Abstract
Insulin plays a critical role in whole-body energy homeostasis by regulating lipid and glucose metabolism. In fat and liver tissues, ADD1/SREBP1c is a key transcription factor to mediate insulin-dependent regulation of gene expression. Although transcriptional and proteolytic activation of ADD1/SREBP1c has been studied intensively, the mechanism by which insulin regulates expression of its target genes with ADD1/SREBP1c at the chromatin level is unclear. Here, we reveal that SWI/SNF chromatin remodeling factors interact with the ADD1/SREBP1c and actively regulate insulin-dependent gene expression. Insulin enhanced recruitment of SWI/SNF chromatin remodeling factors to its target gene promoters with concomitant changes in the chromatin structures as well as gene expression. Furthermore, in vivo overexpression of BAF155/SRG3, a component of the SWI/SNF complex, substantially promoted insulin target gene expression and insulin sensitivity. Taken together, our results suggest that the SWI/SNF chromatin remodeling complexes confer not only insulin-dependent gene expression but also insulin sensitivity in vivo via interaction with ADD1/SREBP1c.
Insulin is a key hormone for whole-body energy homeostasis and serves as a nutritional switch of metabolic programs. Postprandially, increased insulin facilitates energy storage into liver, muscle, and adipose tissues and turns off hepatic glucose production. Insulin stimulates lipogenesis and glycolysis by activating the expression of a subset of genes, such as fatty acid synthase (FAS), acetyl coenzyme A (acetyl-CoA) carboxylase, stearoyl-CoA desaturase 1, and glucokinase. Conversely, insulin suppresses glucose production by repressing gluconeogenic genes, including phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase, in liver and fat tissues where insulin signaling exerts its tissue-specific effects. Accumulating evidence proposes that ADD1/SREBP1c, a basic helix-loop-helix transcription factor, orchestrates fatty acid and glucose metabolism by mediating regulation of insulin-dependent gene expression. For example, even in the absence of insulin, adenoviral overexpression of ADD1/SREBP1c enhances the expression of lipogenic genes as well as the glucokinase gene, whereas dominant-negative ADD1/SREBP1c reverses such effects (8, 9, 12, 32). Additionally, ADD1/SREBP1c has been implicated in insulin-dependent suppression of the PEPCK and insulin receptor substrate-2 genes by directly binding to their promoters (2, 3, 13). In accord with these findings, hepatic responses to fasting and refeeding are diminished in ADD1/SREBP1c-deficient mice (25).
ADD1/SREBP1c is highly expressed in white adipose, brown adipose, and liver tissues (47). Its expression is regulated by nutritional status and insulin and, consequently, controls the transcription of insulin-dependent genes (2, 9, 16, 24). Insulin not only stimulates the mRNA level of ADD1/SREBP1c by an auto-regulatory mechanism but activates the proteolytic maturation of ADD1/SREBP1c, resulting in its nuclear accumulation (1, 8, 51). Furthermore, insulin signaling is crucial in regulating the transcriptional activity of ADD1/SREBP1c by modulating its phosphorylation level (8, 16, 18, 19, 35). Thus, it is likely that ADD1/SREBP1c is well adapted for acute response for insulin-dependent gene regulation to coordinate energy metabolism.
Epigenetic regulation of chromatin structure, by changing the accessibility of DNA-binding protein complexes to template DNA, is critical for eukaryotic gene expression. In eukaryotic cells, two major classes of chromatin remodeling complexes have been identified: ATP-independent and ATP-dependent chromatin remodeling complexes. ATP-independent chromatin-modifying complexes change chromatin structure by covalent modifications of histones, including acetylation, phosphorylation, and methylation, which are usually associated with activation or repression of gene expression (34, 36, 54). ATP-dependent chromatin remodeling complexes, such as SWI/SNF complexes, utilize the energy from ATP hydrolysis, disrupting or altering nucleosome conformation to affect gene expression (31, 50). Recent studies indicate that SWI/SNF complex-dependent chromatin remodeling is actively involved in cell growth and differentiation by regulating several transcription factors, such as p53, MyoD, and glucocorticoid receptor (6, 10, 22, 43).
SWI/SNF chromatin remodeling complexes are heterogeneous complexes, containing BRG1 or Brm ATPase in addition to another 8 to 15 BRG1-associated factors (BAFs), including BAF170, BAF155/SRG3, and SNF5, as defined by reconstitution of chromatin remodeling activity with recombinant proteins in vitro (33). Several lines of evidence indicate that SWI/SNF chromatin remodeling complexes are implicated in both transcriptional activation and repression. For example, transcriptional activity of p53 is increased by overexpression of BRG1 or hSNF5 and reduced by dominant-negative BRG1 or dominant-negative hSNF5 (22). However, transcription of CYP7A1 is substantially suppressed in response to bile acid by recruiting SHP to the CYP7A1 promoter, where SHP associates with Brm- and mSin3A-containing chromatin remodeling complexes (15).
Although transcriptional and posttranslational regulation of ADD1/SREBP1c by insulin has been intensively studied, the mechanism by which ADD1/SREBP1c controls insulin-dependent gene expression by interacting with coregulators at the chromatin level is unknown. Here, we report the first evidence that SWI/SNF chromatin remodeling complexes, through their association with ADD1/SREBP1c, are involved in insulin-dependent gene regulation. We found that insulin augmented the recruitment of both ADD1/SREBP1c and SWI/SNF chromatin remodeling factors to the promoters of insulin target genes. Moreover, overexpression of BAF155/SRG3 in C57BL/6J mice increased the expression of ADD1/SREBP1c and FAS and decreased PEPCK expression, which was accompanied by an increase of insulin sensitivity in vivo. Together, our data suggest that SWI/SNF chromatin remodeling factors would affect insulin sensitivity by regulating insulin-dependent gene expression via interaction with ADD1/SREBP1c.
MATERIALS AND METHODS
Cell culture.
3T3-L1 preadipocytes were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine calf serum. Two days postconfluence, 3T3-L1 cells were incubated with DMEM containing 10% fetal bovine serum, methyl-isobutylxanthine (500 μM), dexamethasone (1 μM), and insulin (5 μg/ml) for 48 h. Culture medium was changed every other day with DMEM containing 10% fetal bovine serum and insulin (1 μg/ml).
Endonuclease accessibility assays.
Endonuclease accessibility assays have been described previously (45). Briefly, 3T3-L1 adipocytes were harvested with RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, 5 mM butyrate, 10 mM NaF, and 1 mM NaVO7, supplemented with protease inhibitors) and incubated for 20 min on ice. The cell pellets were homogenized through 27-gauge syringes, centrifuged at 2,000 rpm at 4°C for 5 min, and resuspended in 50 μl of fresh RSB buffer. Resuspended genomic DNAs were digested with 100 U of several restriction endonucleases. Reactions were stopped by adding proteinase K and 2% sodium dodecyl sulfate (SDS) for 16 h at 45°C. Then chromosomal DNA was extracted with phenol-chloroform twice, precipitated with isopropanol, and resuspended in distilled water. The precipitated DNA was amplified by PCR. PCR amplifications consisted of 0.25 M concentrations of each primer, 0.1 mM concentrations of each deoxynucleoside triphosphate (dNTP), 1× PCR buffer, and 1 U Nova Taq polymerase (Genenmed, Korea) in 20-μl reaction volumes. The PCR products were resolved through 10% polyacrylamide in 0.5× Tris-borate-EDTA gels. Primer sequences are available upon request.
Coimmunoprecipitation and Western blot analysis.
Cell pellets from two 100-mm dishes of differentiated 3T3-L1 adipocytes were suspended in 1 ml of lysis buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1.5 mM MgCl2, and 0.1% [vol/vol] Nonidet P-40) containing protease inhibitors. Lysates were precleared by incubation with 20 μl of preimmune serum and excess protein A-Sepharose beads (Amersham) for 3 h at 4°C. Precleared lysates were incubated with 5 μl of either preimmune serum or anti-ADD1/SREBP1c serum together with 20 μl of protein A-Sepharose beads for 1 h at 4°C. After centrifugation, the pelleted beads were washed two times with 1 ml of lysis buffer for 15 min at 4°C, resuspended in 20 μl of distilled water, and mixed with 5× SDS loading buffer (200 mM Tris-HCl [pH 6.8], 50% glycerol, 5% SDS, 10% β-mercaptoethanol, and 0.05% bromophenol blue). Supernatant and pellets were boiled for 5 min, resolved by SDS-polyacrylamide gel electrophoresis, and analyzed by immunoblotting. For Western blot analysis, isolated immunoprecipitates or nuclear extracts were resolved by SDS-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. The blots were incubated with each of the antibodies, visualized by using the ECL kit (iNtRON, South Korea), and quantified with a LuminoImager, LAS3000, and Science Lab 2001 Image Gauge software (Fuji Photo Film).
Chromatin immunoprecipitation.
Fully differentiated 3T3-L1 adipocytes were incubated with or without insulin (100 nM). The cells were cross-linked in 1% formaldehyde at 37°C for 10 min and resuspended in 200 μl of NP-40-containing buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 8.0), 85 mM KCl, and 0.5% NP-40]. Crude nuclei were precipitated and lysed in 200 μl of lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl [pH 8.1]). Nuclear lysates were sonicated and diluted 10-fold with IP buffer (16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, and 1.1% Triton X-100). Then lysates were incubated with protein A-Sepharose CL-4B (Amersham-Pharmacia) and each antibody for 2 h at 4°C. These immunoprecipitates were sequentially washed for 5 min with 1 ml of TSE 150 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl), 1 ml of TSE 500 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 500 mM NaCl), 1 ml of buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.1]), and 1 ml of TE (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). Immune complexes were eluted with 2 volumes of 250 μl elution buffer (1% SDS and 0.1 M NaHCO3), and 20 μl of 5 M NaCl was added to reverse formaldehyde cross-linking. DNA was extracted with phenol-chloroform and precipitated with isopropyl alcohol and 80 μg of glycogen. Precipitated DNA samples were amplified by PCR in mixture with the following: 0.25 μM concentrations of each primer, 0.1 mM concentrations of each dNTP, 1× PCR buffer, and 1 U Nova Taq polymerase (Genenmed, South Korea) in 20-μl reaction volumes. PCR products were resolved through 10% polyacrylamide-0.5× Tris-borate-EDTA gels. Primers used were as follows: −572 adiponectin-forward (f), 5′-GGTGCTGGGAATTGAACTCA-3′; −213 adiponectin-reverse (r), 5′-CCTGTTTCCAGGCTTTGGCC-3′; −247 ADD1/SREBP1c-f, 5′-AGC CAC CGG CCA TAA ACC AT-3′; +56 ADD1/SREBP1c-r, 5′-GGT TGG TAC CAC AGT GAC CG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-f, 5′-GTGTTCCTACCCCCAATGTG-3′; GAPDH-r, 5′-CTTGCTCAGTGTCCTTGCTG-3′; PEPCK-f, 5′-GAGCAGGGGTCAGTATGT-3′; PEPCK-r, 5′-GACCGTGACTGTTGCTGATGC-3′.
Quantitative real-time RT-PCR.
cDNA was synthesized using Moloney leukemia virus reverse transcriptase with dNTPs and oligo(dT) primers (Invitrogen). These cDNAs were templates for PCR with specific primers at annealing temperatures ranging between 54°C and 60°C in the presence of dNTPs and Taq DNA polymerase. Real-time reverse transcription (RT)-PCR amplification mixtures were brought to a final volume of 20 μl and contained 20 ng of reverse-transcribed total RNA, 0.25 μM concentrations of the forward and reverse primers, and Cybergreen (Bio-Rad). The MyiQ real-time PCR detection system (Bio-Rad) was used for PCR amplifications in 96-well plates. All reactions were done in triplicate and repeated at least three times. The relative amounts of each mRNA were calculated by using the comparative threshold cycle method. GAPDH or β-actin mRNA was used as the invariant control. The primer sequences used for PCR are available upon request. Primers used were as follows: ADD1/SREBP1c-f, 5′-GGGAATTCATGGATTGCACATTTGAA-3′; ADD1/SREBP1c-r, 5′-CCGCTCGAGGTTCCCAGGAAGGGT-3′; adiponectin-f, 5′-ATGCTACTGTTGCAAGCT CTC-3′; adiponectin-r, 5′-GTTGGTATCATGGAAGAGAAG-3′; FAS-f, 5′-TGCTCCCAGCTGCAGGC-3′; FAS-r, 5′-GCCCGGTAGCTCTGGGTGTA-3′; PEPCK-f, 5′-GTCACCATCACCTCCTGGAAGA-3′; PEPCK-r, 5′-GGTGCAGAATCTCGAGTTG-3′;GAPDH-f, 5′-TGCACCACCAACTGCTTAG-3′; GAPDH-r, 5′-GGATGCAGGGATGATGTTC-3′.
Plasmid constructs.
The FAS-luciferase plasmid containing a −220- to +25-bp fragment of the FAS promoter in front of a luciferase reporter gene was described previously (23). ADD1/SREBP1c-luciferase reporter DNA contains the ADD1/SREBP1c promoter spanning −2.7 kb to +1 bp from the first ATG codon in front of the luciferase gene (23). The ADD1/SREBP1c expression vectors encoded amino acids (aa) 1 to 403 (A403), aa 1 to 153 (A153), aa 153 to 308 (A15308), or aa 308 to 403 (A308/403) of rat ADD1 and were cloned in frame into the pcDNA3.1-Myc/HisA plasmid and pGEX4T-1 vector (Pharmacia) (23). SREBP1a cDNA (aa 1 to 490) was cloned into the pGEX4T-1 vector (Pharmacia). The mSin3A expression vector was gift from J. H. Choe. The PEPCK promoter-luciferase vector was provided by K. Chakravarty. The mouse BAF155/SRG3 expression vector was cloned into the pCAGGS vector (22). The SRG3-small interfering RNA (siRNA) construct for reporter assays was generated by cloning annealed short interfering DNA oligomers sense, 5′-CGTGACAGAACAGACCAATTTCAAGAGAATTGGTCTGTTCTGTCACGTTTTT T-3′, and antisense, 5′-AATTAAAAAACGTGACAGAACAGACCAATTCTCTTGAAATTGGTCTGTTCTGTCACGGGCC-3′, into the pSilencer 1.0-U6 vector (Ambion, Inc.) digested with ApaI and EcoRI enzymes.
Gene transduction into 3T3-L1 adipocytes.
Eight to 10 days postdifferentiation of 3T3-L1 adipocytes, transient transfections were performed by using LipofectAMINE 2000 reagent (Invitrogen) or electroporation with Microporator (Digitalbiotechnology, South Korea). Double-stranded RNA corresponding to mouse BAF155/SRG (CGTGACAGAACAGACCAAT) was used for BAF155/SRG3 knockdown experiments. siRNAs for mouse ADD1/SREBP1c were purchased from Dharmacon (SMARTpool).
Animals and treatments.
Male C57BL/6J, ob/ob, and db/db mice were housed in colony cages in 12-h light/12-h dark cycles. For fasting experiments, mice were fasted for 48 h. For refeeding experiments, mice were fasted for 48 h and then refed a normal chow for 6 h before study. Experiments were staggered such that all mice were sacrificed at the same time, which was at the end of the dark cycle. For the streptozotocin (STZ) experiments, mice were treated daily with STZ (Sigma) by four intraperitoneal injections of approximately 0.2 to 0.3 ml of 100 mM sodium citrate solution (pH 4.5) containing STZ (100 mg/kg of body weight). Control mice were injected with 100 mM sodium citrate solution (pH 4.5). After injection, animals were fasted for an additional 24 h, and plasma glucose levels were tested to confirm diabetic condition (glucose level > 300 mg/dl). The animals were fed a chow diet for 12 h, after which insulin was administered to the STZ plus insulin group. The animals were injected subcutaneously with human neutral protamine hagedorn insulin (1.5 units) in 0.2 ml of phosphate-buffered saline (PBS). Mice in the control and STZ groups were injected subcutaneously with 0.2 ml of PBS. After injection of insulin or PBS, the animals were fed a chow diet for 3 h and then sacrificed by halothane anesthesia. For glucose tolerance and insulin tolerance tests, C57BL/6J mice were fasted for 16 h and 6 h, respectively, and basal blood samples were taken, followed by intraperitoneal injection of glucose (1.5 g/kg) or insulin (0.85 U/kg, Humulin R; Eli Lilly and Company). Blood samples were drawn at 15, 30, 60, 90, and 120 min or at 15, 30, 45, 60, 90, and 120 min after injection.
Generation of BAF155/SRG3 Tg mice.
For constructing the pCAGGS-BS expression vector, the 2.3-kb fragment of the pCAGGS vector containing the human cytomegalovirus (CMV) immediate-early enhancer linked to the chicken β-globin poly(A) was inserted into the SalI and PstI sites of the pBluescript vector. For constructing transgenic (Tg) mice, the full-length mouse BAF155/SRG3 cDNA (3.3 kb) fused in frame with the Myc tag was subcloned into the EcoRI site of the pCAGGS-BS expression vector. Expression of the transgene was driven by the human CMV immediate-early enhancer linked to the chicken beta-actin promoter. The inserted fragment was excised from the vector with XhoI and NotI and purified by agarose gel electrophoresis. The transgenic mice were generated by microinjection of the purified DNA into pronuclei of fertilized eggs of FVB/N mice. Transgenic animals were identified by PCR amplification and/or Southern blot analysis of tail DNA. Three founders were established and characterized further. The genetic background was changed by backcrossing the transgenic mice with C57BL/6J mice at least for 7 generations. For most experiments, animals were between 5 and 10 weeks of age. All experiments were carried out in an AAALAC-certified facility in compliance with approved animal policies by the Sungkyunkwan University School of Medicine.
RESULTS
Chromatin remodeling is involved in regulating expression of insulin-responsive genes.
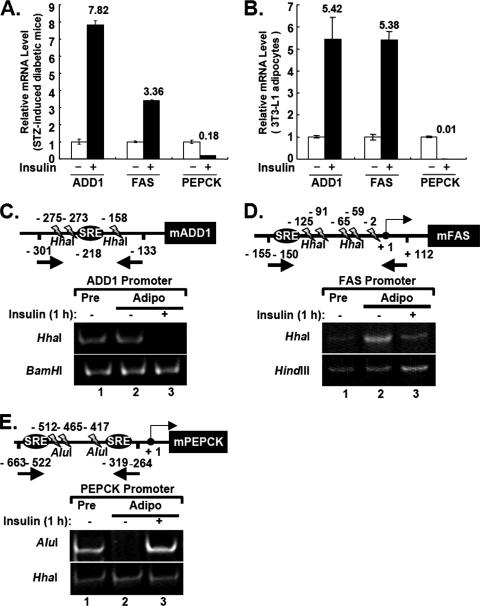
To observe the in vivo effect of insulin on gene expression, we measured the endogenous mRNA levels of several insulin target genes in fat tissues of insulin-deficient diabetic mice that had been induced with STZ in the absence or presence of acute insulin treatment. Consistent with previous studies, insulin stimulated mRNA levels of ADD1/SREBP1c and FAS by 7.8- and 3.4-fold, respectively, while insulin drastically decreased the level of PEPCK mRNA (Fig. 1A). Similar results were observed when cultured 3T3-L1 adipocytes were treated with insulin, indicating that fully differentiated 3T3-L1 adipocytes could provide a suitable system to study insulin-dependent gene regulation (Fig. 1B).
FIG. 1.
Insulin-dependent regulation of ADD1/SREBP1c target genes in fat tissues and 3T3-L1 adipocytes. (A) Insulin regulation of ADD1/SREBP1c, FAS, and PEPCK mRNA expression in epididymal fat tissues from STZ-induced insulin-deficient diabetic mice. Total RNA was isolated from fat tissues and analyzed by quantitative RT-PCR with primer sets specific for mouse ADD1/SREBP1c, FAS, and PEPCK. (B) mRNA level of ADD1/SREBP1c target genes after insulin treatment in 3T3-L1 adipocytes. Total RNA was isolated from 3T3-L1 adipocytes and analyzed by quantitative RT-PCR. Standard errors of the mean are indicated by the error bars (n = 3). (C to E) For restriction endonuclease accessibility assays to ADD1/SREBP1c, FAS, and PEPCK promoters, 3T3-L1 adipocytes were incubated with (+) or without (−) insulin (100 nM) for 1 h. 3T3-L1 preadipocytes were incubated in DMEM supplemented with 10% bovine calf serum. Nuclei were isolated and digested with each restriction enzyme as indicated. Chromosomal DNA was purified and amplified by PCR with specific primers indicated by each arrow. (C) PCR amplification produced a 169-bp fragment from −301 to −133 nucleotides relative to the ATG site for the mouse ADD1/SREBP1c promoter. (D) PCR amplification produced a 268-bp fragment from −155 to +112 nucleotides relative to the transcription start site (TSS) for the FAS promoter. (E) PCR amplification produced a 400-bp fragment from −663 to +264 nucleotides relative to the TSS for the PEPCK promoter. Pre, preadipocytes; Adipo, adipocytes; SRE, sterol regulatory element (ADD1/SREBP1c binding site). Reproducible results were obtained from more than three independent studies.
These positive and negative transcriptional regulations by insulin prompted us to examine whether chromatin remodeling is involved in regulating insulin-dependent gene expression. To directly address this issue, we performed the endonuclease accessibility assays (6, 45). We incubated 3T3-L1 adipocytes with or without insulin, and the isolated nuclei were digested with several restriction enzymes which specifically cut promoter regions of ADD1/SREBP1c, FAS, or PEPCK. The ADD1/SREBP1c promoter spanning −301 to −133 nucleotides upstream from the first ATG codon has three restriction sites for HhaI. The FAS promoter spanning −155 to +122 nucleotides upstream of the transcription start site has five restriction sites for HhaI. While the region of the PEPCK promoter spanning −663 to −264 upstream of the transcription start site has no recognition site for HhaI, it has three sites for AluI. We used BamHI, HindIII, and HhaI restriction enzymes as negative controls for the ADD1/SREBP1c, FAS, and PEPCK promoters, respectively (Fig. 1C to E). Serum starvation conditions reduced the endonuclease accessibility at the ADD1/SREBP1c and FAS promoters in 3T3-L1 adipocytes to a level comparable to that in preadipocytes, while this condition significantly increased the endonuclease accessibility of the PEPCK promoter (Fig. 1C and D). Interestingly, we observed that insulin treatment remarkably increased HhaI endonuclease accessibility to the ADD1/SREBP1c and FAS promoters (Fig. 1C and D), whereas insulin decreased AluI accessibility to the PEPCK promoter (Fig. 1E). These results indicate that insulin changes the chromatin structure of its target gene promoters to affect gene expression in a positive or negative fashion.
SWI/SNF chromatin remodeling factors are recruited to the promoters of insulin's target genes.
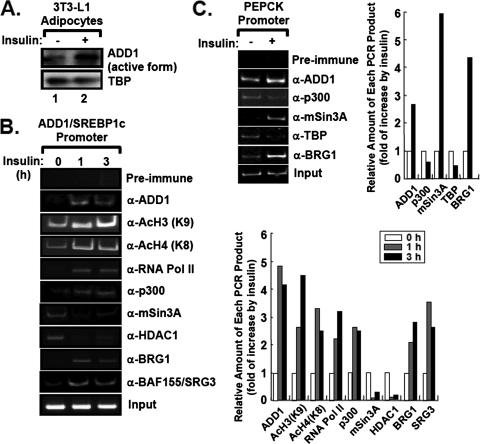
To examine which cofactors associate with or dissociate from the promoters of insulin's target genes upon insulin stimulation, we conducted chromatin immunoprecipitation (ChIP) assays with 3T3-L1 adipocytes in the absence or presence of insulin. As reported previously (11), insulin promoted nuclear accumulation of the active form of ADD1/SREBP1c (Fig. 2A) and significantly enhanced recruitment of ADD1/SREBP1c to its own promoter (Fig. 2B and 3B) (1). Insulin also augmented recruitment of RNA polymerase II to the ADD1/SREBP1c promoter, with increased levels of acetylation at lysine 9 of histone H3 and lysine 8 of histone H4 (Fig. 2B). This increase was accompanied by a concomitant increase of p300 binding (Fig. 2B). Furthermore, insulin stimulated the association of BRG1 and BAF155/SRG3, which are core components of the SWI/SNF chromatin remodeling complex, with the ADD1/SREBP1c promoter (Fig. 2B), suggesting that insulin activates the ADD1/SREBP1c promoter by increasing recruitment of the SWI/SNF chromatin remodeling complex as well as the basal transcription machinery.
FIG. 2.
Insulin increases recruitment of the chromatin remodeling complexes to the promoters of insulin target genes. (A) Western blot analysis of the protein level of the nuclear active form of ADD1/SREBP1c in 3T3-L1 adipocytes with (+) or without (−) insulin (100 nM) for 1 h. (B and C) ChIP analyses of the ADD1/SREBP1c promoter (B) and PEPCK promoter (C). 3T3-L1 adipocytes were incubated with or without insulin (100 nM). The amount of each PCR product was quantitated for the graph. For each experiment, 1% of input was used. α, anti.
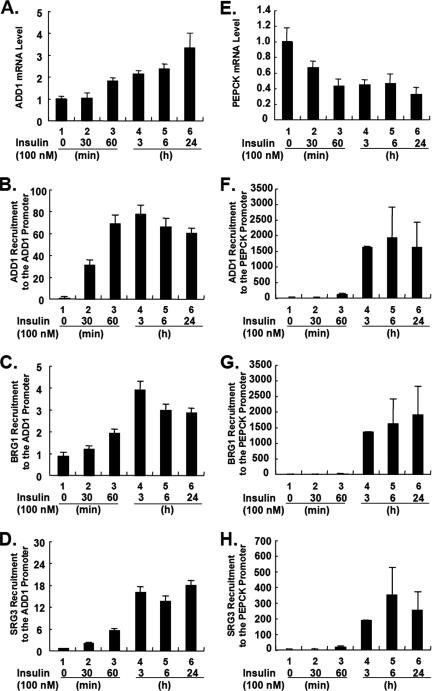
FIG. 3.
Insulin sequentially increases the recruitment of ADD1/SREBP1c and the SWI/SNF chromatin remodeling complex to its target gene promoters. 3T3-L1 adipocytes were treated with insulin for 0 min, 30 min, 60 min, 3 h, 6 h, and 24 h. (A and E) mRNA levels of ADD1/SREBP1c (A) and PEPCK (E) were analyzed by Q-PCR. (B to D) Time-dependent recruitments of ADD1/SREBP1c (B), BRG1 (C), and BAF155/SRG3 (D) onto the ADD1/SREBP1c promoter were analyzed with ChIP assays. (F to H) Time-dependent recruitments of ADD1/SREBP1c (F), BRG1 (G), and BAF155/SRG3 (H) onto the PEPCK promoter were analyzed with ChIP assays. Standard errors of the mean are indicated by the error bars (n = 2). The results are representatives of at least three independent experiments.
Insulin tightly regulates gluconeogenesis via suppressing PEPCK and glucose-6-phosphatase gene expression. Recently, it has been reported that ADD1/SREBP1c mimics the negative effects of insulin on PEPCK gene expression by directly binding to the PEPCK promoter (2). Consistent with this report, insulin treatment modestly enhanced recruitment of ADD1/SREBP1c and BAF155/SRG3 to the PEPCK promoter in 3T3-L1 adipocytes (Fig. 2C). Moreover, we observed that more mSin3A proteins were bound to the PEPCK promoter upon insulin treatment (Fig. 2C). Similar results were obtained from ChIP analysis of the insulin receptor substrate-2 promoter, which has been reported to be repressed by ADD1/SREBP1c in response to insulin signaling as a negative feedback regulation (data not shown) (13). Since the interaction between mSin3A and histone deacetylases (HDACs) is well established for the repression of gene expression (20), enhanced association of mSin3A with the PEPCK promoter by insulin challenge implies that insulin-dependent target gene suppression might be, at least partly, mediated by mSin3A and/or HDACs with ADD1/SREBP1c. Promoter-specific recruitment of mSin3A in response to insulin was also observed in the FAO hepatomic cell line (data not shown).
After insulin treatment in 3T3-L1 adipocytes, we determined the ordering of events of the protein recruitment to the ADD1/SREBP1c and PEPCK promoters with ChIP assays. At the same time, we performed quantitative real-time RT-PCR (Q-PCR) analyses for both genes. Upon insulin stimulation, mRNA levels of ADD1/SREBP1c and PEPCK were upregulated and downregulated, respectively, within several hours (Fig. 3A and E). More interestingly, we observed that ADD1/SREBP1c appeared to be recruited to the ADD1/SREBP1c and PEPCK promoters prior to the recruitment of SWI/SNF chromatin remodeling factors by insulin.
ADD1/SREBP1c physically interacts with SWI/SNF chromatin remodeling factors.
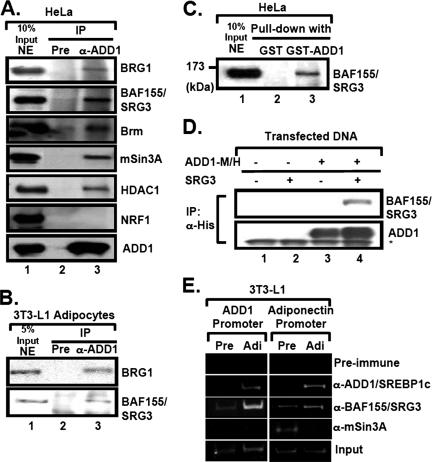
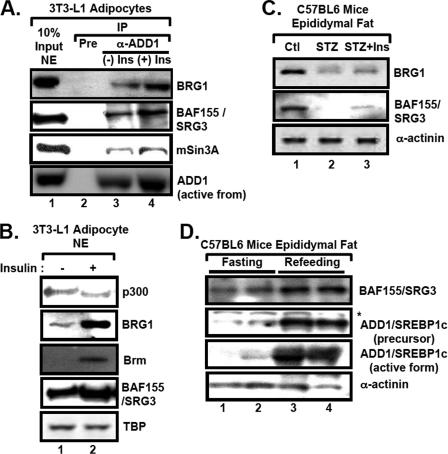
To determine whether ADD1/SREBP1c directly interacts with chromatin remodeling factors, we used the antibody against ADD1/SREBP1c for coimmunoprecipitation (Co-IP) experiments. In HeLa nuclear extracts where SREBP proteins have been originally identified (49), ADD1/SREBP1c formed a protein complex with SWI/SNF chromatin remodeling factors, including BRG1, Brm, and BAF155/SRG3 (Fig. 4A). Also, ADD1/SREBP1c associated with mSin3A/HDAC1 (Fig. 4A). Similar results were obtained with 3T3-L1 adipocytes (Fig. 4B). Interaction between ADD1/SREBP1c and BAF155/SRG3 was further confirmed by glutathione S-transferase (GST) pull-down assays and transient-transfection analysis (Fig. 4C and D).
FIG. 4.
ADD1/SREBP1c associates with BAF155/SRG3, a component of the SWI/SNF chromatin remodeling complex. (A) HeLa nuclear extracts were immunoprecipitated (IP) with preimmune or anti-ADD1/SREBP1c antibodies (α-ADD1). Association of the SWI/SNF complex in the ADD1/SREBP1c immunocomplex was monitored by Western blot analyses with antibodies against BRG1, BAF155/SRG3, Brm, mSin3A, and HDAC1. NRF-1 was used as a negative control for the interaction with ADD1/SREBP1c. Input, 10% of lysates. NE, nuclear extract. (B) 3T3-L1 adipocyte nuclear extracts were immunoprecipitated with preimmune serum or anti-ADD1/SREBP1c antibodies. Pre, preimmune serum. (C) Nuclear extracts from HeLa cells were incubated with glutathione-coated beads containing GST or GST-ADD1/SREBP1c recombinant protein. After washing beads, bound proteins were analyzed by Western blot analyses with the anti-BAF155/SRG3 antibody. In lane 1, 10% input protein was used. (D) HEK293 cells were transfected as indicated with ADD1-Myc/His, Myc-BAF155/SRG3, or both expression vectors by the calcium phosphate method, and total lysates were subjected to Co-IP. The association of BAF155/SRG3 with anti-His precipitates (ADD1/SREBP1c) was detected by Western blot analyses with anti-BAF155/SRG3 and anti-ADD1/SREBP1c antibodies. The asterisk indicates the immunoglobulin heavy chain used for Co-IP. (E) The association of ADD1/SREBP1c, BAF155/SRG3, and mSin3A with the promoters of ADD1/SREBP1c target genes was determined by ChIP analysis. Pre, preadipocytes; Adi, Adipocytes.
To elucidate whether the binding of BAF155/SRG3 to ADD1/SREBP1c target gene promoters is regulated upon the level of ADD1/SREBP1c, we conducted ChIP analysis with preadipocytes and adipocytes. Compared with preadipocytes, ADD1/SREBP1c is highly expressed in adipocytes (17). As target genes of ADD1/SREBP1c, proximal promoter regions of adiponectin and ADD1/SREBP1c genes were examined (1, 38). In adipocytes, recruitment of both ADD1/SREBP1c and BAF155/SRG3 was increased at the promoters of both ADD1/SREBP1c and adiponectin genes in adipocytes (Fig. 4E). However, the recruitment of mSin3A to the adiponectin and ADD1/SREBP1c promoter notably decreased during adipogenesis, although both mSin3A and HDAC1 associated with ADD1/SREBP1c in vitro (Fig. 4A and E). These results suggest that insulin directs the recruitment of different subsets of SWI/SNF complexes and chromatin remodeling factors in a promoter-specific manner to up- or downregulate its target genes.
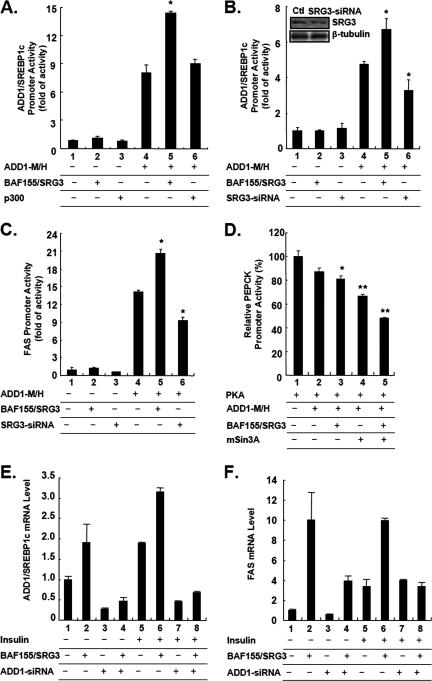
BAF155/SRG3 enhances the transcriptional activity of ADD1/SREBP1c.
To investigate the consequences of the interaction between ADD1/SREBP1c and BAF155/SRG3, we examined the transcriptional activity of ADD1/SREBP1c by luciferase reporter assays in the presence or absence of BAF155/SRG3. Ectopic expression of BAF155/SRG3 enhanced the transcriptional activity of ADD1/SREBP1c (Fig. 5A to C), while reduced expression of BAF155/SRG3 by the siRNA system, which suppressed its expression by 43%, decreased the transcriptional activity of ADD1/SREBP1c at its own and FAS promoters (Fig. 5B and C). In addition, the repressive activity of ADD1/SREBP1c at the PEPCK promoter was further augmented by overexpression of BAF155/SRG3 and/or mSin3A (Fig. 5D). We also examined whether the effects of BAF155/SRG3 overexpression require ADD1/SREBP1c. To address this, we used ADD1/SREBP1c siRNA. 3T3-L1 adipocytes were transiently transfected with BAF155/SRG3 expression vector in combination with siRNA for ADD1/SREBP1c. ADD1-siRNA reduced the endogenous ADD1/SREBP1c mRNA level up to 25% in the basal state (Fig. 5E, lane 1 versus 3). As expected, overexpression of BAF155/SRG3 increased both basal and insulin-stimulated ADD1/SREBP1c and FAS mRNA, while ADD1-siRNA abolished such effects (Fig. 5E and F). Thus, it is likely that the SWI/SNF chromatin remodeling complex requires ADD1/SREBP1c to elicit its effect on insulin-dependent regulation of gene expression. Therefore, it appears that the interaction between ADD1/SREBP1c and BAF155/SRG3 is specific and regulates insulin-dependent gene expression by affecting the transcriptional activity of ADD1/SREBP1c.
FIG. 5.
BAF155/SRG3 modulates the transcriptional activity of ADD1/SREBP1c. (A) HEK293 cells were cotransfected with the ADD1/SREBP1c promoter-luciferase reporter (200 ng) and either empty vector or combinations of ADD1-M/H (100 ng), p300 (100 ng), and BAF155/SRG3 (100 ng) expression vectors as indicated. (B to C) NIH 3T3 mouse fibroblast cells were cotransfected with combinations of empty vector, ADD1-M/H (100 ng), BAF155/SRG3 (100 ng), and SRG3 siRNA (250 ng) expression vectors along with either the ADD1/SREBP1c promoter-luciferase reporter (200 ng) (B) or FAS promoter-luciferase reporter (200 ng) (C) as indicated. The level of endogenous BAF155/SRG3 was assessed by Western blot analysis. (D) HEK293 cells were cotransfected with the PEPCK promoter-luciferase reporter (500 ng) and either empty vector or combinations of PKA (500 ng), ADD1-M/H (50 ng), BAF155/SRG3 (500 ng), and mSin3A (500 ng) expression vectors as indicated. (A to D) the pCMV-lacZ construct was cotransfected as an internal control, and the values plotted are the ratios of luciferase activity to β-galactosidase activity. Standard errors of the mean are indicated by the error bars (n = 2). The results are representatives of at least five independent experiments. *, P < 0.05; **, P < 0.01. (E and F) ADD1/SREBP1c is required for the effects of BAF155/SRG3 on insulin-dependent gene expression. 3T3-L1 adipocytes were transiently transfected with combinations of BAF155/SRG3 expression vector and ADD1 siRNA. Two days after transfection, cells were preincubated in DMEM supplemented with 0.2% bovine serum albumin for 24 h, and cells were treated with insulin for another 24 h. mRNA levels were analyzed by Q-PCR. Standard errors of the mean are indicated by the error bars (n = 2). The results are representatives of at least three independent experiments. +, present; −, absent.
BAF155/SRG3 is necessary for insulin-dependent gene regulation.
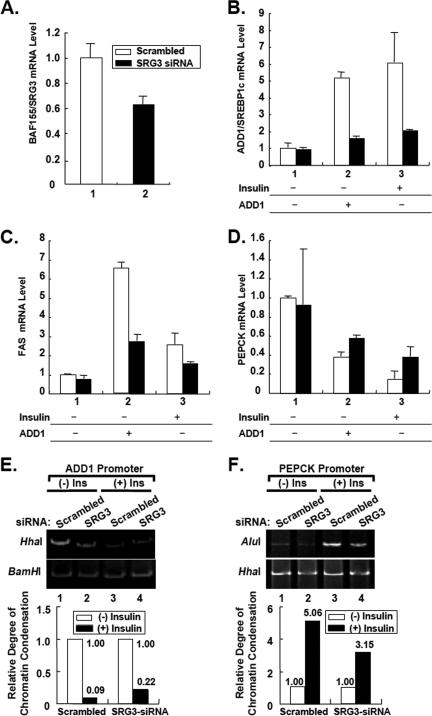
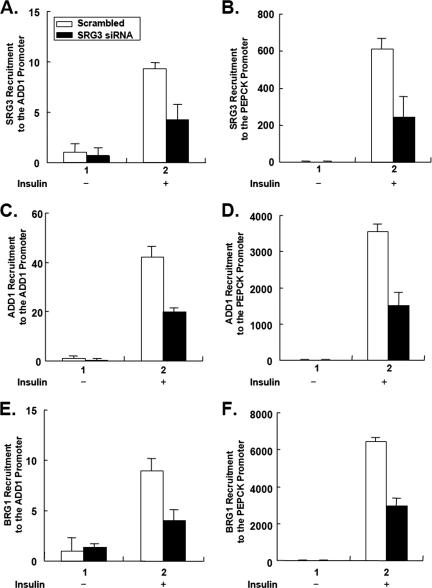
To determine whether BAF155/SRG3 is necessary for the insulin-dependent regulation of gene expression and the transcriptional activity of ADD1/SREBP1c, we transfected 3T3-L1 adipocytes with combinations of BAF155/SRG3 siRNA (SRG3-siRNA) and ADD1/SREBP1c expression vector in the presence or absence of insulin. SRG3-siRNA reduced endogenous BAF155/SRG3 expression about 40% (Fig. 6A). Additionally, BAF155/SRG3 knockdown in 3T3-L1 adipocytes decreased the recruitment of ADD1/SREBP1c and BRG1 at the ADD1/SREBP1c and PEPCK promoters (Fig. 7). Consistently, Q-PCR analyses showed that BAF155/SRG3 knockdown reduced insulin- and ADD1/SREBP1c-dependent expression of ADD1/SREBP1c and FAS mRNAs, while it interfered with insulin- and ADD1/SREBP1c-dependent suppression of the PEPCK gene (Fig. 6B to D). More importantly, endonuclease accessibility assays revealed that BAF155/SRG3 knockdown disturbed insulin-induced chromatin decondensation of the ADD1/SREBP1c promoter and condensation of the PEPCK promoter (Fig. 6E and F). Taken together, these data suggest that BAF155/SRG3 might be a necessary factor for insulin-dependent regulation of gene expression by inducing chromatin remodeling at its target gene promoters.
FIG. 6.
BAF155/SRG3 is necessary for insulin-dependent gene regulation. 3T3-L1 adipocytes were transfected with BAF155/SRG3 siRNA in combination with the active form of ADD1/SREBP1c expression vector. Two days after transfection, cells were preincubated in DMEM supplemented with 0.2% bovine serum albumin for 24 h. Then cells were treated with insulin for another 24 h. (A to D) mRNA levels of BAF155/SRG3 (A), ADD1/SREBP1c (B), FAS (C), and PEPCK (D) were analyzed by Q-PCR. Standard errors of the mean are indicated by the error bars (n = 2). The results are representatives of at least three independent experiments. Open bars indicate scrambled siRNA; filled bars indicate BAF155/SRG3 siRNA. (E and F) Restriction endonuclease accessibility assays with BAF155/SRG3 knockdown adipocytes. The relative band intensity of each lane was quantitated and normalized to the negative control. The graphs indicate change in chromatin accessibility after insulin treatment in cells with (+) or without (−) SRG3 siRNA. HhaI and AluI were used for restriction enzymes for specific digestion for ADD1/SREBP1c promoter and PEPCK promoter, respectively. BamHI and HhaI were used as negative controls for the ADD1/SREBP1c promoter and PEPCK promoter, respectively. Ins, Insulin.
FIG. 7.
BAF155/SRG3 is necessary for insulin-dependent recruitment of the SWI/SNF chromatin remodeling complex into insulin target gene promoters. In BAF155/SRG3 knockdown 3T3-L1 adipocytes, recruitment of BAF155/SRG3, ADD1/SREBP1c, and BRG1 was analyzed by ChIP analysis with (+) or without (−) insulin (100 nM) for 1 h. Relative amounts of PCR product were analyzed by quantitative real-time PCR analysis. Open bars indicate scrambled siRNA. Filled bars indicate BAF155/SRG3 siRNA.
Insulin promotes the interaction between ADD1/SREBP1c and BAF155/SRG3 as well as their protein levels.
To determine whether insulin also affects the association between ADD1/SREBP1c and the SWI/SNF complex, we isolated nuclear extracts from 3T3-L1 adipocytes with or without insulin and immunoprecipitated the extracts with the ADD1/SREBP1c antibody. In the presence of insulin, the interaction between ADD1/SREBP1c and components of the SWI/SNF complex such as BRG1, BAF155/SRG3, and mSin3A was promoted (Fig. 8A). In addition, we observed that protein levels of SWI/SNF complex components, such as BRG1, Brm, and BAF155/SRG3, were increased by insulin treatment in adipocytes (Fig. 8B). In accordance with these cell line experiments, protein levels of the BRG1 and BAF155/SRG3 in vivo were decreased in fat tissues of STZ-treated mice compared with levels in control or STZ-treated mice injected with insulin (Fig. 8C). Next, we examined the protein level of BAF155/SRG3 under fasting and refeeding conditions because insulin plays a key role in refeeding states. Similar to the above results, the level of BAF155/SRG3 was increased in fat tissues (Fig. 8D) as well as in the liver and muscle of refed mice (data not shown). Taken together, these results suggest that the expression level of the SWI/SNF complex is enhanced by insulin, at least in adipose tissue, to confer regulation of insulin-mediated gene expression.
FIG. 8.
Insulin increases the association between ADD1/SREBP1c and BAF155/SRG3 as well as the protein level of SRG3. (A) Nuclear extracts from the 3T3-L1 adipocytes incubated with or without insulin (100 nM) for 1 h were immunoprecipitated (IP) with preimmune serum or anti-ADD1/SREBP1c antibodies. Association of BRG1, BAF155/SRG3, and mSin3A with ADD1/SREBP1c was detected by Western blot analyses. Input, 10% of lysates. (B) The protein level of each protein was assessed by Western blot analyses with anti-p300, anti-Brm, anti-BRG1, and anti-BAF155/SRG3 antibodies. +, present; −, absent. (C) Western blot analyses of fat tissues from insulin-deficient STZ mice. Epididymal fat tissues of control (Ctl), STZ, and insulin-injected STZ mice (STZ+Ins) (n = 5 for each) were analyzed by Western blot analyses with antibodies against BRG1, BAF155/SRG3, ADD1/SREBP1c, and α-actin. (D) Fasting/refeeding response of BAF155/SRG3 protein. C57BL/6J mice were fasted for 48 h and refed for 4 h. Epididymal fat tissues of the mice were analyzed by Western blot analyses with antibodies against BAF155/SRG3, ADD1/SREBP1c, PPARγ, and α-actinin. Asterisks denote nonspecific protein bands. (C and D) The results are representative data from at least three independent experiments (n = 3 or n = 5).
BAF155/SRG3-overexpressing transgenic mice show increased insulin sensitivity.
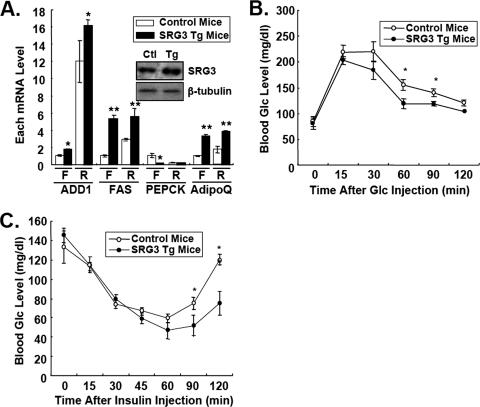
To explore the role of the SWI/SNF chromatin remodeling complex in regulating expression of insulin target genes in vivo, we analyzed transgenic mice overexpressing BAF155/SRG3 driven by the β-actin promoter (SRG3 Tg mice). SRG3 Tg mice showed an almost normal phenotype in metabolic parameters such as body weight, plasma insulin, and plasma lipid contents (data not shown). As shown in Fig. 9A, mRNA levels of ADD1/SREBP1c and FAS were substantially increased, while PEPCK mRNA level was significantly reduced in epididymal tissues. And such effects of SRG3 overexpression in regulation of ADD1/SREBPc target genes were further enhanced by refeeding (Fig. 9A). Next, we tested glucose and insulin tolerance to examine whether altered expression of insulin target genes affected insulin sensitivity in vivo. Remarkably, SRG3 Tg mice exhibited improved glucose tolerance compared to wild-type mice (Fig. 9B). Furthermore, insulin tolerance tests showed that SRG3 Tg mice were significantly more sensitive to the exogenous insulin for its glucose-lowering effect (Fig. 9C). Taken together, these results suggest that BAF155/SRG3 might participate in regulating insulin-dependent gene expression and could affect insulin sensitivity in vivo.
FIG. 9.
BAF155/SRG3 transgenic mice show increased insulin sensitivity. (A) Relative mRNA levels of ADD1/SREBP1c, FAS, adiponectin (AdipoQ), and PEPCK in response to fasting (F) and refeeding (R). C57BL/6J mice were fasted for 16 h and refed for 6 h. Total RNA was isolated from epididymal fat tissues and analyzed by quantitative real-time RT-PCR. The relative mRNA level was determined by normalization of each mRNA level to β-actin. BAF155/SRG3 protein levels in epididymal fat tissues from control (Ctl) and SRG3 Tg mice were analyzed by Western blot analysis. (B and C) Glucose tolerance (B) and insulin tolerance (C) tests in wild-type and BAF155/SRG3 transgenic mice. Values are means ± standard errors of the means (n = 8). *, P < 0.05; **, P < 0.01 for control mice versus SRG3 Tg mice, indicating a significant difference between genotypes.
DISCUSSION
Abnormal expression of ADD1/SREBP1c is associated with metabolic diseases including obesity and diabetes mellitus. In the livers and islets of diabetic animal models such as ob/ob and db/db mice and Zucker diabetic rats, the level of ADD1/SREBP1c mRNA is increased, whereas in fat tissues of those obese animals and patients, its mRNA level is decreased (14, 26, 39, 40, 48). Transgenic mice overexpressing the active form of ADD1/SREBP1c exhibit severe insulin resistance, hyperglycemia, and liver steatosis, whereas the absence of SREBP1 ameliorates liver steatosis in ob/ob leptin-deficient diabetic mice (41, 52). Furthermore, polymorphisms of the ADD1/SREBP1c gene are associated with obesity and type II diabetes (7, 21). Therefore, understanding the molecular mechanism by which ADD1/SREBP1c regulates its target genes would provide a clue for the mechanism-based drug design to treat and improve metabolic disorders.
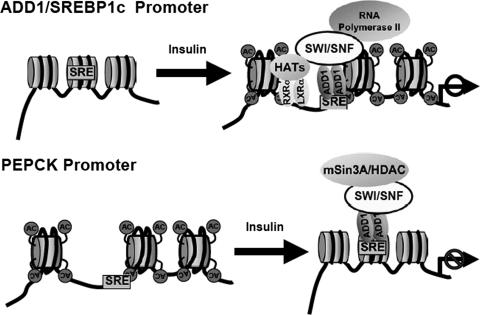
In the present study, we reveal that the process of chromatin remodeling is involved in regulating insulin-dependent gene expression through the interaction between SWI/SNF chromatin remodeling factors and ADD1/SREBP1c. Insulin augmented recruitment of p300 and mSin3A to the ADD1/SREBP1c promoter and PEPCK promoter, respectively, along with increased recruitment of the SWI/SNF chromatin remodeling complexes to both promoters (Fig. 2). This finding implies that regulation of insulin-dependent gene expression requires both ATP-dependent and ATP-independent chromatin modifications. Apparently, ADD1/SREBP1c associated with SWI/SNF chromatin remodeling factors and mSin3A/HDAC complex (Fig. 4) through its basic helix-loop-helix domain (data not shown). However, we failed to observe significantly increased transcriptional activity of ADD1/SREBP1c with p300 (Fig. 5A). The GST pull-down assay also showed that ADD1/SREBP1c barely interacted with p300 (data not shown). ADD1/SREBP1c shares almost all amino acid sequence identity with SREBP1a but for its 4 unique N-terminal amino acids that correspond to the 28 amino acids in the activation domain of SREBP1a (42, 53) (data not shown). Through this region, SREBP1a interacts with p300 and the TRAP/DRIP mediator complex (29, 30). This might explain why ADD1/SREBP1c has relatively low binding affinity to p300. Thus, insulin-dependent recruitment of p300 and histone hyperacetylation at the ADD1/SREBP1c promoter (Fig. 2B) might be rendered by cooperative interactions of ADD1/SREBP1c with other transcription factors, such as Sp1 and NF-Y. Consistent with this model, it has been demonstrated that the SREBP protein increases the DNA-binding ability of Sp1, which interacts with p300 (37). Furthermore, Sp1 and NF-Y physically associate with ADD1/SREBP1c and synergistically activate ADD1/SREBP1c target gene promoters (28, 37). On the other hand, ADD1/SREBP1c binds to the PEPCK promoter competitively with Sp1, resulting in repression of Sp1-dependent activation of PEPCK gene (3). Therefore, upon insulin signaling, ADD1/SREBP1c might regulate the expression of its target genes by changing the accessibility of other transcription factors or coregulators to the template DNA (Fig. 10). Accordingly, a recent study reported that disruption of the binding site for SREBPs drastically lowered basal transcription at the ADD1/SREBP1c promoter in primary hepatocytes, indicating that SREBP1 plays permissive roles in regulating ADD1/SREBP1c expression (4).
FIG. 10.
Schematic model of regulation of insulin-dependent gene expression by ADD1/SREBP1c and the SWI/SNF complex.
This feature of ADD1/SREBP1c seems to be advantageous to function as a molecular switch for positive and negative regulation of target genes in a promoter-specific manner. As described above, ADD1/SREBP1c is able to associate with the mSin3A/HDAC complex, which would repress certain target gene expression (Fig. 4 and 8). These results suggest that ADD1/SREBP1c acts as a molecular switch for insulin action by recruiting different subsets of the SWI/SNF chromatin remodeling complexes, probably in cooperation with other neighboring sequence-specific transcription factors (Fig. 10). Indeed, hBRG1- and hBrm-based hSWI/SNF complexes have been purified either alone or in combination with mSin3A/HDAC, clearly indicating the presence of different pools of SWI/SNF complexes (44).
Because insulin increased protein levels of SWI/SNF chromatin remodeling factors (Fig. 8), it is plausible to speculate that insulin stimulates the transcriptional activity of ADD1/SREBP1c through increasing coregulators of ADD1/SREBP1c. This insulin effect was attenuated by a phosphatidylinositol 3-kinase inhibitor, wortmannin or LY294002, and enhanced by the calpain inhibitor II ALLN, which inhibits proteolytic activity of both calpain I and the 26S proteasome, suggesting that insulin might increase the stability of those proteins through phosphatidylinositol 3-kinase-dependent pathways (Y. S. Lee and J. B. Kim, unpublished data). Of course, we cannot rule out the possibility that posttranslational modifications also influence the insulin-dependent increase in association between ADD1/SREBP1c and chromatin remodeling factors, since insulin-dependent phosphorylation of ADD1/SREBP1c affects the transcriptional activity of ADD1/SREBP1c (8, 16, 18, 19, 35). Similarly, posttranslational modifications of the SWI/SNF chromatin remodeling factors could affect their recruitment to target loci by increasing their affinity to transcription factors such as MyoD (45). Thus, the relationship between insulin-dependent posttranslational modifications of ADD1/SREBP1c and the SWI/SNF chromatin remodeling factors and their avidity to each other remains to be elucidated.
Accumulating data indicate that a substantial component of gene expression regulation is directed at the level of coactivators in diverse biological pathways (46). For example, quantitative changes in PGC-1 allow the functional integration of multiple transcription factors, such as PPARγ, LXRα, and SREBPs. Recently, Lin et al. reported that in response to a high fat diet, PGC-1β physically interacts with ADD1/SREBP1c and enhances its transcriptional activity in the liver (27). Surprisingly, overexpression of PGC-1β could increase the expression of lipogenic genes, such as FAS and stearoyl-CoA desaturase 1, which are the targets of ADD1/SREBP1c. However, overexpression of PGC-1β does not increase the accumulation of triglycerides in the liver by increasing the rate at which lipids are pumped out into plasma by potentiating the transcriptional activity of LXRα.
Overexpression of BAF155/SRG3 in vivo also increased mRNA levels of lipogenic genes, such as ADD1/SREBP1c and FAS, in fat tissue (Fig. 9A) without any substantial increase in fat cell size (Y. S. Lee and J. B. Kim, unpublished data). Strikingly, SRG3 Tg mice showed elevated insulin sensitivity with increased adiponectin expression in adipose tissue, implying that enhanced regulation of expression of insulin target genes could affect insulin sensitization (Fig. 9). However, this insulin-sensitizing effect of BAF155/SRG3 overexpression is not solely due to the promoted transcriptional activity of ADD1/SREBP1c because ADD1/SREBP1c-overexpressing transgenic mice exhibit increased insulin resistance due to lipoatrophy (39). It is possible that SWI/SNF chromatin remodeling complexes might confer insulin sensitivity by serving as an integrator for the metabolic signals, including insulin, which are mediated by several transcription factors, since BRG1 associated with other transcription factors, such as LXRα and PPARγ (data not shown).
Recently, it has been reported that overexpression of BAF155 can increase the protein stability of BAF57 (5). Moreover, we observed that overexpression of BAF155/SRG3 can increase the protein stability of BRG1 and other BAFs, such as SNF5, and total chromatin remodeling activity (P. H. Sohn et al., unpublished data; K. Lee et al., unpublished data). Thus, we cannot rule out the possibility that the effects of overexpression of BAF155/SRG3 might be due to increased levels of other BAFs or stimulated total chromatin remodeling activity. However, because BAF155/SRG3, but not BRG1, could directly interact with ADD1/SREBP1c, it is eligible to propose that BAF155/SRG3 would contribute to the insulin-dependent increase in the recruitment of SWI/SNF factors into insulin target gene promoters. However, detailed mechanisms of insulin-dependent stabilization of SWI/SNF chromatin remodeling factors remain to be elucidated.
Here, we have described for the first time that chromatin remodeling is actively involved in regulating insulin-dependent gene expression and that ADD1/SREBP1c associates with the SWI/SNF chromatin remodeling complex. Another important avenue to explore is whether the SWI/SNF chromatin remodeling complex activates the transcriptional activity of ADD1/SREBP1c cooperatively with PGC-1β in fat cells for energy homeostasis.
Acknowledgments
We thank Joonho Choe for BRG1, Brm, and mSin3A expression vectors and Kaushik Chakravarty for PEPCK-luciferase and PKA expression vectors.
This work was supported by grants from the Molecular and Cellular Biodiscovery Research Program, Science Research Center Program, and the National Research Laboratory Program of Korea Science and Engineering Foundation. Y.S.L., D.H.S., D.H., R.H.S., and J.B.K. were supported by a BK21 Research Fellowship from the Ministry of Education and Human Resources Development.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Amemiya-Kudo, M., H. Shimano, T. Yoshikawa, N. Yahagi, A. H. Hasty, H. Okazaki, Y. Tamura, F. Shionoiri, Y. Iizuka, K. Ohashi, J. Osuga, K. Harada, T. Gotoda, R. Sato, S. Kimura, S. Ishibashi, and N. Yamada. 2000. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 275:31078-31085. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarty, K., P. Leahy, D. Becard, P. Hakimi, M. Foretz, P. Ferre, F. Foufelle, and R. W. Hanson. 2001. Sterol regulatory element-binding protein-1c mimics the negative effect of insulin on phosphoenolpyruvate carboxykinase (GTP) gene transcription. J. Biol. Chem. 276:34816-34823. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarty, K., S. Y. Wu, C. M. Chiang, D. Samols, and R. W. Hanson. 2004. SREBP-1c and Sp1 interact to regulate transcription of the gene for phosphoenolpyruvate carboxykinase (GTP) in the liver. J. Biol. Chem. 279:15385-15395. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G., G. Liang, J. Ou, J. L. Goldstein, and M. S. Brown. 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. USA 101:11245-11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., and T. K. Archer. 2005. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol. Cell. Biol. 25:9016-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 7.Eberle, D., K. Clement, D. Meyre, M. Sahbatou, M. Vaxillaire, A. Le Gall, P. Ferre, A. Basdevant, P. Froguel, and F. Foufelle. 2004. SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes 53:2153-2157. [DOI] [PubMed] [Google Scholar]
- 8.Eberle, D., B. Hegarty, P. Bossard, P. Ferre, and F. Foufelle. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86:839-848. [DOI] [PubMed] [Google Scholar]
- 9.Foufelle, F., and P. Ferre. 2002. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem. J. 366:377-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 11.Hegarty, B. D., A. Bobard, I. Hainault, P. Ferre, P. Bossard, and F. Foufelle. 2005. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc. Natl. Acad. Sci. USA 102:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton, J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 109:1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ide, T., H. Shimano, N. Yahagi, T. Matsuzaka, M. Nakakuki, T. Yamamoto, Y. Nakagawa, A. Takahashi, H. Suzuki, H. Sone, H. Toyoshima, A. Fukamizu, and N. Yamada. 2004. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol. 6:351-357. [DOI] [PubMed] [Google Scholar]
- 14.Kakuma, T., Y. Lee, M. Higa, Z. Wang, W. Pan, I. Shimomura, and R. H. Unger. 2000. Leptin, troglitazone, and the expression of sterol regulatory element binding proteins in liver and pancreatic islets. Proc. Natl. Acad. Sci. USA 97:8536-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemper, J. K., H. Kim, J. Miao, S. Bhalla, and Y. Bae. 2004. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell. Biol. 24:7707-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J. B., P. Sarraf, M. Wright, K. M. Yao, E. Mueller, G. Solanes, B. B. Lowell, and B. M. Spiegelman. 1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Investig. 101:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J. B., and B. M. Spiegelman. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10:1096-1107. [DOI] [PubMed] [Google Scholar]
- 18.Kim, K. H., M. J. Song, E. J. Yoo, S. S. Choe, S. D. Park, and J. B. Kim. 2004. Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c. J. Biol. Chem. 279:51999-52006. [DOI] [PubMed] [Google Scholar]
- 19.Kotzka, J., D. Muller-Wieland, A. Koponen, D. Njamen, L. Kremer, G. Roth, M. Munck, B. Knebel, and W. Krone. 1998. ADD1/SREBP-1c mediates insulin-induced gene expression linked to the MAP kinase pathway. Biochem. Biophys. Res. Commun. 249:375-379. [DOI] [PubMed] [Google Scholar]
- 20.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 21.Laudes, M., I. Barroso, J. Luan, M. A. Soos, G. Yeo, A. Meirhaeghe, L. Logie, A. Vidal-Puig, A. J. Schafer, N. J. Wareham, and S. O'Rahilly. 2004. Genetic variants in human sterol regulatory element binding protein-1c in syndromes of severe insulin resistance and type 2 diabetes. Diabetes 53:842-846. [DOI] [PubMed] [Google Scholar]
- 22.Lee, D., J. W. Kim, T. Seo, S. G. Hwang, E. J. Choi, and J. Choe. 2002. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 277:22330-22337. [DOI] [PubMed] [Google Scholar]
- 23.Lee, Y. S., H. H. Lee, J. Park, E. J. Yoo, C. A. Glackin, Y. I. Choi, S. H. Jeon, R. H. Seong, S. D. Park, and J. B. Kim. 2003. Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. Nucleic Acids Res. 31:7165-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Lay, S., I. Lefrere, C. Trautwein, I. Dugail, and S. Krief. 2002. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1C) regulation of gene expression in 3T3-L1 adipocytes. Identification of CCAAT/enhancer-binding protein beta as an SREBP-1C target. J. Biol. Chem. 277:35625-35634. [DOI] [PubMed] [Google Scholar]
- 25.Liang, G., J. Yang, J. D. Horton, R. E. Hammer, J. L. Goldstein, and M. S. Brown. 2002. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 277:9520-9528. [DOI] [PubMed] [Google Scholar]
- 26.Lin, H. Z., S. Q. Yang, C. Chuckaree, F. Kuhajda, G. Ronnet, and A. M. Diehl. 2000. Metformin reverses fatty liver disease in obese, leptin-deficient mice. Nat. Med. 6:998-1003. [DOI] [PubMed] [Google Scholar]
- 27.Lin, J., R. Yang, P. T. Tarr, P. H. Wu, C. Handschin, S. Li, W. Yang, L. Pei, M. Uldry, P. Tontonoz, C. B. Newgard, and B. M. Spiegelman. 2005. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell 120:261-273. [DOI] [PubMed] [Google Scholar]
- 28.Magana, M. M., S. H. Koo, H. C. Towle, and T. F. Osborne. 2000. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J. Biol. Chem. 275:4726-4733. [DOI] [PubMed] [Google Scholar]
- 29.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 31.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 32.Osborne, T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275:32379-32382. [DOI] [PubMed] [Google Scholar]
- 33.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 34.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 35.Roth, G., J. Kotzka, L. Kremer, S. Lehr, C. Lohaus, H. E. Meyer, W. Krone, and D. Muller-Wieland. 2000. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J. Biol. Chem. 275:33302-33307. [DOI] [PubMed] [Google Scholar]
- 36.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez, H. B., L. Yieh, and T. F. Osborne. 1995. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J. Biol. Chem. 270:1161-1169. [DOI] [PubMed] [Google Scholar]
- 38.Seo, J. B., H. M. Moon, M. J. Noh, Y. S. Lee, H. W. Jeong, E. J. Yoo, W. S. Kim, J. Park, B. S. Youn, J. W. Kim, S. D. Park, and J. B. Kim. 2004. Adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element-binding protein 1c regulates mouse adiponectin expression. J. Biol. Chem. 279:22108-22117. [DOI] [PubMed] [Google Scholar]
- 39.Shimomura, I., Y. Bashmakov, and J. D. Horton. 1999. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 274:30028-30032. [DOI] [PubMed] [Google Scholar]
- 40.Shimomura, I., R. E. Hammer, S. Ikemoto, M. S. Brown, and J. L. Goldstein. 1999. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature 401:73-76. [DOI] [PubMed] [Google Scholar]
- 41.Shimomura, I., R. E. Hammer, J. A. Richardson, S. Ikemoto, Y. Bashmakov, J. L. Goldstein, and M. S. Brown. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12:3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimomura, I., H. Shimano, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Investig. 99:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sif, S. 2004. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell. Biochem. 91:1087-1098. [DOI] [PubMed] [Google Scholar]
- 44.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simone, C., S. V. Forcales, D. A. Hill, A. N. Imbalzano, L. Latella, and P. L. Puri. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36:738-743. [DOI] [PubMed] [Google Scholar]
- 46.Spiegelman, B. M., and R. Heinrich. 2004. Biological control through regulated transcriptional coactivators. Cell 119:157-167. [DOI] [PubMed] [Google Scholar]
- 47.Tontonoz, P., J. B. Kim, R. A. Graves, and B. M. Spiegelman. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13:4753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unger, R. H., Y. T. Zhou, and L. Orci. 1999. Regulation of fatty acid homeostasis in cells: novel role of leptin. Proc. Natl. Acad. Sci. USA 96:2327-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X., M. R. Briggs, X. Hua, C. Yokoyama, J. L. Goldstein, and M. S. Brown. 1993. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J. Biol. Chem. 268:14497-14504. [PubMed] [Google Scholar]
- 50.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 51.Yabe, D., R. Komuro, G. Liang, J. L. Goldstein, and M. S. Brown. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. USA 100:3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yahagi, N., H. Shimano, A. H. Hasty, T. Matsuzaka, T. Ide, T. Yoshikawa, M. Amemiya-Kudo, S. Tomita, H. Okazaki, Y. Tamura, Y. Iizuka, K. Ohashi, J. Osuga, K. Harada, T. Gotoda, R. Nagai, S. Ishibashi, and N. Yamada. 2002. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lep(ob)/Lep(ob) mice. J. Biol. Chem. 277:19353-19357. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama, C., X. Wang, M. R. Briggs, A. Admon, J. Wu, X. Hua, J. L. Goldstein, and M. S. Brown. 1993. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75:187-197. [PubMed] [Google Scholar]
- 54.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]