Abstract
Mouse Grb10 is a tissue-specific imprinted gene with promoter-specific expression. In most tissues, Grb10 is expressed exclusively from the major-type promoter of the maternal allele, whereas in the brain, it is expressed predominantly from the brain type promoter of the paternal allele. Such reciprocally imprinted expression in the brain and other tissues is thought to be regulated by DNA methylation and the Polycomb group (PcG) protein Eed. To investigate how DNA methylation and chromatin remodeling by PcG proteins coordinate tissue-specific imprinting of Grb10, we analyzed epigenetic modifications associated with Grb10 expression in cultured brain cells. Reverse transcriptase PCR analysis revealed that the imprinted paternal expression of Grb10 in the brain implied neuron-specific and developmental stage-specific expression from the paternal brain type promoter, whereas in glial cells and fibroblasts, Grb10 was reciprocally expressed from the maternal major-type promoter. The cell-specific imprinted expression was not directly related to allele-specific DNA methylation in the promoters because the major-type promoter remained biallelically hypomethylated regardless of its activity, whereas gametic DNA methylation in the brain type promoter was maintained during differentiation. Histone modification analysis showed that allelic methylation of histone H3 lysine 4 and H3 lysine 9 were associated with gametic DNA methylation in the brain type promoter, whereas that of H3 lysine 27 regulated by the Eed PcG complex was detected in the paternal major-type promoter, corresponding to its allele-specific silencing. Here, we propose a molecular model that gametic DNA methylation and chromatin remodeling by PcG proteins during cell differentiation cause tissue-specific imprinting in embryonic tissues.
Genomic imprinting in mammals describes the situation where there is nonequivalence in expression between the maternal and paternal alleles at certain gene loci, depending on the parental origin. Genomic imprinting plays essential roles in development, growth, and behavior (6, 30, 31). Such parental origin-specific gene regulation is caused by epigenetic modifications that occur during gametogenesis without any nucleic acid changes. One of the well-known epigenetic modifications is DNA methylation. In the imprinted loci, differentially methylated regions between the maternal and paternal alleles are often found and associated with parental allele-specific expression (7). Another well-known epigenetic modification is histone modification, which represents the determinant of epigenetic features associated with imprinted genes. It has been reported that parental origin-specific gene expression on some imprinted genes is determined by DNA methylation and/or histone modifications (12, 13, 16, 23, 29, 40). Polycomb group (PcG) proteins also play an important role in various epigenetic phenomena (3), such as maintaining the silent state of the homeotic genes, maintaining X-chromosome inactivation (36), and silencing imprinted genes in mammals (24, 33). PcG protein complexes are thought to maintain long-term gene silencing during development through alterations of local chromatin structure (3, 27).
Mouse Grb10 encoding the growth factor receptor-bound protein 10 (Grb10) is an imprinted gene with tissue-specific and promoter-specific expression. In most tissues, the major-type transcript of Grb10 is expressed exclusively from the major-type promoter of the maternal allele, whereas in the brain, the brain type transcript is expressed predominantly from the brain type promoter of the paternal allele (1, 17). DNA methylation analysis has revealed that the CpG island (CGI) in the brain type promoter (CGI2) was gametically methylated in the oocyte as a primary imprint and remained methylated exclusively on the maternal allele in somatic tissues, while the CpG island in the major-type promoter (CGI1) was biallelically hypomethylated in somatic tissues (see Fig. 1 and 4) (17). Hikichi et al. proposed the model for tissue-specific imprinting of Grb10 that the major-type transcript is regulated by DNA methylation-sensitive insulator (CTCF) binding in CGI2 and the brain type transcript is regulated by putative brain-specific activators (17). They suggested that allelic DNA methylation in CGI2 can orchestrate reciprocal imprinting of the two promoters of the Grb10 gene. This model was partially supported by the imprinting analysis of knockout mice of the Dnmt3L gene, encoding a factor for acquisition of maternal methylation imprint in germ cells (14, 18). In the embryos (Dnmt3Lm−/−), produced from Dnmt3L−/− females, maternal chromosome-specific DNA methylation in CGI2 was lost and null expression of the major-type transcript was detected (2). Recently, the PcG protein Eed (embryonic ectoderm development) was identified as a member of a new class of trans-acting factors, which regulate the expression of some paternally repressed imprinted genes, Cdkn1c, Ascl2, Meg3, and Grb10 (24). In Eed−/− embryos, the major-type transcript of Grb10 was biallelically expressed from the major-type promoter without major alteration of DNA methylation in gametically methylated CGI2, albeit various hypomethylated patterns were observed on the paternal allele (24). The expression analysis of these knockout mice suggests that DNA methylation and chromatin remodeling by PcG proteins represent the epigenetic factors that are necessary for establishing and/or maintaining the imprinted expression of Grb10. It remains unknown how they coordinate the tissue-specific and promoter-specific imprinting of Grb10.
FIG. 1.
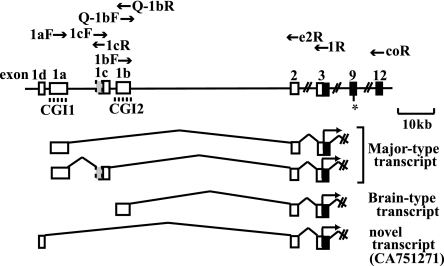
Tissue-specific transcripts of Grb10. Filled boxes, open boxes, and shaded boxes represent protein-coding regions, 5′ untranslated regions, and extended exons 1c, respectively. The dashed lines indicate the CpG islands (CGI1 and CGI2) in the promoters. The primers used for RT-PCR are shown. The asterisk indicates the polymorphic site (G/A) between the C57BL/6 and PWK strains.
FIG. 4.
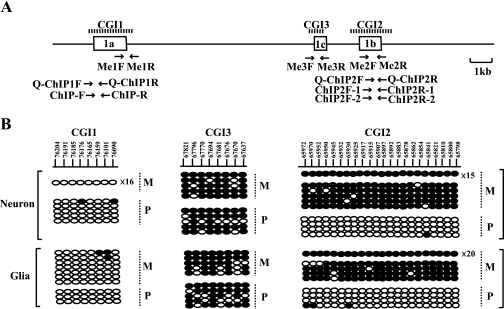
Methylation status of CpG islands in neurons and glial cells. (A) Schematic structure of CpG islands. The dashed lines indicate the registered regions of CGI1, CGI2, and CGI3 (1, 17). Open boxes and arrows represent exons and primers used for methylation analysis and ChIP analysis, respectively. (B) Allele-specific DNA methylation analysis of cultured cells by bisulfite PCR and sequencing. Numbers on the line in the upper panel represent nucleotide positions, given according to GenBank accession no. AL663087. Each line shows an individual clone, and each oval represents a CpG nucleoside; the filled and open ovals indicate hypermethylated and hypomethylated CpGs, respectively. The numbers with “×” given at the right end of the clone lines represent the number of individual clones that show the same pattern of DNA methylation. Parental alleles (M, maternal; P, paternal) are distinguished by DNA polymorphisms between the C57BL/6 and PWK strains.
Recently, mouse genes with brain-specific imprinting patterns were reported. They are Ube3a and Murr1, with neuron-specific and brain developmental stage-specific expressions, respectively. Ube3a is biallelically expressed in most tissues but expressed exclusively from the maternal allele only in neurons, leading to apparent partial imprinting with predominant maternal Ube3a expression in the whole brain (38). Murr1 is imprinted in the adult brain, especially in mature neurons, but not in embryonic and neonatal brains (37). These lines of evidence suggest that brain-specific imprinting may be regulated in part by epigenetic modifications, depending on specification and maturation of cell lineages in the developing brain (9, 19).
Since Grb10 is a tissue-specific imprinted gene, we hypothesized that tissue-specific reciprocal imprinting of Grb10 also depends on cell-specific epigenetic modifications acquired during cell differentiation. To examine our hypothesis, we performed an epigenetic analysis of brain cells with the aid of primary cortical cell cultures, in which neurons or glial cells were cultured separately from products of reciprocal crosses between the C57BL/6 and PWK strains (divergent strains of Mus musculus). In each cultured brain cell, Grb10 expression and epigenetic factors such as DNA methylation and histone modifications were analyzed to investigate how DNA methylation and chromatin remodeling by PcG proteins establish and maintain the tissue-specific and promoter-specific imprinting of Grb10.
MATERIALS AND METHODS
Mice.
All procedures were performed with approval from the Nagasaki University Institutional Animal Care and Use Committee. F1 hybrid mice were obtained by mating C57BL/6 females with PWK males [(C57BL/6 × PWK)F1] and vice versa [(PWK × C57BL/6)F1]. Telencephalon/cerebral cortices and embryonic fibroblasts were prepared from embryonic day 10 (E10) to E15. Tissues were used for RNA and DNA extraction or primary cultures. Brain tissue for reverse transcriptase (RT) PCR was dissected at E10, E16, postnatal day 1, postnatal day 5, 2 weeks, 4 weeks, 6 weeks, and 14 months.
Primary culture.
Methods of primary cultures of cortical neurons, glial cells, and embryonic fibroblasts have been described elsewhere (38). In brief, E15 cerebral cortices without meninges were trypsinized to dissociate brain cells. For neuronal culture, dissociated cells were cultured in neurobasal medium (Gibco BRL, Carlsbad, CA) with B27 supplement (Gibco BRL). Cultures were maintained in 5% CO2 at 37°C for 5 days. For the long culture, half of the culture medium was changed every 3 to 4 days. For glial cell culture, dissociated brain cells were cultured overnight in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) supplemented with 10% fetal calf serum, and then the medium was changed to Neurobasal medium (Gibco BRL) with G5 supplement (GIBCO BRL). After 5 to 7 days in the primary culture, cultured glial components were subcultured. Cultures were maintained in 5% CO2 at 37°C for a total of 14 days. For embryonic fibroblast culture, embryonic fibroblasts derived from E15 embryonic skin were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
cDNA synthesis.
Total RNA was isolated from cultured cells and tissues with RNeasy (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. The RNA was treated with amplification grade DNase I (Invitrogen, Carlsbad, CA) to degrade any genomic DNA present in the sample. The cDNA was generated from total RNA by SuperScript II reverse transcriptase (Invitrogen) primed with oligo(dT)12-18 primers. The first-strand cDNA was synthesized at 42°C for 50 min. Then, mRNA-cDNA chains were denatured and the reverse transcriptase activity was arrested by heating at 70°C for 15 min. An identical reaction was carried out without reverse transcriptase as a negative control.
RT-PCR for expression analysis.
The cDNA obtained was used to perform RT-PCR for expression analysis. The expression of each Grb10 transcript was analyzed using primers 1aF and 1R for the major-type transcript and using primers 1bF and 1R for the brain type transcript. Other transcripts, including exon 1c, were amplified by primer sets 1cF/e2R and 1aF/1cR. PCR amplification with primers 1aF and 1R was performed for 32 to 35 cycles of 15 s at 96°C, 20 s at 60°C, and 60 s at 72°C, with primers 1bF and 1R for 32 to 38 cycles of 15 s at 96°C, 20 s at 60°C, and 60 s at 72°C, and with primer sets 1cF/e2R and 1aF/1cR for 35 cycles of 15 s at 96°C, 20 s at 60°C, and 60 s at 72°C. The primers for Map2, Gfap, and Gapdh used for evaluation of the cultured cells have been described elsewhere (38). For a semiquantitative RT-PCR, optimal template cDNA concentrations were determined according to Gapdh amplification. PCR products were amplified for 25 to 30 cycles of 15 s at 96°C, 20 s at 55°C, and 30 s at 72°C.
Quantitative analysis of gene expression by real-time PCR.
cDNA was applied to real-time PCR for quantitative analysis of each transcript using SYBR green and an ABI Prism 7900 (PE Applied Biosystems, Foster City, CA). PCR was performed on samples at least in triplicate according to the manufacturer's protocol to control for PCR variation. To standardize each experiment, the results were represented as a percentage of expression, calculated by dividing the average value of the expression of the target gene by that of an internal control gene, Gapdh (38). The primers used for real-time PCR were primers 1aF and1R for the major-type transcript and primers Q-1bF and Q-1bR for the brain type transcript. Each experiment was repeated with independent RNAs two to three times.
Sequencing for allelic differences.
A sequence chromatogram was used to detect allelic differences of PCR products. Parental expression of major/brain type transcripts in the brain and kidney was analyzed by RT-PCR using primer sets 1aF/coR and 1bF/coR for 35 to 38 cycles of 15 s at 96°C, 20 s at 60°C, and 120 s at 72°C. Parental chromosome-specific histone modifications in the major-type promoter were analyzed by PCR using the primer set ChIP-F/ChIP-R for 30 cycles of 30 s at 95°C, 30 s at 58°C, and 30 s at 72°C. The PCR products were analyzed by direct sequencing with a BigDye Terminator cycle sequencing kit (PE Applied Biosystems) on an automated sequencer, the ABI Prism 3100 genetic analyzer (PE Applied Biosystems).
DNA methylation analysis.
Isolated DNA was treated with sodium bisulfite using a CpGenome DNA modification kit (Chemicon International Inc., Temecula, CA) according to the manufacturer's protocol. Bisulfite-treated DNA samples were subjected to nested PCR amplification using the following first and second primer pairs, respectively, for each CGI; CGI1, Me1F/Me-1R and Me-1F′/Me-1R′; CGI2, Me-2F/Me-2R and Me-2F′/Me-2R′; and CGI3, Me-3F/Me-3R and Me-3F′/Me-3R′. After the first PCR using the first primer set, the products were used as templates for nested PCR using the second primer set. The nested PCR products were cloned into the TA cloning vector (Invitrogen), and at least 32 clones for each sample were sequenced.
ChIP.
A chromatin immunoprecipitation (ChIP) assay was performed with a ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's protocol. In brief, the chromatin of cultured cells was prepared from ∼1.0 × 106 cells and treated with formaldehyde to cross-link DNA to protein in situ, sonicated to an average size of 0.5 kb, and immunoprecipitated with antibodies. Antibodies against acetyl histone H3 (H3Ac; catalog no. 06-599), acetyl histone H4 (H4Ac; catalog no. 09-866), Lys4 dimethylated histone H3 (H3mK4; catalog no. 07-030), Lys9 trimethylated H3 (H3me3K9; catalog no. 07-212), and Lys27 trimethylated H3 (H3mK27; catalog no. 07-449) were obtained from Upstate Biotechnology. The monoclonal antibody against Lys9 dimethylated histone H3 (H3me2K9) was developed previously (26). Immunoprecipitated samples without antibodies or with rabbit immunoglobulin G precipitation were used as negative controls for precipitations with specific antibodies in each experiment.
Quantitative analysis of immunoprecipitated DNA by real-time PCR.
Immunoprecipitated DNA and input DNA were analyzed by real-time PCR using the same protocol as that used for gene expression analysis. For DNA immunoprecipitated with H3Ac, H4Ac, and H3mK4 antibodies, the quantitative value of immunoprecipitated DNA in each CGI was normalized by dividing the average value of each CGI by that of the internal control, Gapdh. For DNA immunoprecipitated with H3me2K9 and H3me3K9 antibodies, the average value of D13Mit55 was used instead of the value of Gapdh. Each normalized value of immunoprecipitated DNA was further divided by the normalized value of the corresponding input DNA. For the evaluation of DNA immunoprecipitated with H3mK27 antibody, the results were presented as a percentage of immunoprecipitation, calculated by dividing the average value of immunoprecipitated DNA by the average value of the corresponding input DNA. Each experiment was performed three times with independent chromatin extracts. The primers used for real-time PCR were primers Q-ChIP1F and Q-ChIP1R for CGI1 analysis and primers Q-ChIP2F and Q-ChIP2R for CGI2 analysis. The primers for Gapdh and D13Mit55 have been described elsewhere (16).
Hot-stop PCR and SSCP analysis.
Hot-stop PCR was performed for the analysis of allele-specific histone modifications as follows. After a number of PCR cycles sufficient to detect a product using primers ChIP2F-1 and ChIP2R-1, primer ChIP2R-1 labeled by [γ-32P]ATP was added to the mixture, and then one cycle of PCR was performed. The PCR products were digested with the restriction endonuclease Hpy188I and electrophoresed in a 4% polyacrylamide gel. Single-strand conformation polymorphism (SSCP) analysis of PCR products was performed for allele-specific histone methylation in the presence of [γ-32P]ATP-labeled primers ChIP2F-2 and ChIP2R-2. PCR products were resolved by electrophoresis in an MDE nondenaturing acrylamide gel (FMC BioProduct, Rockland, ME).
Primers.
The primers used for the analysis are listed in Table 1.
TABLE 1.
Primers used in this study
| Function(s) and primer | Sequence (5′-3′) | Annealing temp (°C) (PCR cycle no.)a |
|---|---|---|
| Expression and imprinting analysis | ||
| 1aFb | CACGAAGTTTCCGCGCA | |
| 1bF | GCGATCATTCGTCTCTGAGC | |
| 1Rb | AGTATCAGTATCAGACTGCATGTTG | |
| 1cF | ATCGCCATCTACAGTTTCTG | |
| 1cR | CAAGGTACAGAGCTAGGACG | |
| e2R | CTGGTTGGCTTCTTTGTTGTGG | |
| coR | TACGGATCTGCTCATCTTCG | |
| ChIP-F | TCACTTTAGAAACCGGGCA | |
| ChIP-R | AAACTCGGGCTTGCTCA | |
| Quantitative analysis | ||
| Q-1bF | TCATTCGTCTCTGAGCGGCA | |
| Q-1bR | ATACGTGTTACATGCGCCAA | |
| Q-ChIP1F | TCACTTTAGAAACCGGGCA | |
| Q-ChIP1R | AAACTCGGGCTTGCTCA | |
| Q-ChIP2F | GATCATTCGTCTCTGAGC | |
| Q-ChIP2R | ATGCGGCAACATGCGCTGACA | |
| Hot-stop PCR and SSCP analysis | ||
| ChIP2F-1 | TCATTCGTCTCTGAGCGGCA | 60 (32) |
| ChIP2R-1 | TCTGGAGCCTAGAGGAGCG | |
| ChIP2F-2 | AAGCGCGTGCTGGTTTGTA | 60 (35) |
| ChIP2R-2 | ATACGTGTTACATGCGCCAA | |
| DNA methylation analysis | ||
| CGI1 1st | 53 (35) | |
| Me-1F | TGGGGTTTAATATTAAGTTTGA | |
| Me-1R | TTACATCTCTTAAATAAAACA | |
| CGI1 2nd | 53 (35) | |
| Me-1F′ | TGGGGTTTAATATTAAGTTTGA | |
| Me-1R′ | AAATCACCTATAACTCTCCTAC | |
| CGI2 1st | 50 (40) | |
| Me-2F | TGGAGTTTAGAGGAG | |
| Me-2R | AATAGTTATTTTAGTAAGGG | |
| CGI2 2nd | 50 (10) | |
| Me-2F′ | TGGAGTTTAGAGGAG | |
| Me-2R′ | TAAGTGAAGTAATATAGTT | |
| CGI3 1st | 53 (40) | |
| Me-3F | AAAGAAGGTTTGGAGAGATTATTT | |
| Me-3R | CAAACCAAAACTTACTATATTTAATTTAAAC | |
| CGI3 2nd | 53 (10) | |
| Me-3F′ | AAGGTTTGGAGAGATTATTTTTGATT | |
| Me-3R′ | TAATTTAAACTTAACACTATTAAATACC |
For expression and imprinting analysis, the annealing temperature and PCR cycle number depend on the combination of primers used for each analysis. See details in Materials and Methods. For quantitative analysis, the PCR conditions were decided according to the manufacturer's protocol.
Also used for quantitative analysis.
RESULTS
Mouse Grb10 has several tissue-specific promoters.
Three different promoters of Grb10 have previously been reported to initiate tissue-specific transcripts (Fig. 1). We first analyzed the expression of each transcript in E16 fetal tissues. The major-type transcript amplified by PCR using primers 1aF and e2R in exons 1a and 2, respectively, was detected in the fetal brain but was less detected in other tissues, while the brain type transcript amplified by primers 1bF and e2R in exons 1b and 2, respectively, was detected exclusively in the fetal brain (Fig. 2A). Another transcript which was previously reported to be brain specific in adult tissues (1) was examined in fetal tissues. PCR using primers 1cF and e2R in exons 1c and 2, respectively, showed that exon 1c was expressed not only in the fetal brain but also in the fetal liver and kidney (Fig. 2A). To assess whether exon 1c is an alternatively spliced exon of the major-type transcript with exon 1a, we performed PCR using primers 1aF and 1cR in exons 1a and 1c, respectively. The PCR product containing exons 1a and 1c was detected in the fetal tissues (Fig. 2A). Sequence analysis of the RT-PCR product revealed that exon 1c was extended 67 bp upstream of the previously published exon 1c with the consensus splicing site (Fig. 1). Any RT-PCR products with both exons 1a and 1b or both exons 1c and 1b were not found (data not shown). Furthermore, we identified another putative exon, 1d, located 1.2 kb upstream of exon 1a in the expressed sequence tag database (GenBank accession no. CA751271). The existence of the novel exon 1d was confirmed by RT-PCR in the embryonic liver but not in other tissues, including the brain (data not shown).
FIG. 2.
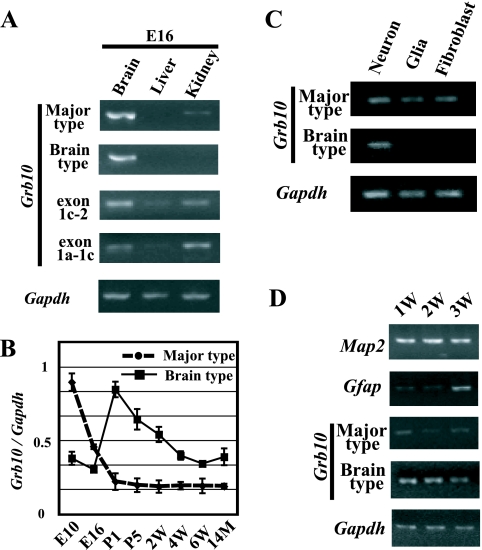
Expression analysis of each transcript in embryonic tissues by RT-PCR. (A) Semiquantitative analysis of tissues from the E16 embryo. Exon 1c-2 and exon 1a-1c represent RT-PCR products amplified by primer sets 1cF/e2R and 1aF/1cR, respectively. The concentration of each cDNA was adjusted for Gapdh amplification as an internal control. (B) Quantitative evaluation of major-type and brain type transcripts in brain tissues from different developmental stages by real-time PCR. The relative amounts of major-type and brain type transcripts are shown. The relative amount of each transcript was calculated by normalizing each value with an internal control, Gapdh. Standard errors of the means are indicated by bars. (C) Expression analysis of major-type and brain type transcripts in the primary cell culture. (D) Evaluation of expression of marker genes and each Grb10 transcript according to the culture period. 1w (1 week), 2w (2 weeks), and 3w (3 weeks) indicate the periods of neuron culture. P1, postnatal day 1; 14M, 14 months.
Expression of Grb10 shifts from the major-type to the brain type transcript during brain development.
To confirm whether the expression level of the brain type transcript changes during brain development, the major-type and brain type transcripts arising from exons 1a and 1b, respectively, were quantitatively analyzed at various developmental stages of the brain. Real-time PCR analysis showed that in the brain, the major-type transcript was highly expressed at E10 and decreased according to brain development, while expression of the brain type transcript was high in the perinatal period and gradually decreased thereafter (Fig. 2B). The result indicates that Grb10 transcripts shift from the major type to the brain type during early brain development.
The brain-specific transcript is expressed in neurons but not in glial cells.
Is the brain type transcript expressed exclusively in the brain restricted to the cell type? To know which type of brain cells, neurons or glial cells, express the brain type transcript, expression analysis of cultured neurons and glial cells was carried out. Prior to the analysis, we confirmed by immunostaining and RT-PCR with the brain precursors, neuronal and glial markers, that over 95% of the two cultured cell types were postmitotic neurons and astrocytes, respectively (data not shown). RT-PCR in cultured cells revealed that the major-type transcript was expressed in all cultured brain cells but that the brain type transcript was expressed only in neurons (Fig. 2C). We next tried to investigate whether these transcripts in the brain were associated with the maturation of neurons. Neurons were cultured for 1, 2, and 3 weeks, and semiquantitative RT-PCR was carried out. Before expression analysis of Grb10, the status of cell proliferation and differentiation by long culture was evaluated by primers for Map2 as a marker for neurons and Gfap as a marker for astrocytes under the normalization of cDNA concentration to Gapdh (Fig. 2D). The expression of Map2 never changed in 3-week-cultured cells, while that of Gfap was detected in the cells cultured for 3 weeks. In these long-culture cells, the brain type transcript was continuously expressed during culture periods, while the major-type transcript was less expressed than the brain type transcript. These results suggest that both types of transcripts are expressed in neurons and that the switching of the promoter from the major type to the brain type is observed during long culture periods.
Promoter-specific paternal expression of Grb10 in the brain.
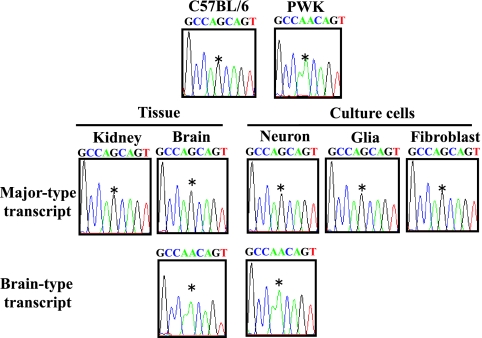
To investigate the imprinted expression of Grb10, we first examined parental expression of the major-type and the brain type transcripts in the brain and kidney from F1 hybrid mice by direct sequencing of the RT-PCR product. A polymorphic site (G/A) in exon 9 between the C57BL/6 and PWK strains was used to determine the paternal allele (Fig. 1). As previously reported by Hikichi et al. (17), the major-type transcript was expressed exclusively from the maternal allele in the kidney and brain, while the brain type transcript was expressed from the paternal allele only in the brain (Fig. 3). We next examined promoter-specific imprinting in neurons, glial cells, and fibroblasts. Expression of the major-type transcript originated exclusively from the maternal allele in all cultured cells, but that of the brain type transcript detected only in neurons originated from the paternal allele (Fig. 3). Thus, predominant paternal Grb10 expression in the brain, as previously described, can be explained by a combination of paternally expressed brain type transcript in neurons and maternally expressed major-type transcript in all cells.
FIG. 3.
Imprinting analysis of promoter-specific expression of Grb10 by sequence chromatograms. Upper panels show the chromatograms of the genomic PCR products from each strain. Middle and lower panels show the chromatograms of the RT-PCR products from tissues and cultured cells of the F1 hybrid, in which alleles were distinguished by the single-nucleotide (G/A) polymorphism (*) at exon 9.
Differentially methylated CGI2 is maintained in cultured neurons and glial cells.
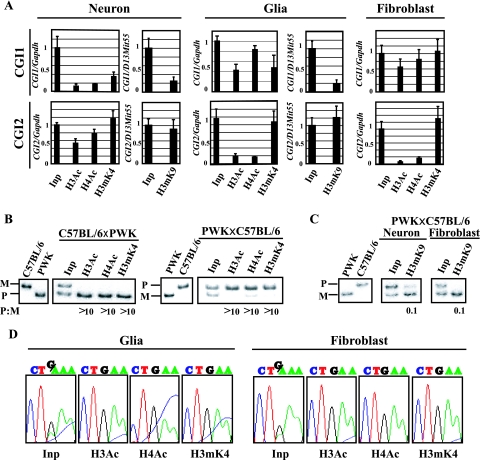
As we found that the brain type transcript was initiated from exon 1b of the paternal allele only in neurons, we analyzed the methylation status of the brain type promoter in neurons and glial cells by the bisulfite method. As shown in Fig. 4A, three promoters are located within different CGIs: exon 1a in CGI1, exon 1b in CGI2, and exon 1c in the “weaker” CpG island, CGI3. The parental origin of the methylated allele was identified by polymorphic sites in F1 hybrids between the C57BL/6 and PWK strains. The methylation analysis of CGI2 showed that the differential methylation established in the germ cells (1, 17) was maintained in neurons and glial cells (Fig. 4B). That in other CpG islands, CGI1 and CGI3, revealed biallelic hypomethylation and hypermethylation, respectively. CGI1 and CGI3 did not show any differential methylation in the cells, although CGI3 was reported to be a putative differentially methylated region in the mouse brain with uniparental disomy for chromosome 11 (1) The methylation status in CGIs, except CGI3, in cultured cells was consistent with that previously reported for tissues (1, 17).
Parental chromosome-specific histone modifications in CGI2 correlate with allele-specific expression of the brain type transcript in neurons.
Parental origin-specific histone modifications are reported to represent the determinant of epigenetic features as well as DNA methylation. Using specific antibodies against acetylated histone H3 (H3Ac), acetylated histone H4 (H4Ac), dimethylated Lys4 histone H3 (H3mK4), and di- and trimethylated Lys9 histone H3 (H3me2K9 and H3me3K9), we performed a ChIP assay with cultured cells. After evaluation of ChIP DNA by allele-specific histone modifications in the Lit1 promoter region as a control (16), histone modifications in CGI1, CGI2, and CGI3 were analyzed by real-time PCR to quantify their precipitated chromatins in these CGIs. To normalize each value, Gapdh and D13Mit55 were used as internal control sequences, where acetylated and methylated histones were known to be biallelically immunoprecipitated, depending on the corresponding antibodies. In CGI2, where the maternal allele-specific DNA methylation was established in the oocyte, H3Ac, H4Ac, H3mK4, and H3me3K9 were clearly immunoprecipitated in neurons, while in glial cells and fibroblasts, although H3mK4 and H3me3K9 were well immunoprecipitated, H3Ac and H4Ac were less precipitated (Fig. 5A). The results obtained with the antibody against H3me2K9 (data not shown) were similar to those obtained with the antibody against H3me3K9.
FIG. 5.
Histone modification analysis of the Grb10 promoter in cultured cells. (A) Quantitative analysis of immunoprecipitated DNA by real-time PCR. Quantitative values of precipitated DNA in CGI1 and CGI2 were normalized by dividing the average value of each CGI by the average value of Gapdh or D13Mit55. Standard errors of the means are indicated by bars. (B) Allele-specific histone modifications in CGI2 in neurons by hot-stop PCR. Digested PCR products of C57BL/6 and PWK genomic DNA are shown as homozygous controls in the first two lanes. M and P represent the products from the maternal allele and the paternal allele, respectively. The ratio of the paternal to maternal (P:M) band intensities, corrected by the ratio in input chromatin (Inp), is indicated below each lane. (C) Allele-specific histone H3K9 methylation in CGI2 by SSCP. PCR products of C57BL/6 and PWK genomic DNA were shown as controls in the first two lanes. (D) Allele-specific histone modifications in CGI1 by sequence chromatograms. Glial cells and fibroblasts derived from F1 hybrids [(C57BL/6 × PWK)F1] were used for analysis. The single-nucleotide (G/A) polymorphism is detected in the input sample (Inp); “G” originated from the maternal allele and “A” from the paternal allele.
To elucidate the parental chromosome-specific histone modifications in CGI2 in neurons, hot-stop PCR was performed (15, 32). The restriction endonuclease Hpy188I was used to recognize the polymorphic site in CGI2. For each of the precipitated samples, the ratio of the paternal to maternal band intensities was determined. These ratios were corrected for the paternal-to-maternal ratios in the input chromatin, because the maternal and paternal alleles were not equally represented in the input chromatin. One of the parental alleles is possibly more sensitive to sonication in these regions because of relaxed chromatin (12, 16, 39). The result revealed that histones H3 and H4 were hyperacetylated and that H3K4 was hypermethylated predominantly on the paternal chromosome (Fig. 5B). To investigate allele-specific histone trimethylation of H3K9 in neurons and fibroblasts, SSCP analysis of PCR products was also performed. In neurons and fibroblasts, H3K9 was hypermethylated on the maternal chromosome (Fig. 5C).
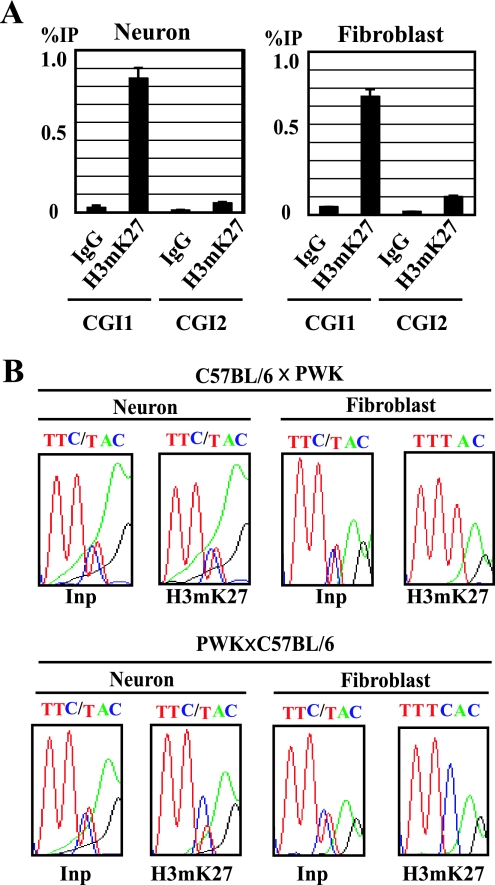
Parental chromosome-specific methylation of histone H3K27 but not H3K9 in CGI1 correlates with allele-specific expression of the major-type transcript.
Histone modifications in CGI1, where CpGs were biallelically hypomethylated in tissues and cultured cells, were next analyzed. In CGI1, H3Ac, H4Ac, and H3mK4 were clearly precipitated in glial cells and fibroblasts, while the precipitations were not observed in neurons (Fig. 5A). H3me3K9 and H3me2K9 in CGI1 were not precipitated in neurons and glial cells (Fig. 5A; data not shown). The maternal chromosome-specific histone H3/H4 acetylation and H3K4 methylation in CGI1 were detected in glial cells and fibroblasts (Fig. 5D). We further analyzed histone H3K27 trimethylation, which is directly regulated by the PcG proteins, because imprinted expression of the major-type Grb10 transcript was reported to be relaxed in the knockout embryos of the PcG gene, Eed (24). In neurons and fibroblasts, H3mK27 was clearly precipitated in CGI1 but not in CGI2 (Fig. 6A). The paternal chromosome-specific methylation of H3K27 was observed in fibroblasts, but a significant allelic difference was not detected in neurons (Fig. 6B). These data suggest that the paternally null expression of the major-type transcript in fibroblasts correlates with paternal chromosome-specific methylation of H3K27 in CGI1. In CGI3, histones H3 and H4 were hypoacetylated and H3K4 was hypomethylated (data not shown). We could not detect significant differences in histone acetylation and methylation in CGI3 between cultured cells.
FIG. 6.
Histone H3K27 methylation analysis of CGI1 and CGI2 in cultured cells. (A) Quantitative analysis of immunoprecipitated DNA by real-time PCR. The percentage of immunoprecipitation (IP) was calculated by dividing the quantitative value of precipitated DNA by that of the corresponding input DNA. Standard errors of the means are indicated by bars. (B) Allele-specific histone modifications in CGI1 by sequence chromatograms. Neurons and fibroblasts derived from F1 hybrids (C57BL/6 × PWK; PWK × C57BL/6) were used for analysis. The single-nucleotide (C/T) polymorphism is detected in the input sample (Inp); “C” originated from the C57BL/6 allele and “T” from the PWK allele. IgG, immunoglobulin G.
DISCUSSION
It has been known that mouse Grb10 shows reciprocal imprinting depending on the tissue-specific promoters. In most tissues, Grb10 is expressed exclusively from the maternal allele, whereas in the brain, it is expressed predominantly from the paternal allele (1, 17). Such reciprocal imprinting of Grb10 in a tissue-specific and promoter-specific manner is a good model to elucidate how promoter-specific imprinting is epigenetically controlled in tissues. In this study, we have developed a cell culture system with which cell-type-specific imprinting of Grb10 can be characterized in the mouse brain. We demonstrated that promoter-specific and developmental stage-specific imprinting of Grb10 expression in the brain is associated with parental allele-specific epigenetic modifications in brain cell lineages.
Two previous reports described that reciprocal imprinting of Grb10 occurs in a tissue-specific and promoter-specific manner (1, 17). Our studies with cultured cortical cells revealed that the brain type transcript containing exon 1b was expressed in neurons but not in glial cells, while the major-type transcript containing exon 1a was expressed in all cultured cells, including neurons (Fig. 2C). These findings indicate that the brain-specific promoter actually implies the neuron-specific promoter and that the major-type promoter works as the common promoter in all tissues. Imprinting analysis of these transcripts clearly showed that the brain type transcript is expressed exclusively from the paternal allele and the major-type transcript is expressed exclusively from the maternal allele (Fig. 3). These results in vitro can explain the previous data that the brain type transcript was not detected in whole embryo at E9.5 (17), when neurogenesis has not yet occurred. In addition, our data on Grb10 expression, i.e., brain development-dependent switching from the major-type to the brain type transcript, can also support the previous report that Grb10 is expressed predominantly from the paternal allele in the adult brain (17), which consists of neurons and glial cells.
In our expression analysis, we detected both brain type and major-type transcripts in cultured neurons (Fig. 2B). Recently, it was reported that the Pcdh (protocadherin) gene was monoallelically expressed in individual neurons (10). The Pcdh gene family (Pcdha, Pcdhb, and Pcdhc) has variable exons and alternative splice forms. Esumi et al. analyzed the expression of transcripts in the variable exons of Pcdha by using a single-cell RT-PCR approach for the determination of the allelic origin for each variable exon at the individual cell level (10). The individual cells showed monoallelic expression for each variable exon. In our analysis of Grb10, the discrepancy between the modifications in CGI1 and the expression of the major-type transcript in neurons was recognized. Similar to a monoallelic expression pattern of variable Pcdha exons in individual neurons, the discrepancy may be explained by the existence of two different cell populations in cultured neurons, each of which expresses either the major-type or the brain type transcript exclusively. As shown in Fig. 2D, the brain type transcript was obviously highly expressed compared to the major-type transcript during long culture periods. The larger population of cells with the brain type transcript may affect the result of histone modifications more than the smaller population of cells with the major-type transcript.
It has been reported that histone modifications and DNA methylation are not synchronized as a transcriptionally active/silent signal in some imprinted genes, such as NDN, Gnas, and Igf2r (21, 22, 23, 33, 35). Our data also showed an epigenetically unsynchronized active/silent signal between DNA methylation and histone modifications in Grb10 (Fig. 7). In this study, we showed that the brain type transcript is expressed in neurons but not in glial cells (Fig. 2C), where both differential methylation in CGI2 and biallelic hypomethylation in CGI1 were maintained regardless of expression (Fig. 4B). The result that allele-specific DNA methylation is not sufficient to direct imprinted expression in brain cells implies that other epigenetic modifications may affect cell lineage-specific imprinting.
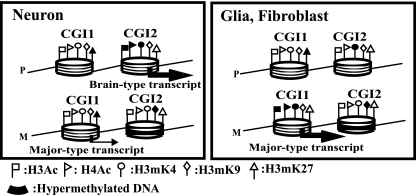
FIG. 7.
Summary of epigenetic modifications across promoter regions of Grb10. M and P represent maternal and paternal chromosomes, respectively. Large and small arrows indicate expression levels. The nucleosome model shows DNA wrapping around a histone octamer with some histone modifications. White and black flags represent hypoacetylated/hypomethylated and hyperacetylated/hypermethylated statuses, respectively.
In our analysis of histone modifications, histone acetylation status correlated with the expression status of the major-type transcript in glial cells and fibroblasts and the brain type transcript in neurons (Fig. 7). Such histone acetylation status in Grb10 expression is consistent with the findings that allele-specific histone acetylation was associated with allelic gene expression in the imprinted gene, NDN (21). Histone acetylation offers the best example of a direct link between tissue-specific gene expression and histone modifications.
Unlike that of histone acetylation, the status of histone methylation has been implicated as an early event for chromatin conformations. Methylation of histones H3K4 and H3K9 is associated with active chromatin and silent chromatin, respectively. According to our results, allele-specific H3K4 and H3K9 methylation in CGI1 and CGI2 did not correlate with allele-specific gene expression in each cultured cell. In glial cells, H3K4 in CGI2 was hypermethylated in the paternal chromosome, which was silent with no brain type transcript. It seems that H3mK4 is maintained during differentiation as an imprint mark with H3mK9 but is not related to promoter activity (28), although histone modifications in oocytes remain unknown. In CGI1, H3me2K9 and H3me3K9 were hypomethylated in both parental chromosomes independent of the expression of the major-type transcript in cultured cells. It is likely that H3K9 methylation in germ cells is maintained as a stable and heritable imprint mark but may not be secondarily acquired during development.
Then, how is maternal chromosome-specific expression of the major-type transcript regulated without differential DNA methylation in CGI1? The PcG protein Eed complex is known to be a part of a memory system that maintains repression of the imprinted X chromosome (36) and silencing of some imprinted genes (24, 33). Grb10 is reported to be one of the imprinted genes that are regulated by the PcG protein Eed complex. Interestingly, in Eed−/− embryos, the major-type transcript was biallelically expressed without major alteration of allelic DNA methylation (24). The Eed/Ezh2 PcG complex possesses histone methyltransferase activity on H3K27 (5, 8, 25) and interacts with histone deacetylases (34). Methylation of H3K27 is a repressive epigenetic mark regulated by the SET domain containing Ezh2/Eed complex (5, 8, 20, 25). In our analysis, H3mK27 was clearly precipitated in neurons and fibroblasts in CGI1 but not in CGI2 (Fig. 6A). The paternal chromosome-specific methylation of H3K27 in CGI1 was observed in fibroblasts but not in neurons (Fig. 6B). These data indicate that the Eed PcG complex can biallelically interact on CGI1 as a trans-acting factor in neurons but paternally in other cells. In the absence of DNA methylation in CGI1, PcG complexes may mediate a nonpermissive chromatin state for transcription, leading to repressive histone modifications. Interestingly, other genes, Cdkn1c and Ascl2, imprinting of which was reported to be regulated by Eed (24), show tissue-specific imprinting, and their imprinted expression in trophoblasts is associated with repressive histone H3K27 methylation rather than DNA methylation (22, 33).
Figure 7 shows the summary of our data. In CGI2, DNA methylation in a gametically methylated CpG island on the maternal allele was maintained throughout development. Allelic methylation of H3K4 and H3K9 associated with gametic DNA methylation was also stable as an epigenetic mark, independent of Grb10 expression. Histone acetylation status was correlated with the expression status of the brain type transcript: histones H3 and H4 were paternally acetylated only in neurons, where the brain type transcript was paternally expressed. H3K27 was not methylated biallelically. In CGI1, biallelic DNA hypomethylation and biallelic hypomethylation of H3K9 were observed. Acetylation of histones H3 and H4 and methylation of H3K4 and H3K27 were allelically detected, corresponding to the allelic expression of the major-type transcript, although the discordance in histone modifications and expression in neurons was detected, probably depending on maturation of neurons. Methylation of H3K9 and H3K27 is thought to be a repressive chromatin marker, but it is not completely clear whether PcG-mediated silencing involves methylation of H3K9 synchronized with H3mK27 in all PcG target genes. We did not observe coexistence of H3mK27 and H3mK9 in both CGI1 and CGI2 of Grb10. Umlauf et al. also reported discordance between localizations of H3mK27 and H3mK9 in some imprinted genes in the Kcnq1 domain (33). Further work should determine how histone modifications, especially methylation of H3K9 and H3K27, are coordinated or uncoordinated as epigenetic determinants in tissue-specific imprinting.
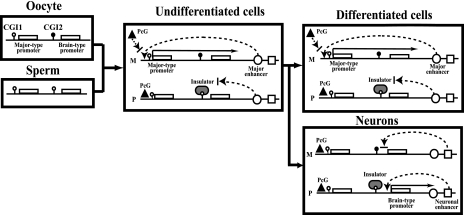
These data about epigenetic modifications analyzed at the cell level, in addition to the evidence for Dnmt3Lm−/− and Eed−/− embryos, lead to a working model for tissue-specific reciprocal imprinting of Grb10 (Fig. 8). The previous model by Hikichi et al. (17) was modified in our model based on the data of DNA methylation and repressive histone modifications mediated by the PcG complex in brain cell lineages. In undifferentiated cells, a DNA methylation-sensitive insulator, CTCF, binds to the paternal CGI2 and blocks the paternal activity of the downstream major enhancer, resulting in silent expression of the major-type transcript on the paternal allele. On the maternal allele, the major enhancer works on the major-type promoter to recruit transcription factors. In CGI1, the Eed/Ezh2 PcG complex binds on the paternal allele, whereas it competes with transcription factors on the maternal allele. The Eed/Ezh2 PcG complex methylates H3K27 and interacts with histone deacetylases, leading to silencing of the chromatin on the paternal CGI1. In Dnmt3Lm−/− embryos, biallelic hypomethylation in CGI2 makes CTCF bind biallelically on CGI2, resulting in null expression of the major-type transcript, regardless of the PcG complex. In Eed−/− embryos, the silent state on the paternal CGI1 regulated by the Eed PcG complex is released to the biallelically active state without major alteration of DNA methylation in maternal CGI2. In neurons, the other molecular mechanism of imprinting works in a promoter-specific manner, different from that in other differentiated cells. During neurogenesis, expression of Grb10 shifts from the major-type to the brain type transcript by switching from the major-type promoter to the brain type promoter. The neuronal enhancer instead of the major enhancer may work on the brain type promoter, depending on DNA methylation in CGI2. The maternally active major-type promoter becomes silent without transcription factors, and consequently, the Eed/Ezh2 PcG complex binds to make the chromatin structure silent. This implies that the PcG complex is necessary to maintain cell-type-specific imprinting. It remains unknown how neuron-specific imprinting is regulated by DNA methylation and/or histone modifications mediated by the PcG complex, because Dnmt3Lm−/− and Eed−/− embryos are lethal by E10.5 (4, 14) and E8.5 (11), respectively, just before neurogenesis.
FIG. 8.
Working models for tissue-specific reciprocal imprinting of Grb10. The previous enhancer/insulator model by Hikichi et al. was modified based on the analysis of DNA methylation and histone modifications mediated by the PcG complex containing Eed (17). Tissue-specific imprinting of Grb10 implies neuron-specific imprinting that is different from imprinting in other undifferentiated and differentiated cells. Black and white lollipops indicate hypermethylated and hypomethylated DNA, respectively. Circles and squares indicate putative major enhancer and neuronal enhancer, respectively, which accelerate Grb10 expression from the major-type promoter and the brain type promoter, respectively. The PcG complex containing Eed is represented by a triangle. CTCF is thought to be a putative insulator (gray oval). M, maternal; P, paternal.
As far as we know, this is the first report of an epigenetic analysis of cultured cells where DNA methylation and chromatin remodeling by PcG proteins establish and maintain cell-type-specific imprinting at one gene locus. Although allelic DNA methylation established in the gamete contributes primarily to tissue-specific imprinting, tissue-specific Grb10 imprinting is directly regulated by the repressive chromatin mediated by the PcG complex during development. Our analysis of promoter-specific and cell-type-specific imprinting of Grb10 gives an important clue for understanding the mechanism of tissue-specific imprinting.
Acknowledgments
We thank F. Ishino for providing information about genomic sequences of Grb10 promoter regions.
T.K. was supported in part by a Grant-in-Aid for Scientific Research (C) and a Grant-in-Aid on Priority Areas (Molecular Brain Science) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Arnaud, P., D. Monk, M. P. Hichins, E. Gordon, W. Dean, C. Beechey, J. Peters, W. Craigen, M. Preece, P. Stanier, G. E. Moore, and G. Kelsey. 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12:1005-1019. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, P., K. Hata, M. Kaneda, E. Li, H. Sasaki, R. Feil, and G. Kelsey. 2006. Stochastic imprinting in the progeny of Dnmt3L−/− females. Hum. Mol. Genet. 15:589-598. [DOI] [PubMed] [Google Scholar]
- 3.Bantignies, F., and G. Cavalli. 2006. Cellular memory and dynamic regulation of Polycomb group proteins. Curr. Opin. Cell Biol. 18:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bester. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 5.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 6.Cattanach, B. M., and C. V. Beechey. 1990. Autosomal and X-chromosome imprinting. Dev. Suppl. 1990:63-72. [PubMed] [Google Scholar]
- 7.Constancia, M., B. Pickard, G. Kelsey, and W. Reik. 1988. Imprinting mechanisms. Genome Res. 8:881-900. [DOI] [PubMed] [Google Scholar]
- 8.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 9.Davies, W., A. R. Isles, and L. S. Wilkinson. 2005. Imprinted gene expression in the brain. Neurosci. Biobehav. Rev. 29:421-430. [DOI] [PubMed] [Google Scholar]
- 10.Esumi, S., N. Kakazu, Y. Taguchi, T. Hirayama, A. Sasaki, T. Hirabayashi, T. Koide, T. Kitsukawa, S. Hamada, and T. Yagi. 2005. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat. Genet. 37:171-176. [DOI] [PubMed] [Google Scholar]
- 11.Faust, C., A. Schumacher, B. Holdener, and T. Magnuson. 1995. The eed mutation disrupts anterior mesoderm production in mice. Development 121:273-285. [DOI] [PubMed] [Google Scholar]
- 12.Fournier, C., Y. Goto, E. Ballestar, K. Delaval, A. M. Hever, M. Esteller, and R. Feil. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory, R. I., T. E. Randall, C. A. Johnson, S. Khosla, I. Hatada, L. P. O'Neill, B. M. Turner, and R. Feil. 2001. DNA methylation is linked to deacethylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol. 21:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 15.Higashimoto, K., H. Soejima, H. Yatsuki, K. Joh, M. Uchiyama, Y. Obata, R. Ono, Y. Wang, Z. Xin, X. Zhu, S. Masuko, F. Ishino, I. Hatada, Y. Jinno, T. Iwasaka, T. Katsuki, and T. Mukai. 2002. Characterization and imprinting status of OBPH1/Obph1 gene: implications for an extended imprinting domain in human and mouse. Genomics 80:575-584. [DOI] [PubMed] [Google Scholar]
- 16.Higashimoto, K., T. Urano, K. Sugiura, H. Yatsuki, K. Joh, W. Zhao, M. Iwakawa, H. Ohashi, M. Oshimura, N. Niikawa, T. Mukai, and H. Soejima. 2003. Loss of CpG methylation is strongly correlated with loss of histone H3 lysine 9 methylation at DMR-LIT1 in patients with Beckwith-Wiedemann syndrome. Am. J. Hum. Genet. 73:948-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hikichi, T., T. Kohda, T. Kaneko-Ishino, and F. Ishino. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31:1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoto, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429:900-903. [DOI] [PubMed] [Google Scholar]
- 19.Kishino, T. 2006. Imprinting in neurons. Cytogenet. Genome Res. 113:209-214. [DOI] [PubMed] [Google Scholar]
- 20.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau, J. C., M. L. Hanel, and R. Wevrick. 2004. Tissue-specific and imprinted epigenetic modifications of the human NDN gene. Nucleic Acids Res. 32:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36:1291-1295. [DOI] [PubMed] [Google Scholar]
- 23.Li, T., T. H. Vu, G. A. Ulaner, Y. Yang, J. F. Hu, and A. R. Hoffman. 2004. Activating and silencing histone modifications from independent allelic switch regions in the imprinted Gnus gene. Hum. Mol. Genet. 13:741-750. [DOI] [PubMed] [Google Scholar]
- 24.Mager, J., N. D. Montgomery, F. P. de Villena, and T. Magnuson. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33:502-507. [DOI] [PubMed] [Google Scholar]
- 25.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawachi, T., H. Soejima, T. Urano, W. Zhao, K. Higashimoto, Y. Satoh, S. Matsukura, S. Kudo, Y. Kitajima, H. Harada, K. Furukawa, H. Matsuzaki, M. Emi, Y. Nakabeppu, K. Miyazaki, M. Sekiguchi, and T. Mukai. 2003. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene 22:8835-8844. [DOI] [PubMed] [Google Scholar]
- 27.Pirrotta, V. 1995. Chromatin complexes regulating gene expression in Drosophila. Curr. Opin. Genet. Dev. 5:466-472. [DOI] [PubMed] [Google Scholar]
- 28.Rougeulle, C., P. Navarro, and P. Avner. 2003. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum. Mol. Genet. 12:3343-3348. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh, S., and T. Wada. 2000. Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader-Willi syndrome. Am. J. Hum. Genet. 66:1958-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surani, M. A., S. C. Barton, and M. L. Norris. 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308:548-550. [DOI] [PubMed] [Google Scholar]
- 31.Tilghman, S. M. 1999. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell 96:185-193. [DOI] [PubMed] [Google Scholar]
- 32.Uejima, H., M. P. Lee, H. Cui, and A. P. Feinberg. 2000. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat. Genet. 25:375-376. [DOI] [PubMed] [Google Scholar]
- 33.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36:1296-1300. [DOI] [PubMed] [Google Scholar]
- 34.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 35.Vu, T. H., T. Li, and A. R. Hoffman. 2004. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 13:2233-2245. [DOI] [PubMed] [Google Scholar]
- 36.Wang, J., J. Mager, Y. Chen, E. Schneider, J. C. Cross, A. Nagy, and T. Magnuson. 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28:371-375. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y., K. Joh, S. Masuko, H. Yatsuki, H. Soejima, A. Nabetani, C. V. Beechey, S. Okinami, and T. Mukai. 2004. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 124:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamasaki, K., K. Joh, T. Ohta, H. Masuzaki, T. Ishimaru, T. Mukai, N. Niikawa, M. Ogawa, J. Wagstaff, and T. Kishino. 2003. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 12:837-847. [DOI] [PubMed] [Google Scholar]
- 39.Yamasaki, Y., T. Kayashima, H. Soejima, A. Kinoshita, K. Yoshiura, N. Matsumoto, T. Ohta, T. Urano, H. Masuzaki, T. Ishimaru, T. Mukai, N. Niikawa, and T. Kishino. 2005. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 14:2511-2520. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y., T. Li, T. H. Vu, G. A. Ulaner, J. F. Hu, and A. R. Hoffman. 2003. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology 144:5658-5670. [DOI] [PubMed] [Google Scholar]