Abstract
In yeast and other eukaryotes, the histone methyltransferase Set1 mediates methylation of lysine 4 on histone H3 (H3K4me). This modification marks the 5′ end of transcribed genes in a 5′-to-3′ tri- to di- to monomethyl gradient and promotes association of chromatin-remodeling and histone-modifying enzymes. Here we show that Ctk1, the serine 2 C-terminal domain (CTD) kinase for RNA polymerase II (RNAP II), regulates H3K4 methylation. We found that CTK1 deletion nearly abolished H3K4 monomethylation yet caused a significant increase in H3K4 di- and trimethylation. Both in individual genes and genome-wide, loss of CTK1 disrupted the H3K4 methylation patterns normally observed. H3K4me2 and H3K4me3 spread 3′ into the bodies of genes, while H3K4 monomethylation was diminished. These effects were dependent on the catalytic activity of Ctk1 but are independent of Set2-mediated H3K36 methylation. Furthermore, these effects are not due to spurious transcription initiation in the bodies of genes, to changes in RNAP II occupancy, to changes in serine 5 CTD phosphorylation patterns, or to “transcriptional stress.” These data show that Ctk1 acts to restrict the spread of H3K4 methylation through a mechanism that is independent of a general transcription defect. The evidence presented suggests that Ctk1 controls the maintenance of suppressive chromatin in the coding regions of genes by both promoting H3K36 methylation, which leads to histone deacetylation, and preventing the 3′ spread of H3K4 trimethylation, a mark associated with transcriptional initiation.
The control of chromatin structure and DNA accessibility is an essential component of eukaryotic gene regulation. Evidence suggests that the association of regulatory factors with DNA is controlled in large part by nucleosome stability (or “occupancy”) at gene promoters (11). Nucleosome stability is, in turn, regulated by the actions of ATP-dependent chromatin-remodeling machines, by histone posttranslational modifications, and by the deposition of histone variants (11, 13, 17).
Histone posttranslational modifications have been shown to influence both nucleosome stability and the organization and function of chromatin (2, 21, 35, 47). These modifications include acetylation, methylation, and ubiquitylation (35, 47). The modifications to the flexible N- and C-terminal “tail” domains of histones are the most thoroughly studied, but a number of newly identified modifications occur in the highly structured globular domains (7). Recent evidence suggests that these modifications can function through their intrinsic physical effects on the chromatin fiber itself and through their ability to act as a docking site for adaptor proteins that recruit additional chromatin modifiers (15, 44, 47).
Histone methylation can occur on lysine and arginine residues. Lysine residues can be methylated up to three times (generating mono-, di-, and trimethylation), and arginine residues can be methylated once or twice, with dimethylation being either symmetric or asymmetric (26). Histone methylation is a relatively stable mark (50), despite the identification of enzymes with histone demethylase activity (8, 42, 49, 51). Histone methylation functions in a broad range of chromatin-mediated processes, from X chromosome inactivation to facultative and constitutive heterochromatin formation and transcriptional regulation (26, 31).
Recently, several histone lysine methylation marks (H3K4me and H3K36me) associated with “active” chromatin have been shown to function in the context of RNA polymerase II (RNAP II) transcription. In the case of H3K4me, the Set1 enzyme that methylates this residue is recruited to the 5′ end of active genes through an interaction with RNAP II, which is mediated by the concerted actions of Kin28 (the RNAP II serine 5 [Ser5] C-terminal domain [CTD] kinase) and the PAF transcription elongation complex (22, 33). In yeast and humans, H3K4 methylation acts, in part, to recruit chromatin-remodeling ATPases and histone-modifying activities. In yeast, H3K4 methylation recruits both the Isw1 and Chd1 ATPases (37, 39). Set1-dependent recruitment of Isw1 was shown to cause chromatin remodeling of several actively transcribing genes in yeast (39). In humans, H3K4 methylation has been found to recruit Chd1 and the ISWI-related remodeling factor NURF (10, 45, 54). NURF association with H3K4 trimethylation occurs through a PHD finger found in this protein, and evidence indicates that this association is required for the proper chromatin remodeling needed to activate Hox gene expression during development (28, 54). H3K4 trimethylation has also been shown to recruit the ING family of PHD finger-containing proteins, including ING2, which is a gene repressor protein that is associated with histone deacetylase activity (41). In addition to the recruitment of effector proteins, H3K4 methylation in humans also promotes hSet1/MLL1/MLL2 association to chromatin through a feed-forward mechanism whereby H3K4 trimethylation by these complexes allows their WDR5 component, a WD40 repeat-containing protein, to bind this histone modification and foster further H3K4 methylation (53).
Set2, on the other hand, associates with elongating polymerase distal to initiation, after dissociation of Set1. Set2 is recruited to RNAP II via its SRI domain and by the actions of the Ctk1 enzyme complex that catalyzes Ser2 CTD phosphorylation within the bodies of genes (12, 19, 43). Set2-mediated H3K36 methylation recruits the RPD3(s) complex to genes via the Eaf3 chromodomain protein (4, 16, 18, 29). In contrast to H3K4 methylation, however, this recruitment leads to hypoacetylation in the bodies of genes, which functions to prevent spurious transcription initiation (4, 16, 18, 29). These data suggest that Set2 functions to create a more suppressive (and possibly more compact) chromatin environment in the bodies of genes. Consistent with this notion, the loss of Set2 bypasses the need for the Bur1 elongation kinase that promotes transcript elongation (6, 18).
To further understand the biological consequences of H3K4 methylation, we sought to identify pathways that selectively regulate distinct H3K4 methylation states. We found that the Ctk1 kinase, which is essential for H3K36 methylation, also regulates H3K4 methylation. However, we found that, instead of causing abolishment of the mark (as it does for H3K36 methylation) (23, 55), Ctk1 loss dramatically decreased H3K4 monomethylation levels while increasing H3K4 di- and trimethylation levels. Furthermore, loss of Ctk1 caused spreading of H3K4 di- and trimethylation across the coding regions of genes. In agreement with the accompanying paper by Ali Shilatifard and colleagues (52), we found that this effect is not simply a result of loss of H3K36 methylation, nor is it due to defects in RNAP II occupancy or transcription. We suggest that Ctk1-catalyzed phosphorylation of the CTD and/or potentially an unknown substrate is required to localize this chromatin modification specifically to the 5′ ends of genes, thereby creating a boundary for H3K4 methylation that prevents a mark associated with transcriptional initiation from spreading into the bodies of genes.
MATERIALS AND METHODS
Yeast strains and plasmids.
Unless otherwise noted, the wild-type (WT) and deletion strains used in our studies were in the BY4741 background (obtained from Research Genetics). The nine-myc-tagged Set1 strain (used in Fig. 1F) was made in the W303 background and was provided by Vincent Geli (9). The WZY42 histone-shuffling strain, containing WT H3 or various histone H3 point mutations (Fig. 1D; see Fig. S1B in the supplemental material), was obtained from Sharon Dent and has been described previously (19). To disrupt CTK1 in the W303-9MycSet1 or WZY42 background (generating YTX045 and YTX046, respectively), high-efficiency transformation was performed with a PCR product amplified from the genomic DNA of a strain in which the CTK1 gene had been replaced with the KanMX cassette (Research Genetics). WT and mutant Ctk1 expression plasmids were a kind gift from Mark Solomon.
FIG. 1.
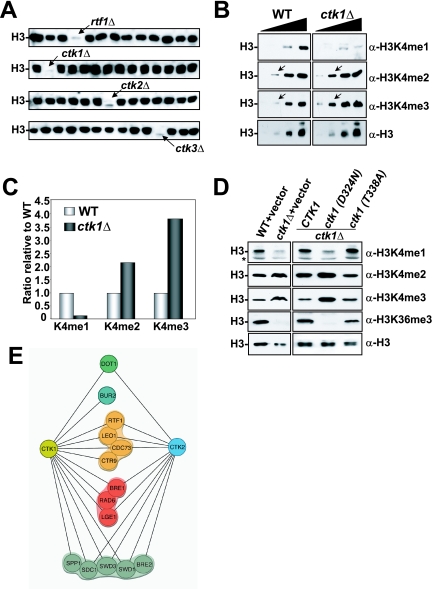
Ctk1 kinase activity regulates H3K4 methylation. (A) Shown is a sample of 56 of the 384 yeast deletion strains screened for H3K4 monomethylation defects. WCEs prepared from asynchronous, logarithmically growing deletion mutants were resolved by 15% SDS-PAGE and transferred onto a PVDF membrane prior to immunoblotting with anti-histone H3 K4 monomethylation-specific antibody (α-H3K4me1). The arrow in the top blot indicates extracts from an rtf1 mutant which encodes a component of the PAF transcription elongation complex previously identified as a regulator of H3K4 mono-, di-, and trimethylation. Arrows in the bottom three blots denote the members of the CTDK-1 complex that were found to regulate H3K4 monomethylation in this study. (B) Characterization of the effect of ctk1Δ on H3K4 methylation states. Increasing amounts (twofold/lane) of either WT (BY4741) or ctk1Δ WCE were resolved by 15% SDS-PAGE and transferred to PVDF prior to immunoblotting with the specified antibodies. Arrows indicate a point in the titration where the H3K4 di- and trimethylation increases are readily observed. (C) Quantification of ctk1Δ effects on the relative proportions of H3 that were H3K4 mono-, di-, and trimethylated. Immunoblots in panel B were quantified with a densitometer, and the results were plotted as the fold changes relative to the WT level. (D) Ctk1 catalytic activity is required to regulate H3K4 methylation. Normalized WCEs from WT (WZY42) or ctk1Δ (YTX045) cells transformed with the empty vector, a CTK1 expression vector, a mutant that produces catalytically dead Ctk1 [ctk1 (D324N)], or a ctk1 mutant that does not abolish its catalytic activity [ctk1 (T338A)] were immunoblotted as described above with the specified antibodies. (E) Synthetic genetic interactions of the CTDK-1 complex. An E-MAP (40) was created by SGA technology (48) and used to cross Natr strains harboring individual deletions of genes encoding two members of the CTDK-1 complex (ctk1Δ and ctk2Δ) with a transcription-targeted array of 384 Kanr deletion strains to create sets of Natr/Kanr haploid double mutants. Growth rates of the double mutants were assessed by automated image analysis of colony size (40), and lines in the diagram connect genes with significant synthetic growth defects. For the degrees of these synthetic interactions, see Table S1 in the supplemental material.
Yeast WCEs and immunoblot analyses.
Yeast whole-cell extracts (WCEs) were prepared as previously described (55). Typically, 10 to 50 μg of proteins (∼2 to 5 μl of WCE) was loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (8% gels for RNAP II and Myc-Set1; 15% gels for histones) and transferred to polyvinylidene difluoride (PVDF) membranes. Western blot analyses were performed by using procedures and secondary detection reagents (ECL Plus) from GE Healthcare. For 6-azauracil studies, cells were grown in synthetic complete medium lacking uracil (SC-Ura) to an optical density at 600 nm (OD600) of 0.8 to 1.0 prior to the addition of 100 μg/ml 6-azauracil for 1 h of incubation at 30°C. For Kin28 and Rpb1 inactivation studies, the WT strain or an appropriate temperature-sensitive strain was grown to an OD600 of 0.8 to 1.0 prior to a shift to a 30°C or 37°C water bath for 2 h before cells were harvested. For detecting H2B monoubiquitylation (a control for Kin28 inactivation), cell pellets corresponding to approximately 106 cells were used for electrophoresis and immunoblotting analysis as described previously (56). Antibodies and their dilutions used in Western blot analyses are as follows. Rabbit antiserum against histone H3 K36 trimethylation (α-H3K36Me3) was purchased from Abcam Inc. and used at a 1:3,500 dilution. Mouse monoclonal anti-FLAG antibody was from Sigma (M2; catalog no. F3165) and was used at 1 μg/ml. Anti-RNAP II CTD antibody H14 (Ser5 phosphorylation specific) was purchased from Covance Inc. and used at a 1:10,000 dilution. Anti-RNAP II CTD antibody for Ser2 phosphorylation was obtained from Upstate Biotechnology Inc. and used at a 1:3,500 dilution. Rabbit anti-glucose-6-phosphate dehydrogenase antibody was purchased from Sigma Inc. (catalog no. A9521) and was used at a 1:80,000 dilution. Mouse monoclonal anti-Myc antibody was purchased from Upstate Biotechnology Inc. (catalog no. 05-724) and used at a 1:3,500 dilution. The rabbit antibodies against histone modifications were purchased from Upstate Biotechnology Inc. and used at the following dilutions: 1:3,500 for anti-H3 monomethyllysine 4 (α-H3K4Me1, 07-436); 1:25,000 for anti-H3 dimethyllysine 4 (α-H3K4Me2, 07-030); 1:10,000 for anti-H3 trimethyllysine 4 (α-H3K4Me3, 07-473); 1:5,000 for anti-H3 acetyllysine 9 (α-H3K9Ac, 07-352) and anti-H3 acetyllysine 14 (α-H3K14Ac, 07-353); 1:30,000 for anti-H27 acetylation (α-H3K27Ac, 07-360); and 1:5,000 for general pan-anti-H2A (07-146), -H2B (07-371), -H3 (05-928), and -H4 (05-858) antibodies.
E-MAP analysis.
E-MAP (epistatic miniarray profile) analysis by synthetic genetic array (SGA) technology (48) was carried out as previously described (40).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (24). Briefly, WCEs were prepared from 1% formaldehyde-fixed WT, ctk1Δ, and set2Δ cells with lysis buffer (50 mM HEPES-KOH, pH 7.5, 300 mM NaCl, 1 mM EDTA, 1% Triton X, 0.1% sodium deoxycholate) and sonicated to shear the chromatin (0.25- to 1.0-kb range). Immunoprecipitation was performed with anti-H3K4me1 (Upstate 07-436), anti-H3K4me2 (Upstate 07-030), anti-H3K4me3 (Upstate 07-473), anti-H3K79me3 (Abcam ab2621), anti-H3K14ac (Upstate 07-353), anti-RNAP II CTD (Covance 8WG16), anti-RNAP II CTD Serine 5P (Covance H14), or anti-H3 (AB1791). All ChIPs utilized protein A-agarose beads, with the exception of the H14 antibody immunoprecipitation, where the protein A beads were preincubated for 30 min at room temperature with a linker immunoglobulin G to the immunoglobulin M antibody (Jackson 315-005-049) before use. After cross-link reversal at 65°C, DNA was extracted according to the manufacturer's instructions with the QIAGEN PCR purification kit. The sequences of the primers used for the locus-specific PCR detection of ChIP enrichment are available upon request.
The value plotted for each fragment in Fig. 2C and F was log2 [a/b], where a = (w/x)/(y/z), where w is the ChIP fragment, x is the ChIP reference, y is the input test fragment, z is the input reference for H3K4me3 ChIP, and all values are the sum of pixel intensities for each band. b is the equivalent value for an H3 ChIP, described above. The log2 [a/b] value was calculated for each experiment separately and averaged. This average was plotted on the y axis. The fold enrichment in Fig. 3C was calculated as the average [(a/b)mutant/(a/b)WT] of three independent replicates. Values in Fig. 3F were calculated as in Fig. 2C and F, except that values were not normalized to H3 levels.
FIG. 2.
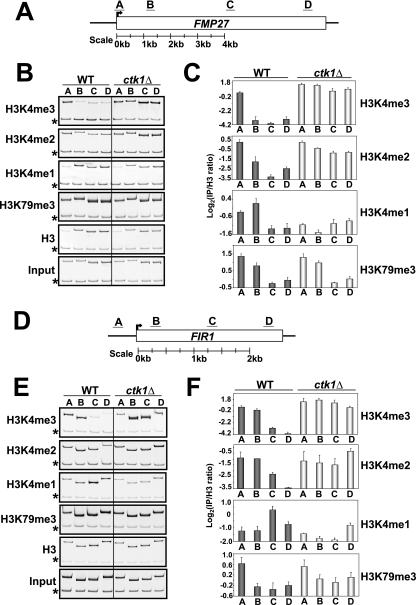
CTK1 deletion results in a spread of H3K4 di- and trimethylation along the coding regions of genes. (A) Schematic representation of the FMP27 locus and the relative locations of the PCR primers used. (B) Results of a ChIP experiment monitoring the distribution of H3K4 mono-, di-, and trimethylation; H3K79 trimethylation; and histone H3 in the WT and ckt1Δ backgrounds. Asterisks denote the location of an internal control band specific to a region of chromosome V lacking an ORF. (C) The degree of enrichment achieved in the ChIP indicated is plotted on the y axis. The primer pair used to assay enrichment is indicated on the x axis. Values are normalized to a histone H3 ChIP to account for differences in underlying nucleosome occupancy (see Materials and Methods). Error bars represent the standard error of the mean of three independent replicates. Note that for this and the subsequent panels, ratio values are log2 transformed to ensure an accurate representation of ratios less than 1. Therefore, every change of 1 U represents a 2-fold change in the abundance of the indicated histone methylation event (a change of 4 U would indicate a 16-fold change and so on). Also, because of differences among the antibodies, the absolute values reported on the y axis apply only to ChIPs performed with the same antibody. (D to F) Same as in A to C except that the FIR1 gene was analyzed.
FIG. 3.
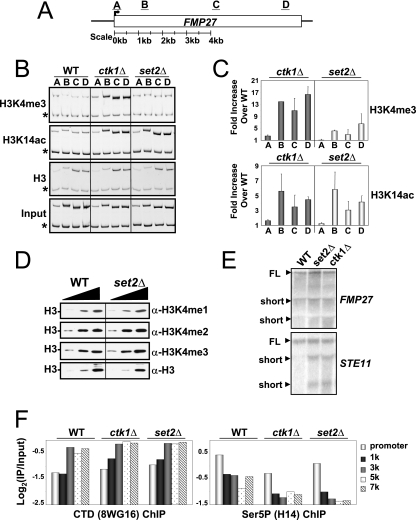
H3K4 methylation spreading in CTK1 deletion strains cannot be explained by defects in Set2/H3K36 methylation, increased cryptic initiation, or defects in RNAP II occupancy. (A) Schematic representation of the FMP27 locus and the relative locations of PCR primers used. (B) Results of a ChIP experiment monitoring the distribution of H3K4me3, H3K14ac, and histone H3 in the WT, ckt1Δ, and set2Δ backgrounds. Asterisks denote the location of an internal control band specific to a region of chromosome V lacking an ORF. (C) The fold enrichment achieved in the ChIP indicated in the mutants over the WT is plotted on the y axis. The primer pair used to assay enrichment is indicated on the x axis. Values are normalized to a histone H3 ChIP to account for differences in underlying nucleosome occupancy (see Materials and Methods). Error bars represent the standard error of the mean of three independent replicates. Results similar to those shown in panels B and C were also obtained with several other genes examined, including PMA1, ADH1, FIR1, and STE11 (data not shown). (D) set2 deletion does not affect H3K4 methylation levels globally. Increasing amounts of either WT or set2Δ WCE were resolved by 15% SDS-PAGE and transferred to PVDF prior to immunoblotting with the specified antibodies. (E) H3K4 methylation spreading observed in ctk1Δ cells cannot be explained by an increase in cryptic initiation or altered levels of transcription. Total RNAs purified from WT, set2Δ, or ctk1Δ cells were used for Northern blot analysis and probed with radiolabeled PCR products corresponding to the 3′ ends of the specified genes. FL denotes the full-length product, and short denotes smaller RNA species that were observed. WT cells naturally exhibit some additional smaller mRNA products for the FMP27 gene. Lanes were loaded with equivalent amounts of total RNA (assayed by ethidium staining [not shown]). STE11 was included as a positive control for cryptic initiation upon deletion of SET2 (4). (F) RNAP II occupancy levels and distribution are not affected by deletion of CTK1 and SET2. A ChIP experiment monitoring the distribution of RNAP II and Ser5 CTD phosphorylated RNAP II in ckt1Δ and set2Δ along the FMP27 gene (see Materials and Methods) was performed. Shown are the fold enrichment values measured at the FMP27 promoter and at increasing distances from the ATG codon (see the code at the far right). The primer pairs used in this panel are distinct from those shown in panel A. RNAP II levels were monitored with the 8WG16 (anti-CTD) antibody; nearly identical results were obtained with a different RNAP II antibody to the N terminus of Rpb1 (sc25758, Santa Cruz; not shown).
An extensive effort was made to analyze Set1 itself by ChIP, but we were unable to obtain reproducible results. We examined Set1 by using all available antibodies to this protein (Santa Cruz rabbit polyclonal antibody and a mouse monoclonal antibody kindly provided by Peter Nagy) or by using Myc antibody with a N-terminally nine-myc-tagged version of this protein. These results prevented us from examining the localization of Set1 on genes in the absence of Ctk1; however, the presence of H3K4 trimethylation along the open reading frames (ORFs) indicates that the enzyme was present to catalyze this event.
ChIP-chip analyses: DNA amplification, labeling, array hybridization and data processing.
ChIP-enriched DNA and reference DNA in all experiments were amplified as previously described (3). Briefly, two initial rounds of DNA synthesis with T7 DNA polymerase with primer 1 (5′-GTTTCCCAGTCACGATCNNNNNNNNN-3′) was followed by 25 cycles of PCR with primer 2 (5′-GTTTCCCAGTCACGATC-3′). Cy3-dUTP or Cy5-dUTP was then incorporated directly by an additional 25 cycles of PCR with primer 2. Microarray hybridizations were performed by standard procedures (14). The arrays were scanned with a GenePix 4000 scanner, and data were extracted with GenePix 5.0 software. Data were normalized such that the median log2 ratio value for all quality elements on each array equaled zero, and the median of the pixel ratios was retrieved for each spot. Only spots of high quality by visual inspection, with at least 50 pixels of quality data (regression R2, >0.6) were used for analysis. Arrayed elements that did not meet these criteria on at least half of the arrays were excluded from analysis. All data were log transformed before further analysis. For ChIP-chip data analysis, the log2 ratio of each spot was transformed to a z score by using the formula zx = (x − μ)/σ, where x is a retrieved spot value, μ is the mean of all retrieved spots from one array, and σ is the standard deviation of all retrieved spots from that same array. z scores from biological replicates were averaged. The same transformation was done for the histone H3 arrays. For normalization with the nucleosome occupancy data, for each probe the average z score from the histone ChIP-chips was subtracted from the average z score from H3K4me3 ChIP-chips. Raw data are available at https://genome.unc.edu/ and through GEO (accession number GSE5572).
Northern blot analyses.
Yeast strains were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% dextrose) to an OD600 of 0.6 to 0.8. Total RNA was prepared by the acid-phenol method as previously described (55). For Northern blotting, 30 μg of each sample was subjected to electrophoresis in a formaldehyde-agarose gel, followed by transfer to a nylon membrane and cross-linked by UV irradiation. Hybridization was carried out in 6× SCP (2.5 M NaCl, 0.7 M Na2HPO4, 0.02 M EDTA)-1% Sarkosyl-0.1 mg/ml salmon sperm DNA. Probes (full-length STE11 and +5051 to +7346 of FMP27) were generated by PCR.
RESULTS
The RNAP II CTD kinase Ctk1 regulates H3K4 methylation.
Given the suggestion that the distinct forms of H3K4 methylation may result in different biological outcomes (38), we initiated a screen to identify regulators of the different H3K4 methylation states. We carried out Western analyses of H3K4 monomethylation with WCEs derived from 384 individual strains of yeast containing single deletions of genes known or predicted to function in some aspect of chromatin function or transcription regulation. The specificity of the H3K4 monomethyl-specific antibody was confirmed (see Fig. S1 in the supplemental material). This screen identified all of the previously established regulators of global H3K4 methylation, including members of the Rad6 complex, the Set1 complex (also known as COMPASS), and the PAF complex (Fig. 1A and data not shown).
In addition to the known regulators, we found that CTK1, CTK2, and CTK3 regulate H3K4 monomethylation (Fig. 1A). All of these genes encode members of the CTDK-1 kinase complex responsible for Ser2 CTD phosphorylation (5, 27, 46) and had not been previously shown to regulate H3K4 methylation. While H3K4 monomethylation initially appeared to be abolished, longer exposures revealed very small amounts of this modification (data not shown). To examine whether the other H3K4 methyl states would be affected by CTK1 deletion (ctk1Δ), we compared WCEs derived from WT or ctk1Δ cells in Western analyses for di- and trimethylation. While H3K4 monomethylation was significantly reduced, di- and trimethylation levels were markedly increased (Fig. 1B and C). It is significant to mention that our initial analyses revealed a slight reduction in the overall histone levels in the ctk1Δ strain, based on the conventional practice of equalizing loading of WCE by total protein (data not shown). Therefore, for the histone modification analyses presented here we normalized loading of WCE to the levels of H3 protein.
The kinase activity of Ctk1 is required for regulation of H3K4 methylation.
The Ctk2 and Ctk3 components of CTDK-1 are necessary for Ctk1 kinase activity (46). Thus, our findings that deletion of each of the members of the CTDK-1 kinase complex resulted in similar effects on H3K4 methylation suggested that the catalytic activity of Ctk1 was required. To examine this further, we analyzed a series of Ctk1 mutants (34). We transformed ctk1Δ cells with an empty vector, a plasmid that expresses WT CTK1, a plasmid that expresses a catalytically dead allele of CTK1 [ctk1 (D324N)], or a plasmid that expresses an allele of CTK1 that encodes an amino acid substitution that does not affect catalytic function [ctk1 (T338A)]. These experiments show that the effects on H3K4 methylation were dependent upon the kinase activity of Ctk1 (Fig. 1D).
CTK1 and CTK2 interact genetically with members of the Set1/COMPASS complex.
To further explore the possibility that Ctk1 regulates H3K4 methylation, we used automated SGA analysis in a high-density E-MAP format (40) that was designed to genetically characterize factors implicated in transcription regulation. Natr strains containing the CTK1 and CTK2 gene deletions were generated and crossed to a set of viable gene deletions selected for their involvement in chromatin and transcription, and the resulting double-mutant strains were examined. As shown in Fig. 1E, the deletions of CTK1 and CTK2 were synthetically slow growing or lethal with members of the Set1, PAF, and Rad6 complexes, as well as with the H3K79 methylase Dot1 and the BUR kinase cyclin subunit Bur2. These results are consistent with Ctk1 regulation of H3K4 methylation, in addition to functions independent of Set1. Indeed, it has been shown that Ctk1 is required for recruitment of the Set2 histone methyltransferase (23, 55).
Deletion of Ctk1 causes loss of H3K4 monomethylation and a 3′ spread of H3K4 di- and trimethylation into the coding regions of genes.
To examine the fate of H3K4 methylation on genes in ctk1Δ cells, we performed ChIP analyses with the different H3K4 methyl-specific antibodies. We found that on actively transcribing genes such as FMP27 and FIR1, the peak of H3K4 monomethylation normally found near the middle to the 3′ region of genes (30, 36) was significantly diminished (Fig. 2A to F). In contrast, H3K4 di- and trimethylation levels had spread 3′ across the ORF on these genes (Fig. 2A to F). We have confirmed these results on six additional genes, PMA1, SCC2, TOM1, ADH1, TAO1, and MDM1 (data not shown). These data suggest that the increased H3K4 di- and trimethylation seen along genes in ctk1Δ comes at the expense of H3K4 monomethylation. Importantly, the level and distribution of H3K79 methylation, a histone modification not affected by Ctk1 (55), was unchanged in ctk1Δ cells. In addition, we did not detect significant changes in histone occupancy across the FMP27 and FIR1 genes in ctk1Δ cells (Fig. 2B and E).
The H3K4 methylation spreading observed in ctk1Δ cells is not caused by defects in H3K36 methylation.
We explored several possible explanations for the spreading of H3K4 methylation on genes in the absence of Ctk1 kinase activity. First, we addressed the possibility that these effects were due simply to a loss of Set2-mediated H3K36 methylation, which is also regulated by Ctk1 (23, 55). Previous studies have shown that a loss of Set2 causes inappropriate transcriptional initiation in the coding regions of genes (termed “cryptic” initiation) and an increase in H3K4 trimethylation in the coding regions of genes (presumably due to Set1 association with RNAP II at sites of aberrant initiation) (4). However, the loss of Set2 resulted in only a minimal spread of H3K4 trimethylation levels into the 3′ coding region of the FMP27 gene (as well as other genes we examined both individually and genome-wide by ChIP-chip; Fig. 3A to C and 5C and D and data not shown). In addition, by Western blot analysis we found that deletion of SET2 did not affect the global levels of H3K4 mono-, di-, and trimethylation (Fig. 3D), in contrast to the changes observed in ctk1Δ cells (Fig. 1B). Furthermore, both set2Δ and ctk1Δ cells, which lack H3K36 methylation, exhibited similar increases of H3 acetylation in the bodies of genes, presumably because of a failure to recruit Rpd3(s) through methyl-H3K36 (Fig. 3A to C) (4, 16, 18). These data imply that Ctk1 regulates H3K4 methylation through a mechanism that is distinct from its regulation of Set2 and H3K36 methylation.
The H3K4 methylation spreading observed in ctk1Δ cells is not caused by inappropriate RNAP II initiation in the bodies of genes.
Next, we tested the hypothesis that ctk1Δ causes high levels of Set1 recruitment to the bodies of genes as a consequence of inappropriate RNAP II initiation in this region. Since cryptic initiation but not spreading of H3K4 methylation is observed in set2Δ strains, we reasoned that if the hypothesis was correct, a higher level of cryptic initiation would be required in ctk1Δ cells to support the downstream spreading of H3K4 methylation observed in that strain. However, the level and pattern of cryptic initiation observed on the STE11 gene were no different between set2Δ and ctk1Δ cells (Fig. 3E). Yet H3K4 trimethylation levels were still significantly increased on the STE11 gene, similar to the results shown for FMP27 (Fig. 3A to C and data not shown). We detected no significant increase in cryptic initiation on the FMP27 gene in either set2Δ or ctk1Δ cells, despite significantly increased H3K4 trimethylation levels (Fig. 3E). These data rule out large increases in RNAP II cryptic transcription initiation as a cause for the spread of H3K4 trimethylation in ctk1Δ strains. Perhaps most significant is the fact that RNAP II levels and Ser5 CTD phosphorylation patterns along the FMP27 gene and all other genes we have examined, including PMA1, FIR1, and ADH1, are generally unaffected by the deletion of CTK1 (Fig. 3F and data not shown). This is in agreement with other reports that RNAP II density, elongation rate, or processivity does not change in ctk1Δ or ctk2Δ cells (1, 25, 32).
The H3K4 methylation spreading observed in ctk1Δ cells is not caused by changes in Set1 protein levels, changes in H2B ubiquitylation, or “transcriptional stress.”
We also tested the hypothesis that loss of Ctk1 kinase activity increases the level of Set1 protein or of its associated proteins. We found, however, that Set1 protein levels and the steady-state levels of mRNA encoding its associated proteins were unaffected in a ctk1Δ strain (Fig. 4A and data not shown). Furthermore, our previous studies have confirmed that the levels of H2B ubiquitylation are unaffected in a ctk1Δ strain (56). These data imply that Ctk1's effects on H3K4 methylation are independent of increased Set1 levels or dramatic changes in H2B ubiquitylation.
FIG. 4.
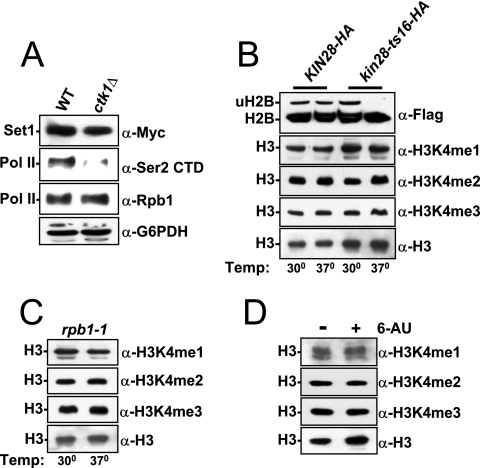
Transcriptional stress does not affect global levels of H3K4 methylation. (A) Set1 levels are not altered in ctk1Δ strains. WT (W303mycSet1) or ctk1Δ (YTX046) WCEs from strains harboring an N-terminal nine-myc tag on Set1 were loaded onto an 8% SDS-PAGE gel and immunoblotted with the specified antibodies. Glucose-6-phosphate dehydrogenase (G6PDH) and RNAP II were probed to ensure equivalent loading, and the anti-Ser2 CTD phosphospecific antibody (Upstate) was used to verify the expected loss of this modification in ctk1Δ cells. (B) Strains containing a Flag-tagged version of H2B that also harbor either a WT or a temperature-sensitive allele of KIN28 (kin28-ts16-HA) were shifted for 2 h at 37°C prior to harvesting cell pellets. A portion of these pellets was directly used for an H2B ubiquitylation assay (a positive control for loss of Kin28 kinase activity), and the remainder of each pellet was used to prepare WCEs to examine the H3K4 methylation status with the specified antibodies. Although H2B ubiquitylation is abolished in the KIN28 mutant, H3K4 methylation is more stable and would be expected to persist during the 2-h heat inactivation (33). (C) An rpb1-1 temperature-sensitive allele was shifted to 37°C for 2 h prior to cell harvesting, cell extraction, and immunoblot analyses with the specified antibodies. (D) WT (BY4741) cells containing the pRS316 vector were grown at 30°C in SC-Ura to an OD600 of 1.0 prior to the addition of 100 μg/ml 6-azauracil (6-AU). These cells were pelleted, and WCE was prepared and used for immunoblot analysis with the specified antibodies. Temp, temperature.
It has recently been reported that loss of CTK1 can cause a specific shift in the 5′ levels of H3K4 trimethylation (and histones) to the 3′ end on a few genes when cells are subjected to transcriptional stress (57). Therefore, we explored the possibility that the spread of H3K4 di- and trimethylation in ctk1Δ strains was due to a transcriptional-stress pathway. We examined RNAP II and elongation factor mutants or chemicals that were previously shown to induce transcriptional stress by affecting elongation. These treatments included inactivation of Rpb1, inactivation of Kin28, and addition of 6-azauracil (57). By Western blot analysis, we found that these treatments had no effect on global H3K4 methylation levels (Fig. 4B to D). These results contrast with those observed for the loss of Ctk1, which nearly abolishes H3K4 monomethylation and considerably increases H3K4 di- and trimethylation (Fig. 1A to D). Furthermore, the accompanying report also shows no defects in the distribution of H3K4 methyl marks under similar conditions of 6-azauracil-induced transcriptional stress (52). Collectively, our results indicate that Ctk1 acts to maintain H3K4 trimethylation at the 5′ end of a gene through a mechanism independent of unregulated RNAP II initiation within coding regions, observable elongation defects, or transcriptional stress.
H3K4 methylation spreading in ctk1Δ cells occurs genome-wide and is restricted to the coding regions of RNAP II transcribed genes.
We used a ChIP-chip approach to examine the genome-wide profiles of H3K4 trimethylation in the WT strain and a CTK1 deletion strain. We used arrays that tiled continuously over the entire genome at a resolution of ∼1 kb and that additionally contained probes that covered all of chromosome III at a resolution of 200 bp (and at 100-bp resolution for one-third of the chromosome). We probed these arrays with DNA recovered from each of the three independent ChIPs represented in Fig. 2. We observed a significant change in the global patterns of H3K4 trimethylation in ctk1Δ cells compared to WT cells (Fig. 5).
FIG. 5.
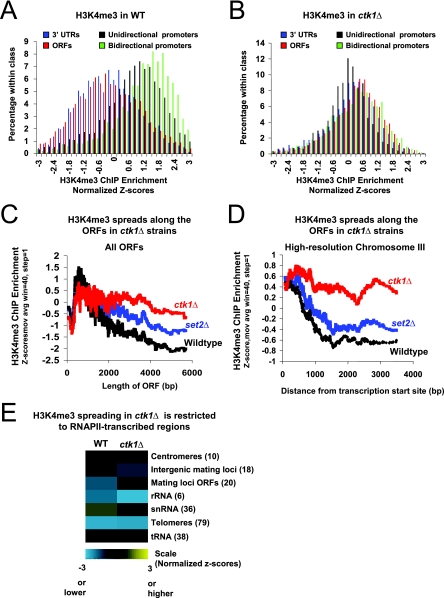
H3K4me3 spreading in ctk1Δ cells occurs genome-wide on the ORFs of RNAP II-regulated genes. (A) Distribution of z scores following H3K4me3 ChIPs from WT WCE. Shown are ORFs (red), 3′ UTRs (blue), unidirectional promoters (black), and bidirectional promoters (green). z scores were normalized to histone H3 distribution as described in Materials and Methods. Promoters are clearly distinguished from ORFs and 3′ UTRs. (B) Same as in panel A, but H3K4me3 ChIPs were performed with WCE from ctk1Δ cells. (C) A moving average (window size = 40, step size = 1) of normalized z scores of H3K4me3 ChIP-chip data in WT (black), set2Δ (blue), and ctk1Δ (red) cells plotted as a function of ORF length. (D) A moving average (window size = 40, step size = 1) of H3K4me3 ChIP-chip z scores in WT (black), set2Δ (blue), and ctk1Δ (red) cells plotted as a function of the distance from the transcription start site among genes with coding regions greater than 1 kb in length and for which data were available on chromosome III (69 genes). A high-resolution microarray covering chromosome III was used for this analysis (see Materials and Methods), and the data plotted in this case were not normalized to histone occupancy. (E) Colors (scale at bottom) represent the median z scores of all data points recorded from all arrayed elements in the indicated Saccharomyces Genome Database functional class (labeled on the right, number of arrayed elements analyzed in parentheses). Data were normalized to histone H3 distribution and derived from three independent ChIP experiments (see Materials and Methods).
In WT cells, we found that trimethylation of H3K4 was generally localized to the 5′ end of loci transcribed by RNAP II (Fig. 5A), consistent with work by others (30, 33, 36). However, in ctk1Δ cells, ORFs are enriched to a degree similar to that of promoters, consistent with a general spread of the mark into coding regions (Fig. 5B). Note that the magnitude of relative enrichment for any class of genomic element is reduced in the ctk1Δ ChIPs, which is also consistent with a more uniform distribution of H3K4me3 caused by large-scale spreading.
We also examined directly if the H3K4me3 signal spread into ORFs. The probes on our microarrays cover each ORF from the start codon to the stop codon. Since H3K4me3 in WT cells extends into the ORF by a fairly stereotypic distance regardless of ORF length, lower ratios will be reported for longer ORFs. This is because for increasingly long ORFs, an increasingly smaller proportion of the ORF will harbor the H3K4me3 modification. This is precisely what is observed in the WT strain (Fig. 5C, black) and to a lesser extent in set2Δ cells, where some minor spreading occurs (Fig. 5C, blue). However, in ctk1Δ cells (Fig. 5C, red) this inverse correlation is not seen except for the longest ORFs, consistent with the spread of H3K4me3 well 3′ of its normal boundary.
We further examined the spreading by using the high-resolution microarray coverage of chromosome III. In WT cells (and to a similar degree in set2Δ cells), the H3K4me3 signal drops sharply as a function of distance from transcriptional initiation, whereas in ctk1Δ mutants H3K4me3 levels remain uniform with increasing distance (30, 36) from the site of initiation (Fig. 5D). These results confirm our findings on individual genes and show that the phenomenon occurs genome-wide.
H3K4 methylation levels in regions not transcribed by RNAP II are not affected by ctk1 deletion.
To further test the relationship between RNAP II transcription and the spread of H3K4me3, we examined if H3K4me3 spreads in regions of the genome that are not transcribed by RNAP II. No spreading of H3K4 trimethylation was observed on Pol I- and Pol III-transcribed genes or on other regions not transcribed by RNAP II, including the Telomeres, HMR, and rRNA gene loci (Fig. 5E). High-resolution analysis of a telomeric region on the right end of chromosome VI confirmed the microarray-based findings and showed that coding regions of RNAP II-regulated genes directly adjacent to these nontranscribed regions exhibited the expected spreading of H3K4 trimethylation in the ctk1 deletion (data not shown). These data help to establish that in the absence of Ctk1, the major effects on the distribution of H3K4 methylation are restricted to RNAP II-transcribed genes.
DISCUSSION
In this report, we describe the results of a screen for regulators of H3K4 monomethylation. Of 384 gene deletions whose products are associated with chromatin and transcription, we identified only one complex, CTDK-1, as a newly described regulator of this modification. In the absence of this kinase, we also observed a sharp increase in global H3K4 di- and trimethylation levels and a corresponding 5′ spread of these methyl marks into the bodies of transcribed genes.
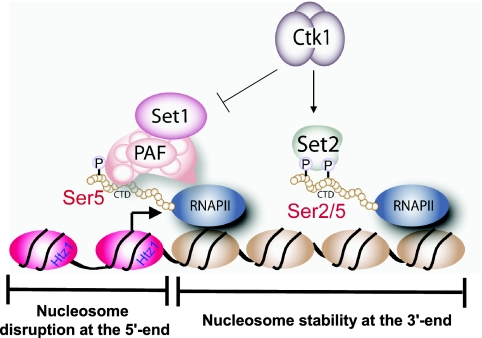
Our results show that the spread of H3K4 methylation in the ctk1Δ strain is independent of Set2/K36 methylation, increased levels of cryptic initiation, RNAP II elongation defects, or transcriptional stress (Fig. 3 and 4). Rather, our results suggest that Ctk1 plays a direct role in restricting H3K4 trimethylation to the 5′ end of a gene. Given that the kinase activity of Ctk1 is required, the data suggest that this effect is mediated through Ser2 CTD phosphorylation on RNAP II. Consistent with this notion, ChIP-chip data show that the spread of H3K4 trimethylation in the ctk1 mutant is coincident with the established pattern of Ser2 CTD phosphorylation on genes (1, 20). We interpret these data to mean that Ser2 CTD phosphorylation acts to prevent the association of Set1 with RNAP II along the transcribed region of genes (Fig. 6). Since it is established that Set1 recruitment depends on Ser5 phosphorylation, it is reasonable to imagine that Set1's initial recruitment is not disrupted in a CTK1 deletion; however, the Ser2 CTD phosphorylation, which in WT cells would normally prevent further association between Set1 and the CTD, does not occur. We suggest that Ctk1 establishes a Ser2/Ser5 CTD phosphorylation pattern that is required to recruit Set2 while establishing, at the same time, a CTD surface that is no longer ideal for Set1 association (Fig. 6). In doing so, Ser2 CTD phosphorylation acts as a brake for the spread of Set1/H3K4 methylation.
FIG. 6.
Role of Ctk1 in the control of chromatin structure. Shown is a model that describes a novel function for Ctk1 in the regulation of histone modifications in the coding regions of RNAP II-regulated genes. In addition to Ctk1 directing the association of Set2 to RNAP II, which results in the establishment of H3K36 methylation and deacetylated chromatin in the bodies of genes, our data reveal that this kinase also acts to prevent the spread of H3K4 trimethylation from its defined position at the 5′ end. We speculate that while Set1 is normally recruited to the 5′ end in a Ser5 CTD phosphorylation- and PAF-dependent manner, the phosphorylation of Ser2 on the CTD by Ctk1 acts to block persistent Set1 association along genes. While direct phosphorylation of Ser2 on the CTD is likely responsible, we do not rule out other possible mechanisms such as direct phosphorylation of COMPASS by Ctk1 or RNAP II kinetic changes upon initial transcription.
It is still unknown which, if any, Set1 complex members interact with the Ser5 phosphorylated epitope of the CTD directly. It may be that Set1's association with the PAF complex is stabilized by Ser5 CTD phosphorylation and then destabilized by a changing CTD phosphorylation pattern. It is also possible that Ctk1's phosphorylation of the CTD results in the recruitment of a specific CTD binding protein that competes for Set1 association with RNAP II. While these effects are most likely mediated by Ser2 phosphorylation of the CTD, other possibilities exist, such as phosphorylation of Set1 or COMPASS members directly by Ctk1 or alterations in the kinetic properties of RNAP II elongation at the onset of transcriptional induction. Further studies are required to determine the precise mechanism by which Ctk1/Ser2 CTD phosphorylation prevents Set1 association with RNAP II.
In summary, our data uncover a novel pathway of regulation of H3K4 methylation. We find that Ctk1 plays a genome-wide function in creating a boundary of H3K4 methylation along the coding regions of genes. The consequence of this spreading is likely to be significant, since H3K4 di- and trimethylation are marks associated with transcriptional initiation and have been found to recruit chromatin-modifying activities. Mislocalization of these activities to downstream regions may cause inappropriate chromatin changes in RNAP II-transcribed regions. Together with the fact that Set2/H3K36 methylation mediates deacetylation in the bodies of genes, our data argue that Ctk1 is directly required for maintenance of a suppressive chromatin structure distal of transcriptional initiation in the ORFs of genes. By recruiting H3K36 methylation and preventing H3K4 methylation, Ctk1 is critical for the creation of appropriate chromatin boundaries along RNAP II-transcribed loci.
Supplementary Material
Acknowledgments
We thank Mark Solomon for providing CTK1 expression plasmids, Vincent Geli for providing the 9xmyc Set1 strain, Bill Marzluff for the use of equipment, and Ali Shilatifard for communicating unpublished results. We are extremely grateful for the advice, suggestions, and encouragement given by Scott Briggs and Zu-Wen Sun. We also thank Mary Bryk, Steve Buratowski, Ceylan Cakit, Adam Cockrell, Sevinc Ercan, Sean Hanlon, Michael Keogh, Kelby Kizer, Michael Shales, and Jeff Smith for technical help and suggestions.
This work was supported by NIH grants to B.D.S. and J.D.L. and an American Heart Association grant to B.D.S. B.D.S. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 3.Bohlander, S. K., R. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 5.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, Y., A. Sutton, R. Sternglanz, and G. Prelich. 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26:3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. [DOI] [PubMed] [Google Scholar]
- 8.Cuthbert, G. L., S. Daujat, A. W. Snowden, H. Erdjument-Bromage, T. Hagiwara, M. Yamada, R. Schneider, P. D. Gregory, P. Tempst, A. J. Bannister, and T. Kouzarides. 2004. Histone deimination antagonizes arginine methylation. Cell 118:545-553. [DOI] [PubMed] [Google Scholar]
- 9.Dehé, P. M., M. Pamblanco, P. Luciano, R. Lebrun, D. Moinier, R. Sendra, A. Verreault, V. Tordera, and V. Geli. 2005. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 353:477-484. [DOI] [PubMed] [Google Scholar]
- 10.Flanagan, J. F., L. Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines, Y. Kim, W. Minor, F. Rastinejad, and S. Khorasanizadeh. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181-1185. [DOI] [PubMed] [Google Scholar]
- 11.Giresi, P. G., M. Gupta, and J. D. Lieb. 2006. Regulation of nucleosome stability as a mediator of chromatin function. Curr. Opin. Genet. Dev. 16:171-176. [DOI] [PubMed] [Google Scholar]
- 12.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff, S., T. Furuyama, and K. Ahmad. 2004. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 20:320-326. [DOI] [PubMed] [Google Scholar]
- 14.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 17.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19:295-310. [DOI] [PubMed] [Google Scholar]
- 18.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 19.Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf, and B. D. Strahl. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25:3305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 22.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 23.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 25.Laribee, R. N., N. J. Krogan, T. Xiao, Y. Shibata, T. R. Hughes, J. F. Greenblatt, and B. D. Strahl. 2005. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 15:1487-1493. [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. Y., C. Teyssier, B. D. Strahl, and M. R. Stallcup. 2005. Role of protein methylation in regulation of transcription. Endocrine Rev. 26:147-170. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka, C. D. Allis, and D. J. Patel. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442:91-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieb, J. D., and N. D. Clarke. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 123:1187-1190. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat Rev. Mol. Cell. Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 32.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 34.Ostapenko, D., and M. J. Solomon. 2003. Budding yeast CTDK-I is required for DNA damage-induced transcription. Eukaryot. Cell 2:274-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 36.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 37.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 39.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 40.Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, J. S. Weissman, and N. J. Krogan. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507-519. [DOI] [PubMed] [Google Scholar]
- 41.Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita, T. Hung, D. Carney, P. Pena, F. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Shi, J. Cote, K. F. Chua, and O. Gozani. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 43.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 44.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844-847. [DOI] [PubMed] [Google Scholar]
- 45.Sims, R. J., III, C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel, and D. Reinberg. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280:41789-41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sterner, D. E., J. M. Lee, S. E. Hardin, and A. L. Greenleaf. 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 48.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 49.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 50.van Holde, K. E. 1989. Chromatin. Springer, New York, N.Y.
- 51.Wang, Y., J. Wysocka, J. Sayegh, Y. H. Lee, J. R. Perlin, L. Leonelli, L. S. Sonbuchner, C. H. McDonald, R. G. Cook, Y. Dou, R. G. Roeder, S. Clarke, M. R. Stallcup, C. D. Allis, and S. A. Coonrod. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279-283. [DOI] [PubMed] [Google Scholar]
- 52.Wood, A., A. Shukla, J. Schneider, J. S. Lee, J. D. Stanton, T. Dzuiba, S. K. Swanson, L. Florens, M. P. Washburn, J. Wyricky, S. R. Bhaumik, and A. Shilatifard. 2007. Ctk complex-mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 27:709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121:859-872. [DOI] [PubMed] [Google Scholar]
- 54.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 55.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.