, Afg3l1-, and Afg3l2-specific polyclonal antibodies. Preimmune antiserum was used as a negative control. Immunoprecipitates (IP) were analyzed by SDS-PAGE and immunoblotting.
m-AAA protease assembly in a paraplegin-deficient mouse model for HSP.
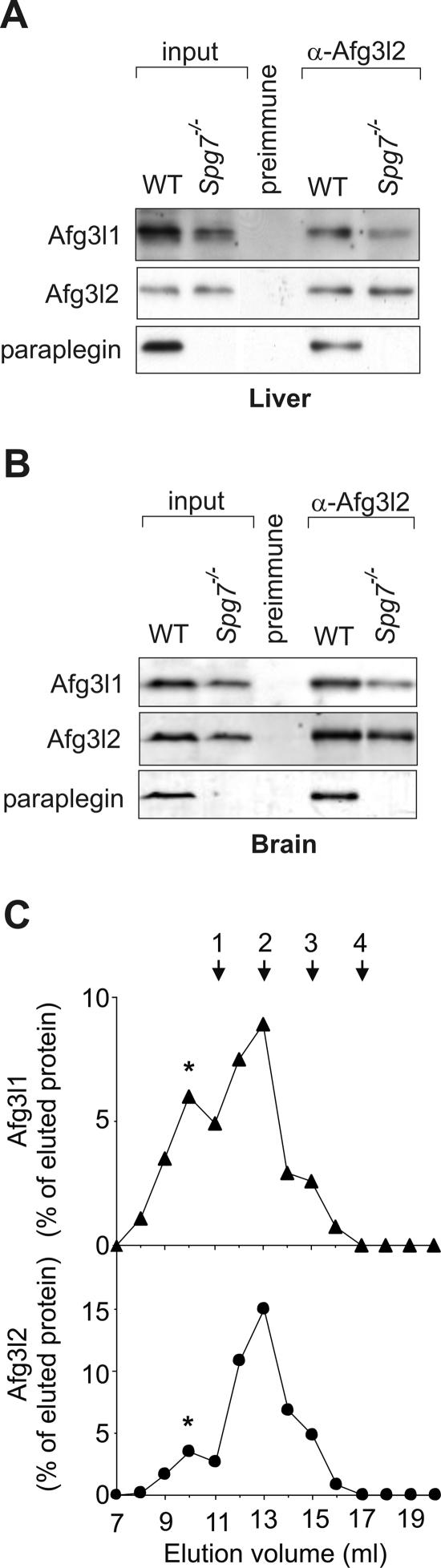
To monitor the consequences of the loss of paraplegin on the formation of murine m-AAA protease complexes, we examined the assembly of Afg3l1 and Afg3l2 in mitochondria of Spg7−/− mice. Mitochondria were isolated from liver and brain, solubilized in digitonin, and subjected to coimmunoprecipitation using affinity-purified Afg3l2-specific antibodies and preimmune serum (Fig. 3). Afg3l1 was precipitated with Afg3l2 but not with preimmune serum when detergent extracts of Spg7−/− liver or brain mitochondria were analyzed (Fig. 3A and B). Consistently, gel filtration experiments detected Afg3l1 and Afg3l2 as part of a high-molecular-mass complex in the inner membrane of Spg7−/− liver mitochondria (Fig. 3C). Thus, the loss of paraplegin does not prevent the assembly of Afg3l1 and Afg3l2 in the mitochondrial inner membrane. These findings point to variability in the assembly of murine m-AAA proteases which appears to be largely dependent on the availability of the individual subunits.
FIG. 3.
A hetero-oligomeric Afg3l1/Afg3l2 complex in mitochondria of Spg7−/− mice. Digitonin extracts of mitochondria (400 μg mitochondrial protein) from (A) liver and (B) brain of wild-type (WT) and paraplegin-deficient Spg7−/− mice were subjected to coimmunoprecipitations using affinity-purified Afg3l2-specific polyclonal antibodies as described in the legend for Fig. 2. Preimmune serum was used as a negative control. Precipitates were analyzed by SDS-PAGE and immunoblotting using paraplegin-, Afg3l1-, and Afg3l2-specific polyclonal antibodies. The amounts of Afg3l1 in the precipitates derived from wild-type and Spg7−/− mitochondria varied slightly in different experiments. (C) A high-molecular-mass complex containing Afg3l1 and Afg3l2 in Spg7−/− mitochondria. Extracts (1 mg mitochondrial protein) of liver mitochondria isolated from Spg7−/− mice were fractionated by Superose 6 sizing chromatography. Eluate fractions were TCA precipitated and analyzed by SDS-PAGE and immunoblotting using Afg3l1- and Afg3l2-specific polyclonal antibodies. Afg3l1 (filled triangles) and Afg3l2 (filled circles) in the eluate were quantified by laser densitometry and are given as a percent of the respective protein in the total eluate. An asterisk marks a high-molecular-mass complex of Afg3l1 and Afg3l2 which coelutes with the prohibitins Phb1 the prohibitins Phb1 and Phb2, suggesting an interaction of the m-AAA protease complex with prohibitins, as observed in yeast mitochondria (31). The following marker proteins were used for calibration: 1, dimeric ATP synthase (1.5 MDa); 2, monomeric ATP synthase (750 kDa); 3, apoferritin (443 kDa); 4, alcohol dehydrogenase (150 kDa).
Tissue-specific variations in the relative abundance of m-AAA protease subunits.
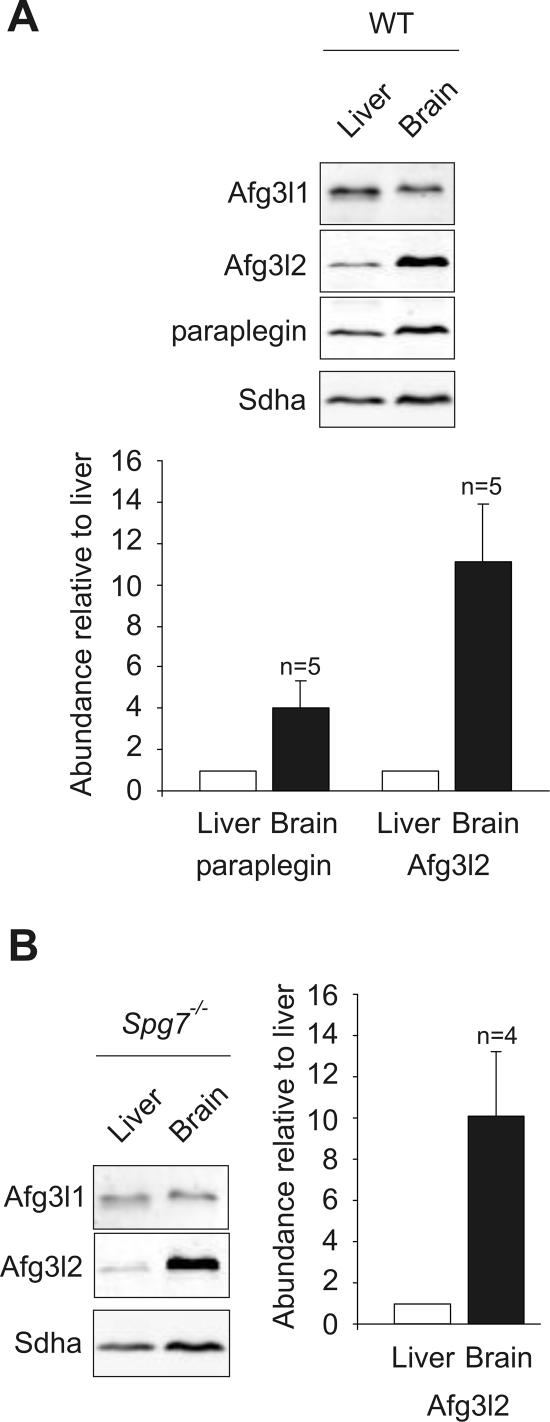
In the course of these experiments, we noticed that steady-state levels of m-AAA protease subunits appear to differ in liver and brain mitochondria. To exclude indirect effects due to differences in the purity of the organellar preparations from different tissues, we compared the relative abundance of Afg3l1, Afg3l2, and paraplegin in mitochondria isolated from liver and brain. The relative steady-state levels of Afg3l2 and paraplegin were determined by the immunoblotting of mitochondrial extracts containing equal amounts of Afg3l1 (Fig. 4A). Strikingly, Afg3l2 accumulated at ∼10-fold-higher levels in brain than in liver (Fig. 4A). Paraplegin was also more abundant in brain than in liver mitochondria and was present at ∼4-fold-higher levels in organelles isolated from murine brain (Fig. 4A). In contrast, Afg3l1 was present in only slightly reduced amounts in brain mitochondria relative to the 70-kDa subunit of succinate dehydrogenase (Fig. 4A).
FIG. 4.
Relative abundance of paraplegin, Afg3l1 and Afg3l2 in liver and brain mitochondria. Mitochondria were isolated from (A) wild-type or (B) Spg7−/− murine liver and brain. Extracts containing equal amounts of Afg3l1 were analyzed by SDS-PAGE and immunoblotting using paraplegin-, Afg3l1-, and Afg3l2-specific antibodies as well as a monoclonal antibody directed against the 70-kDa subunit of succinate dehydrogenase (Sdha). Paraplegin and Afg3l2 present in liver and brain mitochondria were quantified by fluorescence-based imaging using Afg3l1 as a loading control. The ratio of signals detected for Afg3l2 and paraplegin in brain versus liver mitochondria was calculated to monitor the relative abundance of m-AAA protease subunits in both tissues. The average of n independent experiments (± standard error of the mean) using tissues isolated from different, randomly chosen animals is shown.
It is conceivable that the relative abundance of Afg3l1 and Afg3l2 is altered in the absence of paraplegin, leading to a partial compensation for the loss of paraplegin. We therefore compared the relative abundance of Afg3l1 and Afg3l2 in mitochondria isolated from liver and brain of Spg7−/− mice lacking paraplegin (Fig. 4B). When normalized to Afg3l1, Afg3l2 still accumulated at ∼10-fold-higher levels in brain mitochondria of Spg7−/− mice, demonstrating that the relative abundance of both proteins was not altered in the absence of paraplegin.
These experiments reveal unexpected differences in the relative abundance of m-AAA protease subunits in different tissues. Considering the observed variability in their assembly, it appears likely that the subunit composition of the proteolytic complexes varies in a tissue-specific manner.
Homo- and hetero-oligomeric m-AAA proteases in brain mitochondria.
To assess the assembly of m-AAA protease subunits in a more quantitative manner, we performed immunodepletion experiments. Brain mitochondria were solubilized in digitonin, and affinity-purified Afg3l2-specific antibodies were used to completely deplete the extract of Afg3l2 (Fig. 5A). Afg3l2 was immunologically not detectable in the supernatant fraction after immunodepletion. Concomitant with Afg3l2, Afg3l1 and paraplegin were present at only drastically diminished levels (Fig. 5A). The incubation of mitochondrial extracts with preimmune serum affected the steady-state level of neither Afg3l2 nor Afg3l1 or paraplegin (Fig. 5A). We therefore conclude that Afg3l1, Afg3l2, and paraplegin assemble quantitatively with each other in the inner membrane of brain mitochondria.
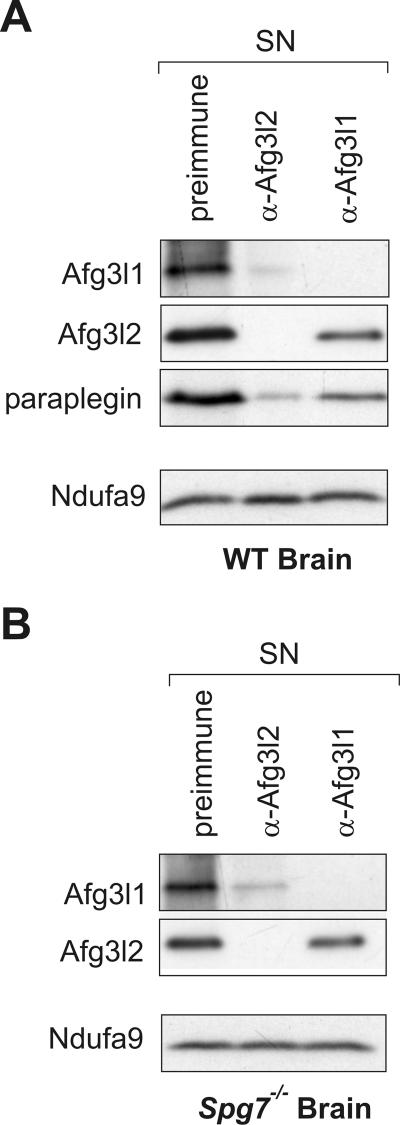
FIG. 5.
Coexistence of m-AAA proteases built up of different subunits in brain mitochondria. Mitochondria (150 μg mitochondrial protein) from (A) wild-type (WT) and (B) Spg7−/− brain were lysed in digitonin and incubated with saturating amounts of preimmune serum or affinity-purified Afg3l2- or Afg3l1C-specific antibodies. After the removal of the precipitate, supernatant fractions (SN) were analyzed by SDS-PAGE and examined for the presence of paraplegin, Afg3l1, and Afg3l2 by immunoblotting. A monoclonal antibody directed against the 39-kDa subunit of complex I (Ndufa9) was used to control for equal gel loading. Whereas Afg3l1 and Afg3l2 could be completely depleted from the supernatant fraction by using affinity-purified Afg3l1- or Afg3l2-specific antibodies, respectively, extracts could not be depleted of paraplegin using available paraplegin-specific antibodies (data not shown).
Notably, when similar experiments were performed with Afg3l1-specific antibodies, Afg3l2 and paraplegin were only slightly depleted from mitochondrial extracts. This most likely reflects the decreased abundance of Afg3l1 compared to that of Afg3l2 and paraplegin. Thus, at least two different m-AAA protease complexes coexist in brain mitochondria: Afg3l2/paraplegin and less abundant Afg3l1 complexes that also contain Afg3l2, paraplegin, or both.
Which m-AAA protease complexes are present in paraplegin-deficient brain mitochondria? To address this question, we isolated brain mitochondria from Spg7−/− mice and analyzed the complex composition of m-AAA protease complexes by immunodepletion using Afg3l1- and Afg3l2-specific antibodies (Fig. 5B). Similar to wild-type mitochondria, Afg3l1 was almost completely depleted from extracts of Spg7−/− brain mitochondria with Afg3l2 antibodies demonstrating quantitative assembly (Fig. 5B). In contrast, the steady-state level of Afg3l2 was hardly affected by depleting Afg3l1 (Fig. 5B). We conclude that Afg3l2 is present in excess relative to Afg3l1 in brain mitochondria of wild-type and Spg7−/− mice. While Afg3l1 is quantitatively assembled with Afg3l2, the majority of Afg3l2 is not part of this structure and most likely forms a homo-oligomeric complex.
Proteolytic activity of homo- and hetero-oligomeric m-AAA proteases.
These experiments demonstrate the existence of m-AAA protease complexes with variable subunit composition in murine mitochondria. To examine the proteolytic activity of the different assemblies, we carried out complementation studies with m-AAA protease-deficient yta10Δ yta12Δ yeast cells.
In initial experiments, paraplegin, Afg3l1, and Afg3l2 were independently expressed in yta10Δ yta12Δ cells and respiratory cell growth was examined (Fig. 6A). Strikingly, the expression of either Afg3l1 or Afg3l2 but not paraplegin restored the growth of yta10Δ yta12Δ cells on glycerol-containing medium (Fig. 6A). This is reminiscent of hAFG3L2, which formed homo-oligomeric, proteolytically active complexes when expressed in yeast (Fig. 1). Gel filtration analysis of mitochondrial extracts indeed revealed homo-oligomerization of Afg3l1 and Afg3l2 in yeast, whereas paraplegin did not form a high-molecular-mass complex in the absence of other AAA protease subunits (data not shown). The expression of proteolytic site variants of Afg3l1 (Afg3l1E567Q) or Afg3l2 (Afg3l2E574Q) did not promote respiratory growth of yta10Δ yta12Δ cells, indicating that the homo-oligomeric complexes exert proteolytic activity (Fig. 6A). Consistently, we observed the maturation of MrpL32 in yta10Δ yta12Δ cells harboring Afg3l1 or Afg3l2, whereas this activity was abolished by mutations in the proteolytic center of each protein (Fig. 6B). Moreover, the processing of Ccp1 was restored upon the expression of Afg3l1 or Afg3l2 but largely inhibited in the presence of proteolytically inactive variants of each protein (Fig. 6C). It should be noted that the respiratory deficiency of yta10Δ yta12Δ cells was not restored upon the expression of paraplegin, nor was mature MrpL32 or Ccp1 present in these cells (Fig. 6). Thus, the ability to form homo-oligomeric complexes appears to be restricted to Afg3l1 and Afg3l2.
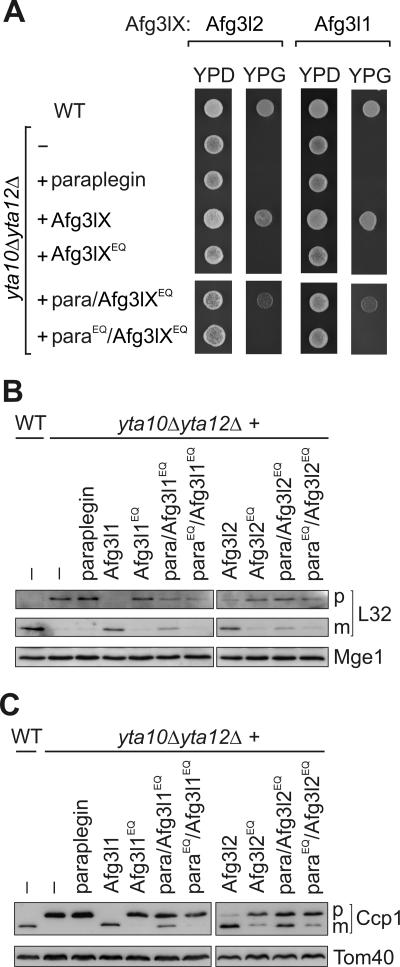
FIG. 6.
Proteolytic activity of homo-oligomeric and hetero-oligomeric m-AAA protease complexes in yeast. (A) Respiratory growth of yta10Δ yta12Δ cells expressing murine m-AAA protease subunits. Wild-type (WT) cells, yta10Δ yta12Δ cells, and yta10Δ yta12Δ cells expressing either paraplegin, Afg3l1 (Afg3lX), Afg3l2 (Afg3lX), or their mutant variants Afg3l1E567Q or Afg3l2E574Q (Afg3lXEQ) were grown at 30°C on glucose-containing (YPD) or glycerol-containing (YPG) media to examine the respiratory competence of the cells. To assess the activity of hetero-oligomeric complexes, the mutant variants Afg3l1E567Q or Afg3l2E574Q were coexpressed with paraplegin (para/Afg3lXEQ) or with parapleginE575Q (paraEQ/Afg3lXEQ) in yta10Δ yta12Δ cells and cell growth was analyzed as above. (B and C) Processing of yeast MrpL32 and Ccp1 by murine m-AAA proteases. Protein processing was analyzed in yta10Δ yta12Δ cells harboring murine m-AAA protease subunits by SDS-PAGE as described for panel A and immunoblotting as described in the legend for Fig. 1 by using MrpL32 (L32)-specific antisera (B) or Ccp1-specific antisera (C). p, precursor forms; m, mature forms. Mge1 and Tom40 were used to control for equal gel loading.
In further experiments, the proteolytic activity of hetero-oligomeric murine m-AAA proteases composed of Afg3l1 and paraplegin or of Afg3l2 and paraplegin was assessed in yeast. After the coexpression of either pair of subunits, their interaction was verified by coimmunoprecipitation experiments (data not shown). Similar to cells containing only Afg3l1 or Afg3l2, the respiratory growth of yta10Δ yta12Δ cells was restored if paraplegin was coexpressed with either protein (data not shown). To examine whether this phenotype is due to hetero-oligomerization with paraplegin rather than homo-oligomerization of Afg3l1 or Afg3l2, respectively, we expressed proteolytically inactive variants of these proteins. Previous studies on yeast m-AAA protease have demonstrated that the protease is only inactivated when all subunits carry point mutations in their proteolytic center (3). In contrast, a hetero-oligomeric AAA protease complex composed of proteolytically active and inactive subunits mediates protein processing and maintains respiratory growth (3). Whereas neither the expression of paraplegin nor that of Afg3l1E567Q allowed yta10Δ yta12Δ cells to grow on glycerol-containing medium, respiratory cell growth was restored upon the coexpression of both proteins (Fig. 6A). In contrast, yta10Δ yta12Δ cells expressing proteolytically inactive Afg3l1E567Q and parapleginE575Q were respiratory deficient. Thus, respiratory growth can be maintained by a proteolytically active Afg3l1/paraplegin complex.
To directly assess the proteolytic activity of the hetero-oligomeric Afg3l1/paraplegin complex, the maturation of Ccp1 and MrpL32 was monitored in these cells (Fig. 6B and C). Mature forms of both Ccp1 and MrpL32 were detected in cells harboring paraplegin and Afg3l1E567Q but not in the presence of parapleginE575Q and Afg3l1E567Q. We therefore conclude that an AAA protease complex composed of Afg3l1 and paraplegin subunits is proteolytically active.
Notably, very similar results were obtained when yta10Δ yta12Δ cells expressing Afg3l2 and paraplegin were analyzed (Fig. 6). Introducing a point mutation into the proteolytic center of Afg3l2 (Afg3l2E574Q) did not abolish respiratory growth if paraplegin was coexpressed. Moreover, Ccp1 and MrpL32 were partially processed in these cells. Upon the coexpression of Afg3l2E574Q and parapleginE575Q, however, the respiratory competence of the cells was not restored, and both Ccp1 and MrpL32 accumulated in the precursor form (Fig. 6). Thus, the Afg3l2/paraplegin complex exerts proteolytic activity in yeast.
DISCUSSION
We demonstrate the assembly of mammalian m-AAA protease subunits into proteolytic complexes which vary in their subunit composition in different tissues and according to subunit availability. Whereas Afg3l2 and murine Afg3l1 can homo-oligomerize, we did not obtain any evidence for the homo-oligomerization of paraplegin. Rather, paraplegin was found only as part of hetero-oligomeric complexes also containing Afg3l2, Afg3l1, or both. These findings reveal that the loss of paraplegin in HSP does not lead to the loss of m-AAA protease activity per se, but rather to the formation of proteolytic complexes with different subunit compositions. In the absence of paraplegin, homo-oligomeric Afg3l2 complexes replace hetero-oligomeric Afg3l2/paraplegin complexes in brain mitochondria. We propose that differences in the substrate specificity of these m-AAA proteases are causative for cell-specific axonal degeneration in HSP.
A homo-oligomeric m-AAA protease built of hAFG3L2 subunits is present in human HSP fibroblasts. This complex exerts proteolytic activity and suppresses the respiratory deficiency of yeast cells lacking both Yta10 and Yta12. Similar to the yeast enzyme, the hAFG3L2 complex specifically recognizes and degrades misfolded polypeptides that are either integral parts of the inner membrane or peripherally associated at its matrix-exposed surface. Moreover, homo-oligomeric hAFG3L2 complexes can substitute for regulatory functions of the yeast orthologue and mediate the proteolytic processing of the ribosomal subunit MrpL32 and of Ccp1. As the maturation of Ccp1 occurs by subsequent processing by the m-AAA protease and the rhomboid protease Pcp1 in the inner membrane of yeast mitochondria (12), it can be concluded that hAFG3L2 complexes can functionally interact with rhomboid. Recent findings indeed indicate an intimate functional interplay of m-AAA and rhomboid protease in yeast. Ccp1 maturation by rhomboid requires its ATP-dependent membrane dislocation, rather than proteolytic processing, by the m-AAA protease (33a). This function can also be carried out by the homo-oligomeric hAFG3L2 complex. It should be noted that similar observations were made upon expression of murine Afg3l2 or Afg3l1 in m-AAA protease-deficient yeast cells, indicating that protein quality control, protein processing, and membrane dislocation represent conserved activities of these homo-oligomeric m-AAA protease complexes.
The formation of homo-oligomeric proteolytic complexes demonstrates variability in the assembly of m-AAA protease subunits and raises the intriguing question of why only some AAA protease subunits have the propensity to homo-oligomerize. Crystal structures of bacterial AAA proteases revealed that intersubunit contacts occur mainly between protease domains of the monomers (7, 32). Although generally highly conserved, m-AAA protease subunits differ significantly in C-terminal regions. It is therefore conceivable that sequence differences of AAA protease subunits in this region determine their ability for homo-oligomerization.
Variability in the assembly is also illustrated by our analysis of m-AAA protease complexes in murine mitochondria which harbor an additional proteolytic subunit, Afg3l1 (20). Coimmunoprecipitation and immunodepletion experiments reveal an almost quantitative assembly of paraplegin, Afg3l1, and Afg3l2 in murine liver and brain mitochondria. It remains to be determined, however, if all subunits are present within one proteolytic complex or if different m-AAA protease complexes coexist with each other in the inner membrane of mitochondria. Evidence for the coexistence of at least two m-AAA proteases was obtained for brain mitochondria. Whereas Afg3l1 and paraplegin were immunodepleted with Afg3l2 and therefore are quantitatively assembled, the depletion of Afg3l1 only slightly affected the presence of Afg3l2 or paraplegin in mitochondrial extracts. These observations indicate that Afg3l1 is present at lower molar concentrations than those of Afg3l2 and paraplegin in murine brain mitochondria and point to the coexistence of various m-AAA protease complexes in the inner membrane: an Afg3l2/paraplegin complex and less abundant Afg3l1 complexes that also contain Afg3l2, paraplegin, or both.
How does the loss of paraplegin in HSP affect the formation of m-AAA complexes in mitochondria? The versatile assemblies of m-AAA protease subunits suggest that it is solely the availability of subunits, i.e., their expression in different tissues, which determines the composition of the proteolytic complexes. Consistently, the loss of paraplegin leads to the accumulation of two m-AAA protease complexes in brain mitochondria in mice, a hetero-oligomeric Afg3l1/Afg3l2 complex and a homo-oligomeric Afg3l2 complex. The observation that the relative abundance of m-AAA protease subunits can strongly vary between different tissues is of central importance for understanding phenotypic consequences of mutations in m-AAA protease subunits. Afg3l2 was found to be ∼10-fold and paraplegin still ∼4-fold more abundant in murine brain than in liver compared to the relative abundance of Afg3l1 in these tissues. Immunodepletion experiments indicate that a homo-oligomeric Afg3l2 complex is indeed the predominant m-AAA protease present in brain mitochondria of mice lacking paraplegin. Similarly, a homo-oligomeric hAFG3l2 complex is formed in HSP fibroblasts and is therefore expected to also be present in human brain mitochondria in the absence of paraplegin, as hAFG3L1 represents a pseudogene in humans. The decreased abundance of Afg3l1 relative to other m-AAA protease subunits in murine brain mitochondria therefore explains why the deletion of paraplegin leads to similar phenotypic consequences in mice and humans, despite the presence of an additional proteolytic subunit in murine mitochondria. Thus, tissue-specific consequences of a paraplegin deficiency can be at least partially rationalized by the differential expression and variable assembly of m-AAA protease subunits. This is reminiscent of mutations in the mitochondrial translation factor EFG1. Recent findings have demonstrated that the resulting tissue-specific oxidative phosphorylation system defects are caused by the varying abundance of several translation factors in different tissues (2).
Homo-oligomeric, proteolytically active hAFG3l2 complexes may partially substitute for the loss of hAFG3L2/paraplegin complexes in HSP mitochondria and lessen the phenotypic consequences of the loss of paraplegin. Indeed, complementation studies with yeast demonstrate that the substrate specificity of these assemblies overlaps with that of hetero-oligomeric m-AAA proteases containing paraplegin. Respiratory growth of yeast cells lacking Yta10 and Yta12 and deficiencies in the processing of MrpL32 and Ccp1 are restored in the presence of homo-oligomeric Afg3l1, Afg3l2, and hAFG3L2 complexes as well as by hetero-oligomeric assemblies of Afg3l1, Afg3l2, or hAFG3L2 with paraplegin. Thus, housekeeping functions of the m-AAA protease can be carried out by proteolytic complexes with variable subunit composition. In view of the overlapping function of these proteolytic complexes formed in the presence or absence of paraplegin, cell-specific axonal degradation in HSP is difficult to reconcile with an impaired proteolysis of common substrates of the different m-AAA proteases. We propose that specific substrates for paraplegin-containing m-AAA proteases exist which are expressed in a tissue-specific manner or whose impaired proteolysis affects only the function of specific cells.
In conclusion, future studies of the phenotypic consequences of the loss of paraplegin in HSP patients must take into account the versatile assembly of m-AAA protease subunits in different tissues and potential differences in the substrate specificity of m-AAA protease complexes formed. Furthermore, our findings predict that mutations in Afg3l2 or Afg3l1 may be linked to phenotypes different from that of HSP. The development of animal models for these subunits of the m-AAA protease will help elucidate this intricate interplay of functions.
Supplementary Material
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft, the European Union (6th framework program), and the German-Israeli-Project (DIP grant F.5.1) to T.L.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Alexander, C., M. Votruba, U. E. Pesch, D. L. Thiselton, S. Mayer, A. Moore, M. Rodriguez, U. Kellner, B. Leo-Kottler, G. Auburger, S. S. Bhattacharya, and B. Wissinger. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211-215. [DOI] [PubMed] [Google Scholar]
- 2.Antonicka, H., F. Sasarman, N. G. Kennaway, and E. A. Shoubridge. 2006. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum. Mol. Genet. 15:1835-1846. [DOI] [PubMed] [Google Scholar]
- 3.Arlt, H., G. Steglich, R. Perryman, B. Guiard, W. Neupert, and T. Langer. 1998. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 17:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlt, H., R. Tauer, H. Feldmann, W. Neupert, and T. Langer. 1996. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875-885. [DOI] [PubMed] [Google Scholar]
- 5.Atorino, L., L. Silvestri, M. Koppen, L. Cassina, A. Ballabio, R. Marconi, T. Langer, and G. Casari. 2003. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 163:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustin, S., M. Nolden, S. Müller, O. Hardt, I. Arnold, and T. Langer. 2005. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J. Biol. Chem. 280:2691-2699. [DOI] [PubMed] [Google Scholar]
- 7.Bieniossek, C., T. Schalch, M. Bumann, M. Meister, R. Meier, and U. Baumann. 2006. The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casari, G., M. De-Fusco, S. Ciarmatori, M. Zeviani, M. Mora, P. Fernandez, G. DeMichele, A. Filla, S. Cocozza, R. Marconi, A. Durr, B. Fontaine, and A. Ballabio. 1998. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973-983. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125:1241-1252. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C. 2006. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 22:79-99. [DOI] [PubMed] [Google Scholar]
- 11.Delettre, C., G. Lenaers, J. M. Griffoin, N. Gigarel, C. Lorenzo, P. Belenguer, L. Pelloquin, J. Grosgeorge, C. Turc-Carel, E. Perret, C. Astarie-Dequeker, L. Lasquellec, B. Arnaud, B. Ducommun, J. Kaplan, and C. P. Hamel. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207-210. [DOI] [PubMed] [Google Scholar]
- 12.Esser, K., B. Tursun, M. Ingenhoven, G. Michaelis, and E. Pratje. 2002. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the m-AAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323:835-843. [DOI] [PubMed] [Google Scholar]
- 13.Ferreirinha, F., A. Quattrini, M. Priozzi, V. Valsecchi, G. Dina, V. Broccoli, A. Auricchio, F. Piemonte, G. Tozzi, L. Gaeta, G. Casari, A. Ballabio, and E. I. Rugarli. 2004. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Investig. 113:231-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forner, F., L. J. Foster, S. Campanaro, G. Valle, and M. Mann. 2006. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell Proteomics 5:608-619. [DOI] [PubMed] [Google Scholar]
- 15.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 16.Ishihara, N., Y. Fujita, T. Oka, and K. Mihara. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhola, M. K., Z. H. Shah, L. A. Grivell, and H. T. Jacobs. 2000. The mitochondrial inner membrane AAA metalloprotease family in metazoans. FEBS Lett. 481:91-95. [DOI] [PubMed] [Google Scholar]
- 18.Karata, K., T. Inagawa, A. J. Wilkinson, T. Tatsuta, and T. Ogura. 1999. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 274:26225-26232. [DOI] [PubMed] [Google Scholar]
- 19.Korbel, D., S. Wurth, M. Kaser, and T. Langer. 2004. Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits. EMBO Rep. 5:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremmidiotis, G., A. E. Gardner, C. Settasatian, A. Savoia, G. R. Sutherland, and D. F. Callen. 2001. Molecular and functional analyses of the human and mouse genes encoding AFG3L1, a mitochondrial metalloprotease homologous to the human spastic paraplegia protein. Genomics 76:58-65. [DOI] [PubMed] [Google Scholar]
- 21.Kwong, J. Q., M. F. Beal, and G. Manfredi. 2006. The role of mitochondria in inherited neurodegenerative diseases. J. Neurochem. 97:1659-1675. [DOI] [PubMed] [Google Scholar]
- 22.Leonhard, K., B. Guiard, G. Pellechia, A. Tzagoloff, W. Neupert, and T. Langer. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 24.Mattiazzi, M., M. D'Aurelio, C. D. Gajewski, K. Martushova, M. Kiaei, M. F. Beal, and G. Manfredi. 2002. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 277:29626-29633. [DOI] [PubMed] [Google Scholar]
- 25.Mootha, V. K., J. Bunkenborg, J. V. Olsen, M. Hjerrild, J. R. Wisniewski, E. Stahl, M. S. Bolouri, H. N. Ray, S. Sihag, M. Kamal, N. Patterson, E. S. Lander, and M. Mann. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115:629-640. [DOI] [PubMed] [Google Scholar]
- 26.Nolden, M., S. Ehses, M. Koppen, A. Bernacchia, E. I. Rugarli, and T. Langer. 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123:277-289. [DOI] [PubMed] [Google Scholar]
- 27.Nolden, M., B. Kisters-Woike, T. Langer, and M. Graef. 2006. Quality control of proteins in the mitochondrion. Top. Curr. Genet. 16:119-147. [Google Scholar]
- 28.Ogura, T., S. W. Whiteheart, and A. J. Wilkinson. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146:106-112. [DOI] [PubMed] [Google Scholar]
- 29.Rugarli, E. I., and T. Langer. 2006. Translating m-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends Mol. Med. 12:262-269. [DOI] [PubMed] [Google Scholar]
- 30.Schägger, H. 2001. Blue-native gels to isolated protein complexes from mitochondria. Methods Cell Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 31.Steglich, G., W. Neupert, and T. Langer. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suno, R., H. Niwa, D. Tsuchiya, X. Zhang, M. Yoshida, and K. Morikawa. 2006. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell 22:575-585. [DOI] [PubMed] [Google Scholar]
- 33.Tatsuta, T., and T. Langer. Studying proteolysis within mitochondria. Curr. Top. Gen., in press. [DOI] [PubMed]
- 33a.Tatsuta, T., S. Augustin, M. Nolden, B. Friedrich, and T. Langer. m-AAA protease-driven membrane dislocation allows intermembrane cleavage by rhomboid in mitochondria. EMBOJ. in press. [DOI] [PMC free article] [PubMed]
- 34.Taylor, R. W., and D. M. Turnbull. 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.