Abstract
In trypanosome RNA editing, uridylate (U) residues are inserted and deleted at numerous sites within mitochondrial pre-mRNAs by an ∼20S protein complex that catalyzes cycles of cleavage, U addition/U removal, and ligation. We used RNA interference to deplete TbMP18 (band VII), the last unexamined major protein of our purified editing complex, showing it is essential. TbMP18 is critical for the U-deletional and U-insertional cleavages and for integrity of the ∼20S editing complex, whose other major components, TbMP99, TbMP81, TbMP63, TbMP52, TbMP48, TbMP42 (bands I through VI), and TbMP57, instead sediment as ∼10S associations. Additionally, TbMP18 augments editing substrate recognition by the TbMP57 terminal U transferase, possibly aiding the recognition component, TbMP81. The other editing activities and their coordination in precleaved editing remain active in the absence of TbMP18. These data are reminiscent of the data on editing subcomplexes reported by A. Schnaufer et al. (Mol. Cell 12:307-319, 2003) and suggest that these subcomplexes are held together in the ∼20S complex by TbMP18, as was proposed previously. Our data additionally imply that the proteins are less long-lived in these subcomplexes than they are when held in the complete editing complex. The editing endonucleolytic cleavages being lost when the editing complex becomes fragmented, as upon TbMP18 depletion, should be advantageous to the trypanosome, minimizing broken mRNAs.
Trypanosomes are early diverging parasitic protozoa that cause debilitating diseases, such as African sleeping sickness, and exhibit numerous interesting biological properties. Notably, many of their mitochondrial transcripts undergo a unique form of processing, termed RNA editing, in which uridylate residues (U's) are inserted and deleted at specific locations within pre-mRNAs (reviewed in references 46, 48, and 49). This editing is essential for parasite viability (44) and is very extensive in certain transcripts, creating up to three-quarters of the codons and frequently the start and stop signals. Editing is directed by numerous short guide RNA molecules (gRNAs), which are complementary to regions of the edited transcript (5) and thus mismatch the unedited mRNA at each editing site. The first gRNA is also complementary to the segment just 3′ of the editing domain, so it can base pair to this region in the pre-mRNA, forming an “anchor duplex” that extends up to the first editing site. In each editing cycle (see Fig. 1A), the pre-mRNA is cleaved just upstream of the anchor duplex (5; see also reference 8), then U's are either added to or removed from the 3′ end of the upstream cleavage fragment by a terminal U transferase (TUTase) (4) or a 3′-U-exonuclease (3′-U-exo) (5; see also reference 38), and finally, the transcript is resealed by an RNA ligase (28, 38, 39, 44). Upon completion of an editing cycle, the anchor duplex extends up to the next mismatch to direct another cycle, and editing thus progresses 3′ to 5′ along the pre-mRNA.
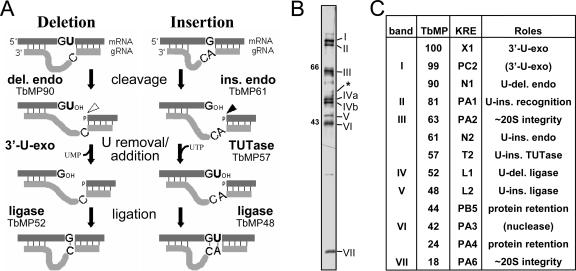
FIG. 1.
Mechanism of RNA editing and the purified editing complex. (A) Editing cycles, as described in the text, with the responsible enzymes indicated. U-deletional endonuclease (del. endo; cleavage shown with a hollow arrowhead) or U-insertional endonuclease (ins. endo; cleavage shown with a solid arrowhead) cleaves when the residue immediately upstream of the anchor duplex is a U or a purine, respectively, and is stimulated or inhibited by adenosine polyphosphates (8), such as AMP-CP. G and A represent either purine, and C represents a pyrimidine. (B) Silver staining of editing complex purified by Q-Sepharose and DNA cellulose chromatography, showing a different representative preparation from 667 cells than the four presented earlier (three in reference 38 and one other in reference 57), with seven major staining proteins that were designated band I (TbMP99) through band VII (TbMP18). (Band V [TbMP48] appears lighter upon photography because it silver stains brown, not black; band IV [TbMP52] has two isoforms in these cells [39]; and band III [TbMP63] exhibits microheterogeneity [20].) The asterisk indicates the position of the TbMP57 TUTase, identified by immunoblotting a comparable gel (57); that study also revealed that TbMP57 and band III (TbMP63) are present in the same relative abundance in total cell extract, indicating that TbMP57 was not lost during this purification. Size markers are in kDa. (C) Various nomenclatures for editing proteins. The columns show the original “band” designations from Rusche et al. (38), the TbMP designations (T. brucei mitochondrial protein, followed by the molecular weight of its cytoplasmic precursor) from Panigrahi et al. (33), and a more recent “KRE” nomenclature (kinetoplastid RNA editing, followed by a letter for the kind of protein and a number indicating ascending size within the group) (49). The literature, in addition to using these nomenclatures, also refers to TbMP52 as REL1 or DREL, TbMP48 as REL2 or IREL, and TbMP57 as TUT2. The final column shows a summary of the demonstrated (and proposed) roles in editing, as referenced in the Introduction; the role of TbMP18 is shown in this study.
The editing cycles are catalyzed by a protein complex, or editosome (22, 37, 38, 45), that has been purified by several different procedures. The first reported purification (38) yielded complex that catalyzes full editing cycles in vitro (9) and reproducibly contains seven major staining proteins (38), temporarily designated bands I through VII (see Fig. 1B). However, subsequently reported purification procedures yield complexes that catalyze precleaved editing especially actively and contain ∼20 noted proteins, designated TbMP# (see the legend to Fig. 1B) (31, 33, 35) and subsequently KREX# (see the legend to Fig. 1B) (49). These proteins, whose relative abundance appears to vary in different preparations, include bands I through VII and several critical enzymes that are minor staining components of the seven major protein complex (e.g., TbMP57 TUTase) (see Fig. 1B legend) (57). Curiously, all these preparations of the editing complex, with quite disparate numbers of major components, are reported to sediment at ∼20S in glycerol gradients. The basis for the differences in the major protein profiles between editing complex purifications remains unclear but does not appear to be due to a loss of important proteins during the former purification, as that purified complex catalyzes full U-deletional and U-insertional editing cycles the most actively of any preparation yet reported (11, 12) and at least TbMP57 is not selectively lost (57). Possibly the differences reflect the different cell lines used for the purifications, 667 in reference 38 and IsTar 1.7a in reference 31, 33, and 35, as 29.13 cells subjected to a latter kind of purification yielded complex with a pattern of major staining proteins (43) that appeared strikingly similar to the former preparations. Figure 1C summarizes various nomenclatures for these key proteins. For uniformity with the bulk of the editing literature, after their initial mention, we will use the TbMP# nomenclature.
Most of the proteins of the editing complex have been examined using biochemical and genetic approaches, almost all utilizing RNA interference (RNAi). Some of these proteins are the requisite enzymes. TbMP90 and TbMP61 are the endonucleases for U deletion and U insertion, respectively (7, 23, 32, 52), TbMP57 is the TUTase that adds the U's in U insertion (2, 15), and TbMP52 (band IV) and TbMP48 (band V) (38, 39, 44; see also references 28 and 40) are the RNA ligases that preferentially seal in U deletion and U insertion, respectively (12; see also references 19 and 43). Additionally, several proteins have been implicated as the 3′-U-exo: TbMP99 (band I) (43; see also reference 56), the related protein TbMP100 (24, 56), and TbMP42 (band VI) (6). Finally, mHel61p is a helicase that can augment editing (29). Several of the other editing proteins have critical structural and auxiliary roles. TbMP81 (band II) is required for protein and/or substrate recognition at each step of U insertion (26), including TbMP48 ligase retention (14, 30), but not for U deletion or for an ∼15S association of the other editing proteins (26, 30). TbMP63 (band III) is required for editing complex integrity, TbMP52 ligase retention, and U-deletional and U-insertional cleavages (20). Also, TbMP24 (42) and TbMP44 (53) appear essential for retention of all other examined editing proteins of the ∼20S complex. Indeed, all these noted structural proteins, as well as TbMP18 (band VII), have potential interaction domains, including homologous oligonucleotide/oligosaccharide binding (OB fold) domains and zinc fingers (reviewed in reference 56). Furthermore, other proteins that are not components of the ∼20S complex can affect editing, including TbMP108 (which adds U tails onto gRNAs) (2, 3), TbgBP21 and TbgBP25 (annealing stimulators) (1), RBP16 (a cytochrome b mRNA factor) (36) and REAP1 (a possible pre-mRNA binding component) (27).
The editing activities within the ∼20S editing complex appear functionally and spatially partitioned. The three parallel steps of U deletion and U insertion utilize distinct enzymes (7, 8, 10, 12, 19, 38, 52) and TbMP63 holds the U-deletional ligase (20, 34, 43) while TbMP81 holds the U-insertional ligase (14, 30, 43) and is also required for the other steps of U insertion (26). Notably, overexpressing affinity-tagged ligases enriches ∼5S to 10S subcomplexes that contain mainly TbMP81, TbMP57, and TbMP48 and catalyze precleaved U insertion or subcomplexes that contain mainly TbMP99, TbMP63, and TbMP52 and catalyze precleaved U deletion (43) (both types of subcomplexes also contain some TbMP18). Yeast two-hybrid analyses (43) also support these associations. Nonetheless, the U-deletional and U-insertional portions of the editing complex appear functionally interconnected, since RNAi of the U insertion-specific TbMP48 ligase allows the U-deletional TbMP52 ligase to also seal U insertion (14, 17, 30), and the U-insertional TUTase can act within the U-deletional cycle (57).
The current study assesses the role of TbMP18 and shows it is essential for viability, integrity of the ∼20S editing complex, and both U-deletional and U-insertional cleavage activities. The other editing activities and major editing proteins are not similarly dependent on TbMP18, and without it, the complex disassociates into subcomplexes reminiscent of those described by Schnaufer et al. (43).
MATERIALS AND METHODS
Cloning the TbMP18 gene.
Tryptic peptides of band VII (TbMP18), isolated from purified Trypanosoma brucei editing complex as shown in Fig. 1B, were subjected to Edman degradation at the Wistar Institute (see references 19 and 39), yielding sequences of 23 and 11 amino acids. PCR of trypanosome genomic DNA with degenerate primers generated a product that included seven additional confirmatory amino acids and was used to probe a cDNA library. The complete cDNA, providing 84 nucleotides (nt) of confirmatory information, contains a 165-codon coding region with 227 and 225 nt of 3′ and 5′ untranslated regions.
RNAi cell lines and growth analysis.
A total of 491 bp of TbMP18 coding region, PCR amplified from genomic DNA using 5′-ATTCTCGAGG CTAGCTCTTA CATCACGCCG-3′ and 5′-TGTAAGCTTT ACGACGGCAC ACCACTCTGG-3′ primers, was inserted between the opposing tetracycline (Tet)-inducible T7 promoters of the pZJM vector (54) and integrated into chromosomal ribosomal DNA of procyclic T. brucei 29.13 cells (as in references 20 and 30), which constitutively express T7 RNA polymerase and Tet repressor proteins (55). After extreme dilution, clonal cell lines that express double-stranded RNA from this TbMP18 segment upon induction were selected (as in references 20 and 30).
Extract preparation.
Traditional trypanosome mitochondrial extracts (18, 41), prepared from ∼4 × 1010 cells, and rapidly prepared small-scale extracts (20, 26, 39), from ∼2 × 108 cells, are here referred to as “traditional” and “rapid” extracts, respectively. Traditional extracts yield ample protein for glycerol gradient sedimentation but require approximately 6 to 8 h postlysis, during which time editing activities can become lost when the cells lack certain editing proteins (see, e.g., reference 26). Rapid extracts reduce preparation time to ∼1.5 h postlysis and minimize such secondary loss of activities (26).
Northern and Western blotting.
Northern blots of RNA from 2.5 × 107 cells (as in reference 30) were probed using the pZJM insert. For quantitative Western analysis, extract dilutions in twofold increments (≤3 μg protein) were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis mini gels (Bio-Rad), transferred to nitrocellulose membranes (Bio-Rad) in buffer (25 mM Tris, 0.2 M glycine, 20% methanol), and then blocked (1 h at 25°C) and probed (overnight at 10°C) in 1 × TNT (10 mM Tris pH 8.0, 150 mM NaCl, and 0.05% Tween 20) with 3% bovine serum albumin (fraction V; Sigma). Membranes for analyzing extracts were incubated first with an antibody to a load control mitochondrial protein, either lipoamide dehydrogenase (lip) (1:10,000; from R. L. Krauth-Siegel) or heat shock protein 70 (hsp) (1:10,000; from P. T. Englund), and then with a mixture of antibodies to (i) TbMP63 (1:2,000), TbMP48 (1:1,000), and TbMP18 (1:500), (ii) TbMP99 (1:500) and TbMP42 (1:1,000), subsequently reanalyzed with antibody to TbMP52 (1:500), or (iii) TbMP57 (1:2,000), subsequently reanalyzed with antibody to TbMP81 (1:500). Antibodies to the editing proteins were described previously (30). Membranes for analyzing glycerol gradient fractions were sliced horizontally and probed with individual antibodies at the dilutions listed above. All membranes were then washed (three times with the same buffer, 20 min each, at 25°C), incubated (1 h at 25°C) with horseradish peroxidase-conjugated goat anti-immunoglobulin G (1:5,000; Santa Cruz), washed again, and developed with the ECL-Plus chemiluminescence detection kit (Amersham). Bands were scanned using a FluorChem 8000 advanced fluorescence, chemiluminescence, and visible light-imaging system with AlphaEaseFC software. For each antibody, the signals from the control extract were graphed against the intended amount of protein loaded and the best-fit equation (all linear or quadratic equations, yielding R2 values of >0.96; Microsoft Excel) was used to calculate the values for the signals from the RNAi extracts, which were then corrected for loading of the lanes as determined by similar analysis used for the load control protein.
Glycerol gradient sedimentation and adenylylation.
Traditional extract (∼1 mg protein) was centrifuged in an SW 41 rotor (38,000 rpm, 14 h) on a 10 to 30% glycerol gradient, with thyroglobulin (19S) and catalase (11S) markers run in parallel (as in reference 19), and 15 (∼0.75-ml) fractions were collected from the bottom. From these fractions, adenylylation assays (following deadenylylation) (38) used 10 μl, Western analyses used 12 μl, and activity assays used 6 μl.
Activity assays.
Substrate RNAs (prepared as described in reference 26) were 3′- or 5′-end labeled and gel isolated (11, 20). The 20-μl reaction mixtures were in 10 mM KCl-MRB buffer [25 mM Tris-HCl (pH 8), 10 mM Mg(OAc)2, 1 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 10 mM KCl] freshly supplemented with 5 mM dithiothreitol and 0.8 U/ml RNasin (Promega), at 27°C for 30 or 45 min, unless noted. RNAs were resolved on 40-cm 9% polyacrylamide-7.5 M urea gels (or 15% polyacrylamide-7.5 M urea gels for experiments for Fig. 6 through 8) and visualized by autoradiography. As described in the next two paragraphs, we used two types of assays, which we refer to as “specific” and “basic.”
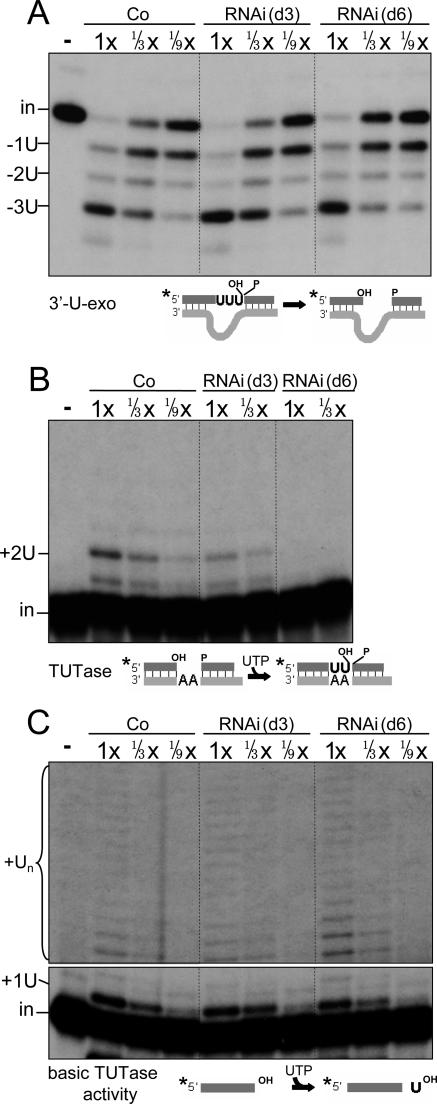
FIG. 6.
TbMP18 is not necessary for the 3′-U-exo activity but augments TUTase activity at an editing site. Assessments of specific 3′-U-exo activity (A), specific TUTase activity (B), and basic TUTase activity (C), using substrates diagramed below, are shown. (In panel C, 1x equals 0.24 μg protein.) In this and all subsequent experiments with precleaved substrates, the position of the labeled input oligo (in), partial (−1U and −2U) and complete (−3U) removal products, and complete addition product (+2U) are indicated. For basic TUTase activity, the input (in) and major product (+1U), as well as longer addition products (Un, likely produced by the p108 TUTase, shown in a darker exposure) are indicated.
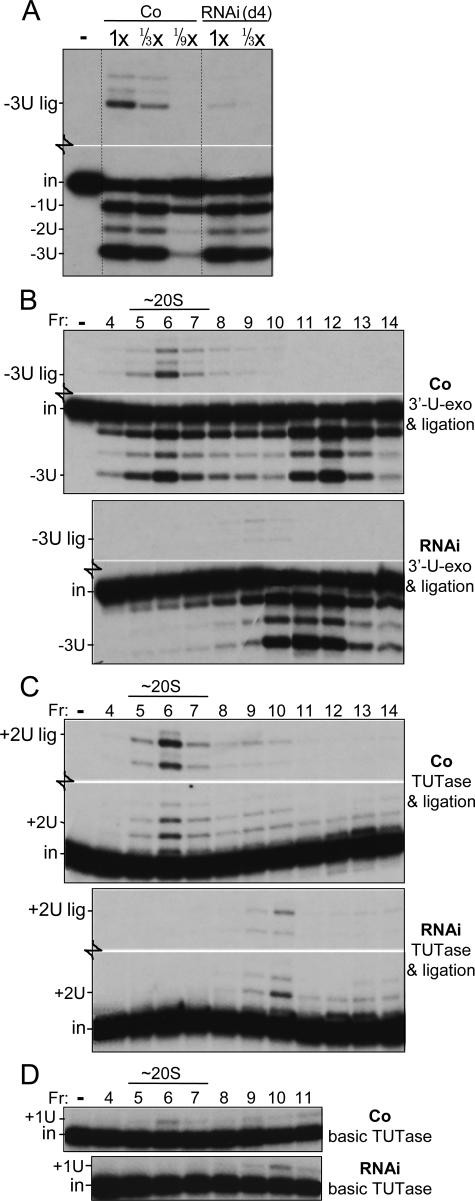
FIG. 8.
Assessment of precleaved editing in traditional extracts and their glycerol gradient fractions. (A and B) Precleaved U deletion activity was assessed as described for Fig. 7B except for use of traditional extracts (1× equals 1.2 μg protein) (A) or the indicated glycerol gradient fractions (B). (Control experiments [data not shown] indicate that ligation in the RNAi gradient fractions in panel B was not limited by inefficient ligase activation.) (C) Precleaved U insertion activity was assessed as described for Fig. 7C. To save space, in panels A through C, the blank middle regions of these gels are not shown. Assessment of the individual 3′-U-exo and TUTase activities (not shown) gave results similar to those shown in the lower gel regions of panels A and B and of panel C, respectively. (D) Basic TUTase activity was assessed as described for Fig. 6C. The TbMP108 TUTase sediments in fractions 12 and higher fractions (not shown).
(i) Specific editing assays.
The steps of the U-deletional and U-insertional cycles were assessed using standard editing substrates, either the 3′ portion of T. brucei ATPase 6 (A6) pre-mRNA m[0,4] (8, 22, 45) annealed to a gRNA (20) or three oligoribonucleotides (oligos) annealed to mimic a precleaved U-insertional editing site (with the RNAs described in reference 21) or a precleaved U-deletional editing site (with the RNAs described in reference 26). The following assays were used. (i) Cleavage assays used 3′-end-labeled A6 RNA and gRNA D33′ (11), which guides the removal of three U's at editing site 1, or gRNA I47G (20), which guides the addition of two U's (+2U) at editing site 2; ∼5% of the input mRNA becomes cleaved in the control reactions. These respective reactions contained potassium pyrophosphate (PPi) (0.25 or 0.3 mM), torula yeast RNA (4.5 ng or 45 pg), and AMP-CP (3 or 0 mM) (8, 12, 20, 26). (ii) The 3′-U-exo and TUTase assays used the precleaved substrates (see above), with the 16-nt (U-deletional) or 18-nt (U-insertional) upstream oligos 5′-end labeled. These respective reaction mixtures also contained PPi (0.25 or 0.5 mM) and UTP (0 or 2.4 mM) (20, 26). The 3′-U-exo assay also contained the single-stranded RNA gPCA6-2A (21) that lacks terminal U's, at an ∼5-fold molar excess over the labeled oligo. (iii) The U-deletional ligase assay used the oligos of the TUTase assay, with 0.3 mM ATP but no UTP or PPi (see references 20 and 26). This assay specifically scores the TbMP52 ligase because the TbMP48 ligase cannot efficiently seal a gapped substrate (12, 21); ∼10% of the input mRNA became ligated in the control reaction. U-insertional ligation could not be assessed individually, due to the overlapping substrate recognition by the U-deletional TbMP52 ligase (14, 17, 30). (iv) The U-deletional ligase was assessed in combination with the 3′-U-exo using the oligos of the 3′-U-exo assay with 0.3 mM ATP and 4.5 ng torula yeast RNA but no PPi (26). The U-insertional ligase was assessed in combination with the TUTase using the substrate of the TUTase assay with 2.4 mM UTP but no PPi. For assessing the combined final two steps of editing using fractions from the glycerol gradients, the U deletion reactions used no torula RNA, and the U insertion reactions used 0.15 mM ATP.
(ii) Basic editing assays.
Editing enzymes were assessed independently of their abilities to act at an editing site, using substrates that retain minimal features required for activity (26). (a) Assays for the basic U deletion-like cleavage and basic U insertion-like cleavage used 3′-end-labeled A6 RNA and a complementary oligo (AncES1 [5′GAUGCCAGGU AAGUAUUCUA UAACUCCA-3′] or D33′ [11]), respectively) that forms an anchor-like duplex directing cleavage just downstream of a U or a purine residue, respectively (26, 47). These reaction mixtures were supplemented as was done for the specific U-deletional cleavage assays except that AMP-CP was omitted for the U insertion-like reaction. (b) The basic TUTase assay, scoring an addition of one U to the 3′ end of a single-stranded RNA (4, 38), used only the upstream oligo of the specific TUTase assay, with 2.4 mM UTP, at 27°C for 15 min.
RESULTS
TbMP18 is essential for trypanosome viability.
The TbMP18 (band VII) protein was identified as the smallest of seven major (and additional minor) proteins that copurify (Fig. 1B) (38) and actively catalyze editing cycles (11, 12). These proteins also remain together and active through other chromatographic resins, velocity centrifugation, and native gel electrophoresis (38, 47). We cloned two similar versions of this gene, one that perfectly matched the subsequently published TbMP18 sequence (34) and one with six silent substitutions and two conservative substitutions, presumably allelic isoforms. Antibodies generated against the recombinant protein show that TbMP18 sediments with the ∼20S peak of other editing proteins (see Fig. 4A), demonstrating that it is almost exclusively in the editing complex.
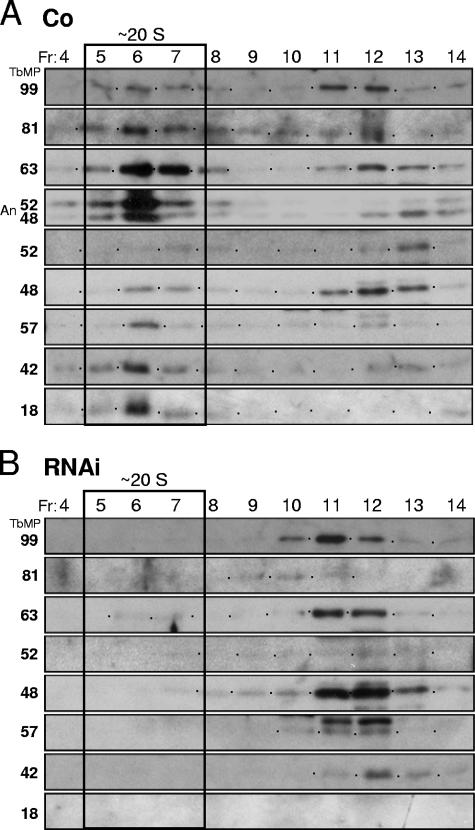
FIG. 4.
TbMP18 is critical for stability of the editing complex. Results of glycerol gradient sedimentation analysis using traditional extracts of control 29.13 cells (Co) (A) and day 4-induced RNAi cells (B), as used in Fig. 2D and 3C, are shown. The fractions (Fr; 15 = top) were assayed by Western blotting, using the indicated antibodies, and by adenylylation (An). The TbMP81 protein reproducibly appears unexpectedly faint in the RNAi fractions, relative to the abundance prior to centrifugation (Fig. 3C).
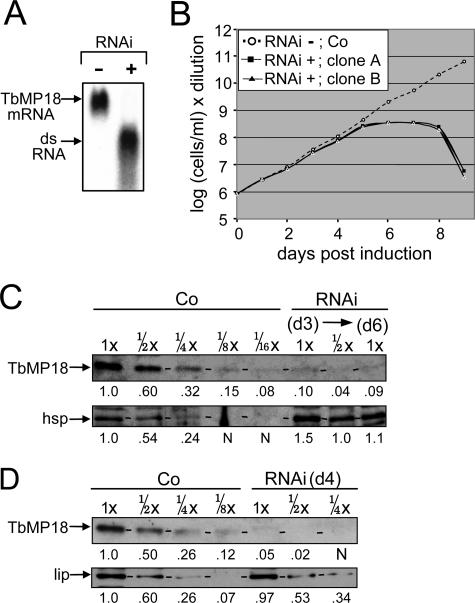
To examine the role of TbMP18, this protein was depleted by using RNAi (see Materials and Methods), and two clonal cell lines (RNAi cell lines) were utilized for all analyses and gave virtually identical results. Northern blotting at 24 h of RNAi induction confirmed efficient depletion of the TbMP18 mRNA as well as production of the double-stranded RNA (Fig. 2A). The induced RNAi cell lines cease expanding after five days and die after eight days, demonstrating TbMP18 is essential for viability (Fig. 2B).
FIG. 2.
RNAi reveals that TbMP18 is essential. (A) Northern blot showing the ∼1-kb TbMP18 mRNA and the ∼0.5-kb double-stranded RNA (dsRNA) transcript from uninduced (−) or 24-h-induced (+) RNAi cells. In this experiment and all subsequent experiments, additional clones gave analogous results. (B) Cell growth profile of two independent, clonal RNAi cell lines (RNAi) and control 29.13 cells (Co) following Tet addition. (C and D) Western blots showing TbMP18 and a load control protein (hsp or lip) from the control cells (Co) and RNAi cells, in rapid cell extracts (26) prepared on day 3 (d3) and day 6 (d6) postinduction (C) and in traditional mitochondrial extracts (18, 41) prepared on day 4 (d4) postinduction (D). The protein amount analyzed is indicated below the extract designation, with 1x equal to 3 μg. Dashes on these and subsequent protein gels indicate the position of the examined protein. Quantitation of the gels (see Materials and Methods) is shown below the lanes (N represents values not significantly above the background level); for the load control protein, the values represent the relative intensities, while for TbMP18, they represent the relative intensities after correcting for the signal from the load control protein.
TbMP18 protein levels were monitored by Western blotting using dilutions of rapid cell extracts (Fig. 2C) prepared at day 3 and day 6 of RNAi induction. The extent of depletion was determined relative to dilutions of control cell extracts prepared in parallel and then corrected for deviations in protein loading as determined from the amount of lip or hsp, mitochondrial proteins whose abundance is unaffected by RNA editing. This quantitation showed that TbMP18 is reduced to ∼1/12 to 1/16 of the control level at day 3 and not appreciably more reduced by day 6 (Fig. 2C). A traditional extract at day 4 of induction confirmed this efficient depletion (Fig. 2D).
TbMP18 is critical for integrity of the editing complex and augments stability of its component proteins.
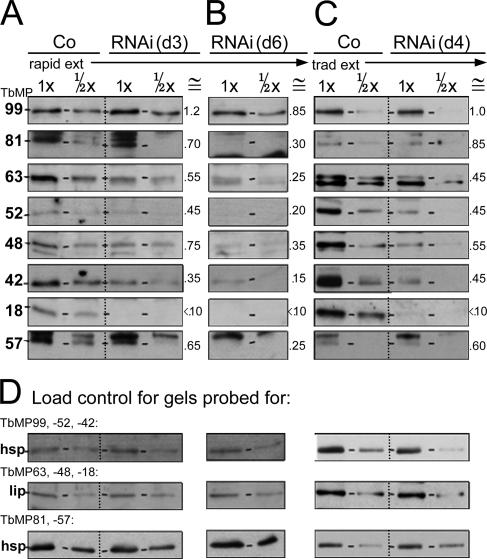
To determine whether other editing proteins are depleted upon TbMP18 RNAi, extracts were probed with antibodies against the six other major proteins of our editing complex (38) and TbMP57 TUTase (Fig. 3A to C). Again, extents of depletion were determined by comparison to dilutions of control extract (Fig. 3A to C) and corrected for loadings using the control protein (Fig. 3D), as done for Fig. 2 and described in its legend. At day 3 of RNAi, when TbMP18 is depleted to <10% of control levels but cell growth is unaffected, these other proteins remain at ≳1/2 of control levels (quantitation is given in the ≅ column of Fig. 3A). At day 6, when cell growth has ceased and therefore secondary effects may well arise, these proteins are reduced another ∼2-fold (Fig. 3B). Traditional extract shows a corresponding amount of these editing proteins (Fig. 3C), confirming that this lengthy extract preparation does not cause further protein loss from the TbMP18 RNAi cell extract. The fact that these proteins are lost to a much smaller extent than TbMP18 implies that their abundance is not directly dependent on this protein. Nonetheless, their more extensive decrease at later times implies they are less stable without TbMP18.
FIG. 3.
Retention of other editing proteins upon depletion of TbMP18. Results of Western analyses of the indicated proteins (left) from control 29.13 cells and RNAi cells, assessed from rapid cell extracts prepared at day 3 (d3) (A) and day 6 (d6) (B) of induction and from traditional extracts prepared at day 4 (d4) of induction (C) are shown. (D) Load controls for the different gels using either hsp or lip antibody, as indicated. For each antibody and type of extract, the gels were run and blotted together. The numbers below the extract designation indicate the relative amounts of extract protein loaded onto the gel, part of a larger analyzed range of extract dilutions. The ≅ columns of panels A through C indicate the approximate remaining abundance of each protein, relative to the corresponding load control band, as determined for TbMP18 in Fig. 2C and D (quantitation not shown for panel D). These numbers represent a combination of the values from quantitation of these bands and of some additional Western blotting analyses and are rounded to the nearest 0.05.
To investigate the association of the other editing proteins without TbMP18, we performed velocity centrifugation on the large-scale traditional extracts made in parallel from day 4-induced TbMP18 RNAi cells and control cells. Western blotting of the resultant fractions showed that the ∼20S complex is prominent in control extract (Fig. 4A) but is lost upon induction of the RNAi (Fig. 4B), implying that TbMP18 is critical for assembly and/or maintenance of this complex. The other editing proteins instead sediment in the ∼10S region (Fig. 4B), indicative of much smaller associations. This observation, in conjunction with the gradual decrease in abundance of these proteins following RNAi induction (Fig. 3A and B), suggests that these editing proteins are somewhat shorter-lived in the cell when the editing complex is disrupted. We conclude that TbMP18 is critical for integrity of the ∼20S editing complex.
Effect of TbMP18 RNAi on editing activities.
To determine how depletion of TbMP18 affects editing, the individual steps of U deletion and U insertion (Fig. 1A) were assessed using rapid extracts prepared at day 3 (before growth inhibition, when primary effects of TbMP18 loss are evident) and at day 6 (after growth inhibition, when secondary effects may become more prominent). Such rapid extracts can retain editing activities that diminish during the lengthy traditional extract preparation (26; see Materials and Methods). All experiments used extract dilutions to ensure that assessment was in the approximately linear range, allowing quantitation by comparison to control extracts, and multiple clonal lines gave confirmatory results.
(i) The cleavage steps of editing.
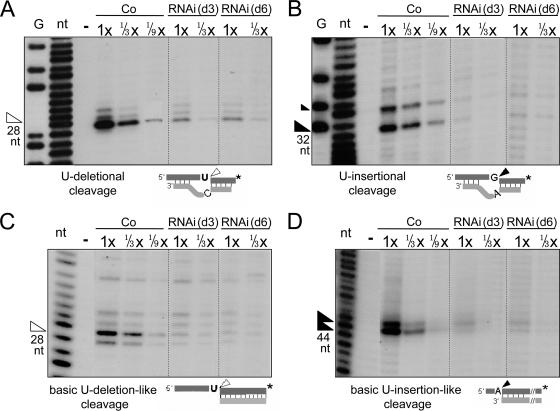
We specifically scored the U-deletional and U-insertional cleavage activities independent of the subsequent editing steps by using 3′-end-labeled mRNA to visualize the downstream cleavage products, which are unaffected by the 3′-U-exo or TUTase steps, and adding PPi to inhibit the ligases. Upon TbMP18 depletion, the U-deletional (Fig. 5A) and U-insertional (Fig. 5B) cleavage activities are greatly reduced, both to ≲1/9 of the control level at day 3 and similarly reduced at day 6. Controls (data not shown) demonstrate that this is an actual decrease in cleavage activity and not an altered response to AMP-CP, which oppositely affects U-deletional and U-insertional cleavages (8). Since the decreases in cleavage activities closely parallel the extent and kinetics of TbMP18 loss, and fragmentation of the ∼20S editing complex, these cleavages appear dependent on TbMP18.
FIG. 5.
TbMP18 is essential for both U-deletional and U-insertional cleavages. (A and B) Assessments of specific editing cleavage activities at U-deletional (A) and U-insertional (B) editing sites. (The fainter 34-nt product above the main band in panel B represents cleavage at the next U-insertional editing site.) The reactions contain and lack AMP-CP, respectively. (C and D) Assessments of the basic activities of the U-deletional (C) and U-insertional (D) endonucleases. The assay for the latter generates two main products (26). These experiments used rapid extracts prepared from control or RNAi cells induced for 3 or 6 days, as indicated. In this and subsequent experiments, the protein amount analyzed is indicated below the extract designation and 2.4 μg was used as 1× (unless otherwise noted). G and nt, G and nucleotide ladders from treatment of this mRNA with NaOH and RNase T1; −, no extract; arrowhead, the expected product (size shown below). In the substrate diagrams of this figure and subsequent figures, the asterisk represents the 32P label of the mRNA strand.
We next scored the basic activity of the U-insertional and U-deletional endonucleases, as there are examples where RNAi cells have lost cleavage at an editing site but retain the basic cleavage activity of that nuclease (26). These basic assays use RNA substrates with simple 5′ single-strand/3′ double-strand junctions, which resemble partial editing sites (diagramed in Fig. 5C and D). These U deletion-like and U insertion-like basic cleavage substrates have U's and purines just upstream of the anchor duplex, respectively, and their cleavage is stimulated or inhibited by AMP-CP (26), like the analogous editing sites (8). Both basic cleavage activities are diminished about as much as the cleavages at the actual editing sites at day 3 as well as day 6 of TbMP18 RNAi induction (Fig. 5C and D). Furthermore, the residual activities retain their normal AMP-CP sensitivities (data not shown). The comparable decreases in these basic cleavages and the editing site cleavages indicate that the U-deletional and U-insertional endonucleases have not simply lost their abilities to recognize editing sites and suggest that they may instead be depleted.
(ii) The 3′-U-exo step of U deletion and the TUTase step of U insertion.
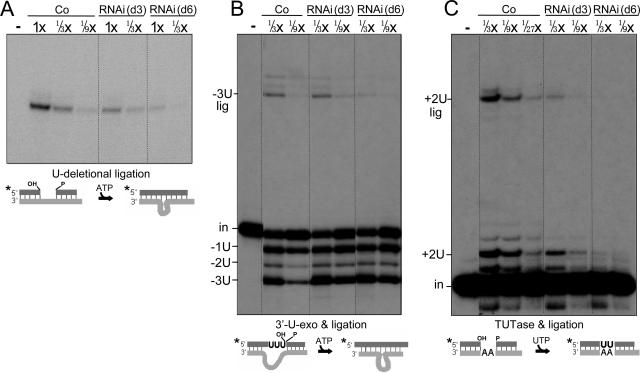
To specifically assess the second step of U deletion, which is U removal from the 3′ end of the upstream cleavage fragment by the 3′-U-exo, we used a precleaved U-deletional substrate (26) and PPi to inhibit ligation. TbMP18 RNAi causes no significant decrease in 3′-U-exo activity (Fig. 6A). In fact, at day 3 of induction, this activity consistently appears increased ∼2-fold (Fig. 6A). Confirming that this activity is U specific, the next residue, a single-stranded A, is retained (Fig. 6A). Therefore, unlike the endonuclease activities, the 3′-U-exo activity does not require TbMP18.
To specifically assess the second step of U-insertional editing, which is U addition to the 3′ end of the upstream cleavage fragment by the TbMP57 TUTase (2, 15), we used a precleaved U-insertional substrate (21) and PPi. At day 3 of RNAi, the guided addition of two U's is reduced to ∼1/4 of the control level (Fig. 6B). This reduction is less extensive than for the cleavage activities (Fig. 5) and the TbMP18 protein (Fig. 2C) but more extensive than that for the TbMP57 protein (Fig. 3A). This result suggests that TbMP18 is not absolutely required for the TUTase step of U insertion but that it stimulates this reaction, and this could be through direct binding or through indirect associations. By day 6, the TUTase activity is further reduced (Fig. 6B), possibly reflecting the instability of the smaller protein associations.
We next used a basic assay for TUTase activity, to address whether loss of TbMP18 affects the TUTase activity itself or its specific action at an editing site. In this basic assay (4), which measures U addition onto the 3′ end of a single-stranded RNA that is not an editing substrate, the TbMP57 TUTase preferentially adds one U (2, 3, 15). (The p108 TUTase, which adds the U tail onto gRNAs, is reported to preferentially add numerous U's onto such substrates [2, 3].) Interestingly, this basic activity that adds preferentially one U residue is not appreciably diminished at day 3 of the RNAi and is only slightly reduced at day 6 (Fig. 6C). This occurs despite the amount of TbMP57 protein being reduced (Fig. 3A and B), suggesting that the basic TUTase activity is somewhat more robust when the ∼20S editing complex is no longer intact (Fig. 4B), as previously proposed from other RNAi analyses (26). Thus, TbMP18 appears to stimulate functional recognition of an editing substrate by TUTase, not its basic activity that adds preferentially one U residue, reminiscent of the role of TbMP81 (26).
(iii) Ligation in U deletion.
In the specific assay for U-deletional ligation (20; see also reference 26), we utilized a precleaved substrate that mimics a U deletion substrate after U removal and requires ligation across a small gap in gRNA base pairing (diagramed in Fig. 7A) (20, 21), which is disfavored by the U-insertional ligase (12). This ligation activity is slightly reduced at day 3 of RNAi and is somewhat more reduced at day 6 (Fig. 7A). This decrease is much less extensive than that of TbMP18 (Fig. 2C) and roughly parallels the slow reduction in abundance of TbMP52, the U-deletional ligase (Fig. 3A and B), suggesting that it is an indirect effect of TbMP18 loss. Furthermore, the basic activity of this ligase, measured by dimerization of a single-stranded RNA (20), and its specific ligation activity are reduced to similar extents (Fig. 7A), indicating that editing substrate recognition by this ligase is not impaired. Thus, TbMP18 is not required for TbMP52 ligase action on a U-deletional substrate.
FIG. 7.
TbMP18 is not critical for the U-deletional and U-insertional ligation activities. Assessments of U-deletional ligation (A), precleaved U deletion (3′-U-exo plus ligation) (B), and precleaved U insertion (TUTase plus ligation) (C) used the substrates diagramed below, as described in the text. (In panel A, 1× equals 1.2 μg protein.) The fully edited −3U lig and +2U lig products are indicated.
To discern whether the remaining activity of the U-deletional ligase is still coordinated with the 3′-U-exo activity when TbMP18 is depleted, we assessed these activities together using the precleaved U deletion substrate of Fig. 6A but omitting PPi and including ATP. This coupled reaction generates approximately equal amounts of edited product from day 3 RNAi and control extracts (Fig. 7B, −3U lig band). This likely reflects the RNAi extracts having somewhat increased 3′-U-exo activity (Fig. 6A and Fig. 7B, −3U band), and decreased ligation efficiency (Fig. 7A), the combination of which generates amounts of edited product similar to those of the control. These data indicate that TbMP18 is not required for coordination of the last two steps of U deletion.
(iv) Ligation in U insertion.
The ligation step of U insertion, although not straightforward to score as an independent reaction (see reference 26), can be meaningfully assessed in conjunction with the TUTase step by using the precleaved substrate of Fig. 6B but omitting PPi (see reference 21). Since the U insertion-specific ligase (TbMP48), but not the U-deletional ligase (TbMP52), is preadenylylated in these extracts (see, e.g., reference 12), performing this assay without added ATP makes it selective for the former ligase (20, 26; see also reference 21). In this assay (Fig. 7C), at day 3 of induction, the edited product from the combined TUTase and U-insertional ligation steps is ∼1/9 of the control level (band +2U lig). However, the product of the TUTase step is reduced to ∼1/5 of the control level (Fig. 7C, sum of the +2U and +2U lig bands), as expected from the individual TUTase assay (Fig. 6B), suggesting that the product of the actual ligation step is reduced to ∼1/2 of the control level. (Both assays are further reduced at day 6 [Fig. 7C].) This is consistent with the slow reduction in TbMP48 abundance (Fig. 3A and B). We thus conclude that TbMP18 is not directly required for effective coordination of the last two steps of U insertion.
(v) Precleaved editing activities in traditional extracts and gradient fractions.
Finally, to assess the sedimentation properties of editing activities remaining upon TbMP18 RNAi, we utilized the larger-scale, traditional extracts. After this multihour preparation of RNAi cell extract, the U-deletional ligase no longer functions normally in conjunction with the 3′-U-exo (Fig. 8A and B), in contrast to what is observed with rapid extracts (Fig. 7A and B), suggesting that TbMP18 helps stabilize this editing activity during the lengthy extract preparation. However, the 3′-U-exo activity remains active in this traditional extract of the RNAi cells (Fig. 8A, lower panel). Upon glycerol gradient centrifugation, this activity sediments at ∼10S (Fig. 8B), along with the bulk of the editing proteins (Fig. 4), where some is also commonly found in control cell gradients (Fig. 8B) (see also reference 57). This is also the approximate region of the gradient where the precleaved U deletion subcomplex has been observed (43). Thus, TbMP18 appears to help stabilize the ligation but not the 3′-U-exo activity of precleaved U deletion.
In precleaved U insertion assays, the traditional extracts of the TbMP18 RNAi cells generate reduced TUTase product and, hence, reduced edited product relative to that generated by the control cell extract (Fig. 8C and data not shown), as expected from the studies of the rapid extracts (Fig. 6B and 7C). Upon glycerol gradient centrifugation, the residual U-insertional TUTase activity (sum of the +2U and +2U lig bands) and the U-insertional ligation activity that generates the edited product (+2U lig band) both peak at ∼12S (Fig. 8C). The basic activity of the TbMP57 TUTase that preferentially adds one U (Fig. 8D) cosediments with the editing-site-specific +2U activity (Fig. 8C) and does not appear diminished in the RNAi gradient relative to the control gradient. These data reinforce that TbMP18 is not required for the basic U addition activity of the TbMP57 TUTase but stimulates its activity at an editing site.
DISCUSSION
This study uses RNAi to address the role of TbMP18, the smallest major protein of our ∼20S editing complex from T. brucei (Fig. 1B and 4A) (see also reference 38). Using dilutions of extract prepared at various time points following RNAi induction, protein levels and individual editing activities were examined. The RNAi reduces TbMP18 to <10% of control levels (Fig. 2C and D), and a few days later the cells stop growing and subsequently die (Fig. 2B), demonstrating that this protein is essential. Prior to eliciting any growth effect, TbMP18 depletion causes a loss of the ∼20S editing complex (Fig. 4). The other major editing proteins are not similarly depleted (Fig. 3) but sediment as smaller associations (Fig. 4B) and slowly decrease in abundance (Fig. 3). Thus, TbMP18 is critical for integrity of the ∼20S editing complex, and without it, the other major editing proteins appear somewhat less long-lived than they are in the native complex.
There is precedent for editing proteins having markedly reduced stability when separate from their binding partners, including the inability to overexpress native TbMP52 ligase in vivo (39), loss of endogenous TbMP52 or TbMP48 upon RNAi of TbMP63 or TbMP81 (14, 20, 30, 53), their binding partners (43), and depletion of TbMP44 (42) and TbMP24 (53), causing the other examined editing proteins to appear almost entirely lost. In the cases of TbMP44 and TbMP24 depletion (42, 53), it remains unclear whether these other proteins were lost to a greater extent than occurs upon TbMP18 RNAi, which could suggest different modes of action, or possibly to a similar extent, as one amount was assayed and antibody responses can be very steep (Fig. 3). Upon TbMP18 RNAi, the gradual decrease of the other editing proteins (Fig. 3A and B) most likely reflects their not being protected in the ∼20S editing complex, indicating that without TbMP18, this complex is disrupted in vivo.
The effect of TbMP18 depletion on editing was determined by assessing the individual editing steps and, for affected steps, also assessing the basic activities of those enzymes, to distinguish loss of enzymatic activity from loss of substrate recognition (as shown can occur in reference 26). Paralleling TbMP18 depletion, the U-deletional and U-insertional cleavage steps as well as the basic activities of those enzymes are reduced to ∼10% of control levels (Fig. 5), indicating that TbMP18 is required for these activities. At the second editing step, the 3′-U-exo remains active (Fig. 6A and 7B). Indeed, it is increased in TbMP18 RNAi cell extracts, which could reflect the enzyme having greater substrate access when not in the ∼20S editing complex; such has been suggested previously, when the editing complex is disassembled by other treatments (26). In contrast, the TUTase step of U insertion is somewhat reduced at day 3 of the RNAi, although less than the TbMP18 protein, and diminishes further after growth inhibition (Fig. 7B), whereas the basic activity of this TbMP57 TUTase is not appreciably diminished (Fig. 8D; see also Fig. 6C), indicating that TbMP18 augments, but is not required for, substrate recognition by this TUTase. At the third editing step, the U-deletional and U-insertional ligases remain coordinated with the previous U addition or U removal step and seal their substrates (Fig. 7B and C) approximately as efficiently as expected from the slightly reduced abundances of those ligases (Fig. 3A and B); thus, TbMP18 is not critical for the ligation steps. Therefore, TbMP18 is required for ∼20S editing complex integrity and the U-deletional and U-insertional cleavage activities but not for the other activities or their coordination, although it augments substrate recognition by the TUTase and stability of the other major editing proteins.
These assessments of editing activities utilized rapidly prepared cell extracts, which we earlier found retain several editing activities of TbMP81 RNAi cells much more efficiently than multihour traditional mitochondrial extracts (26). Similar to the case with TbMP81 RNAi cells, the ability of the U-deletional ligase to seal the 3′-U-exo product, which is seen to be retained at nearly control levels when using rapid extracts (Fig. 7B), becomes lost during the lengthy, traditional extract preparation (Fig. 8A). Nonetheless, when scoring control cells, this precleaved U deletion is catalyzed with impressively similar efficiency by comparable amounts of rapid and traditional extracts. This difference in the retention of ligation efficiency from RNAi cells underscores the value of assessing activities of depleted cells using rapid extracts.
Previous studies depleting TbMP81 by RNAi revealed that this protein is required for U-insertional substrate recognition by the TUTase, as well as for the other two steps of U insertion (26), and this recognition may be facilitated by its OB fold domain (and/or zinc fingers) (15, 26, 43). Since TbMP18 also contains a similar OB fold domain (34, 56), it might aid TUTase recognition by augmenting the action of TbMP81, using this domain.
Upon overexpressing affinity-tagged ligases in trypanosomes, Schnaufer et al. (43) observed accumulation of ∼5S-10S subcomplexes that catalyze the coordinated final two steps of U deletion or U insertion, but not the endonucleolytic cleavage steps, features akin to what we observe upon TbMP18 depletion. Their ∼5S-10S subcomplexes contain mainly TbMP99, TbMP63, and affinity-tagged TbMP52 or mainly TbMP81, TbMP57, and affinity-tagged TbMP48, respectively, and both preparations appear to also include a small amount of TbMP18 (43). Yeast two-hybrid studies showing that TbMP18 interacts with TbMP81 and TbMP63 prompted the hypothesis that TbMP18 helps to hold those subcomplexes together within the ∼20S complex (43). Our current study provides evidence for this, as TbMP18 depletion disrupts the ∼20S editing complex and yields associations that can coordinate the final steps of editing and sediment at ∼10S. TbMP18 may also help to stabilize those subcomplexes, since without it, their protein components slowly diminish in abundance (Fig. 3) and some editing activities appear unstable to the lengthy traditional extract preparation (Fig. 8A and B). Furthermore, both Schnaufer et al. (43) and we (Fig. 4) find that the ratios of the proteins ascribed to each subcomplex vary across the ∼10S region of the gradients, implying there may be multiple kinds of subcomplexes. Suggesting that these various populations have different levels of activity, upon TbMP18 RNAi, the residual precleaved U insertion corresponds to the slower-sedimenting portion of the subcomplexes (Fig. 8C). Furthermore, because TbMP18 augments TUTase activity (Fig. 6B), in the pooled subcomplexes studied by Schnaufer et al. (43), those that contain TbMP18 may be more active than those that do not. Thus, the present study of TbMP18 complements the study of editing subcomplexes by Schnaufer et al. (43).
The U-deletional and U-insertional endonucleolytic cleavages, the first committed steps of the editing cycles, are lost upon RNAi of TbMP18 (this study) or of TbMP63 (20) (proteins that are not the endonucleases [7, 52]) and these cleavages were also not observed from the 5S-10S editing subcomplexes generated in vivo (43). In all of these cases, the ∼20S complex is fragmented into smaller associations, suggesting that maintenance of these cleavage activities in the trypanosome requires a fairly intact editing complex. Nonetheless, cleavage can be catalyzed by less than the full complement of these editing proteins when they are synthesized in vitro (23). We hypothesize that it is biologically advantageous for the trypanosome to restrict these cleavages to occur only when the complex is poised to complete the full editing cycle, as this should minimize accumulation of cleaved mRNAs, which would likely be lost from the productive maturation pathway. Furthermore, to favor rejoining of previously cleaved transcripts, it would seem advantageous if the later steps of editing remain active, or even show increased activity, when the ∼20S complex is dissociated, and that was also observed (Fig. 7 and 8) (see also references 20 and 43). Observations of in vivo editing further support this idea. In healthy cells, the vast majority of the mRNAs are intact, even for COIII transcripts, where most molecules contain substantial misediting (13, 16) (see also references 25, 50, and 51), potentially allowing subsequent reediting to generate a functional mRNA (8). However, cells in which the essential editing ligase has been mutationally inactivated do accumulate mRNAs that are broken at an editing site (19). Thus, the editing machinery may have evolved to help maintain the integrity of the mRNA substrate by constraining the cleavages to occur only when the mRNA can be religated.
Acknowledgments
We thank Paul Englund and present and past members of his and our laboratories for helpful discussions, Laura Rusche for purification of the editing complex, David Reim at the Wistar Institute for sequencing the tryptic peptides, Mike Hemann for cloning the band VII gene, Ken Piller for the procyclic strain 667 cDNA library, Paul Englund for the pZJM vector and heat shock protein 70 antibody, R. Louise Krauth-Siegel for lipoamide dehydrogenase antibody, and the reviewers for helpful comments.
This work was supported by NIH grant GM34231.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, and L. Simpson. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9:62-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev, R., I. Aphasizheva, and L. Simpson. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. USA 100:10617-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. [DOI] [PubMed] [Google Scholar]
- 4.Bakalara, N., A. M. Simpson, and L. Simpson. 1989. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 264:18679-18686. [PubMed] [Google Scholar]
- 5.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Brecht, M., M. Niemann, E. Schluter, U. F. Muller, K. Stuart, and H. U. Goringer. 2005. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell 17:621-630. [DOI] [PubMed] [Google Scholar]
- 7.Carnes, J., J. R. Trotter, N. L. Ernst, A. Steinberg, and K. Stuart. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 102:16614-16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Reyes, J., L. N. Rusche, K. J. Piller, and B. Sollner-Webb. 1998. T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell 1:401-409. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Reyes, J., L. N. Rusche, and B. Sollner-Webb. 1998. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 26:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Reyes, J., and B. Sollner-Webb. 1996. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93:8901-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Reyes, J., A. Zhelonkina, L. Rusche, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Reyes, J., A. G. Zhelonkina, C. E. Huang, and B. Sollner-Webb. 2002. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 22:4652-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker, C. J., and B. Sollner-Webb. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61:1001-1011. [DOI] [PubMed] [Google Scholar]
- 14.Drozdz, M., S. S. Palazzo, R. Salavati, J. O'Rear, C. Clayton, and K. Stuart. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, N. L., B. Panicucci, R. P. Igo, Jr., A. K. Panigrahi, R. Salavati, and K. Stuart. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell 11:1525-1536. [DOI] [PubMed] [Google Scholar]
- 16.Feagin, J. E., J. M. Abraham, and K. Stuart. 1988. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53:413-422. [DOI] [PubMed] [Google Scholar]
- 17.Gao, G., and L. Simpson. 2003. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 278:27570-27574. [DOI] [PubMed] [Google Scholar]
- 18.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 19.Huang, C. E., J. Cruz-Reyes, A. G. Zhelonkina, S. O'Hearn, E. Wirtz, and B. Sollner-Webb. 2001. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 20:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, C. E., S. F. O'Hearn, and B. Sollner-Webb. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igo, R. P., Jr., S. S. Palazzo, M. L. Burgess, A. K. Panigrahi, and K. Stuart. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kable, M. L., S. D. Seiwert, S. Heidmann, and K. Stuart. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189-1195. [DOI] [PubMed] [Google Scholar]
- 23.Kang, X., G. Gao, K. Rogers, A. M. Falick, S. Zhou, and L. Simpson. 2006. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. USA 19:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, X., K. Rogers, G. Gao, A. M. Falick, S. Zhou, and L. Simpson. 2005. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. USA 102:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koslowsky, D. J., G. J. Bhat, L. K. Read, and K. Stuart. 1991. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell 67:537-546. [DOI] [PubMed] [Google Scholar]
- 26.Law, J. A., C. E. Huang, S. F. O'Hearn, and B. Sollner-Webb. 2005. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell. Biol. 25:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison-Antenucci, S., and S. L. Hajduk. 2001. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell 7:879-886. [DOI] [PubMed] [Google Scholar]
- 28.McManus, M. T., M. Shimamura, J. Grams, and S. L. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missel, A., A. E. Souza, G. Norskau, and H. U. Goringer. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Hearn, S. F., C. E. Huang, M. Hemann, A. Zhelonkina, and B. Sollner-Webb. 2003. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell. Biol. 23:7909-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panigrahi, A. K., T. E. Allen, K. Stuart, P. A. Haynes, and S. P. Gygi. 2003. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J. Am. Soc. Mass Spectrom. 14:728-735. [DOI] [PubMed] [Google Scholar]
- 32.Panigrahi, A. K., N. L. Ernst, G. J. Domingo, M. Fleck, R. Salavati, and K. D. Stuart. 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panigrahi, A. K., S. P. Gygi, N. L. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. S. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. D. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., S. P. Gygi, N. L. Ernst, S. S. Palazzo, D. S. Weston, R. Aebersold, R. Salavati, and K. D. Stuart. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panigrahi, A. K., A. Schnaufer, N. L. Ernst, B. Wang, N. Carmean, R. Salavati, and K. Stuart. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier, M., and L. K. Read. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard, V. W., M. E. Harris, and S. L. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rusche, L. N., J. Cruz-Reyes, K. J. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rusche, L. N., C. E. Huang, K. J. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatini, R., and S. L. Hajduk. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 270:7233-7240. [DOI] [PubMed] [Google Scholar]
- 41.Sabatini, R. S., B. K. Adler, S. Madison-Antenucci, M. T. McManus, and S. L. Hajduk. 1998. Biochemical methods for analysis of kinetoplastid RNA editing. Methods 15:15-26. [DOI] [PubMed] [Google Scholar]
- 42.Salavati, R., N. L. Ernst, J. O'Rear, T. Gilliam, S. Tarun, Jr., and K. Stuart. 2006. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA 12:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnaufer, A., N. L. Ernst, S. S. Palazzo, J. O'Rear, R. Salavati, and K. Stuart. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12:307-319. [DOI] [PubMed] [Google Scholar]
- 44.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, Jr., E. Wirtz, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 45.Seiwert, S. D., S. Heidmann, and K. Stuart. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831-841. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, L., S. Sbicego, and R. Aphasizhev. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sollner-Webb, B., L. N. Rusche, and J. Cruz-Reyes. 2001. Ribonuclease activities of trypanosome RNA editing complex directed to cleave specifically at a chosen site. Methods Enzymol. 341:154-174. [DOI] [PubMed] [Google Scholar]
- 48.Stuart, K., and A. K. Panigrahi. 2002. RNA editing: complexity and complications. Mol. Microbiol. 45:591-596. [DOI] [PubMed] [Google Scholar]
- 49.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30:97-105. [DOI] [PubMed] [Google Scholar]
- 50.Sturm, N. R., D. A. Maslov, B. Blum, and L. Simpson. 1992. Generation of unexpected editing patterns in Leishmania tarentolae mitochondrial mRNAs: misediting produced by misguiding. Cell 70:469-476. [DOI] [PubMed] [Google Scholar]
- 51.Sturm, N. R., and L. Simpson. 1990. Partially edited mRNAs for cytochrome b and subunit III of cytochrome oxidase from Leishmania tarentolae mitochondria: RNA editing intermediates. Cell 61:871-878. [DOI] [PubMed] [Google Scholar]
- 52.Trotter, J. R., N. L. Ernst, J. Carnes, B. Panicucci, and K. Stuart. 2005. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell 20:403-412. [DOI] [PubMed] [Google Scholar]
- 53.Wang, B., N. L. Ernst, S. S. Palazzo, A. K. Panigrahi, R. Salavati, and K. Stuart. 2003. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot. Cell 2:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 55.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 56.Worthey, E. A., A. Schnaufer, I. S. Mian, K. Stuart, and R. Salavati. 2003. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 31:6392-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhelonkina, A. G., S. F. O'Hearn, J. A. Law, J. Cruz-Reyes, C. E. Huang, V. S. Alatortsev, and B. Sollner-Webb. 2006. T. brucei RNA editing: action of the U-insertional TUTase within a U-deletion cycle. RNA 12:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]