Abstract
Early differentiation of B lymphocytes requires the function of multiple transcription factors that regulate the specification and commitment of the lineage. Loss- and gain-of-function experiments have provided important insight into the transcriptional control of B lymphopoiesis, whereby E2A was suggested to act upstream of EBF1 and Pax5 downstream of EBF1. However, this simple hierarchy cannot account for all observations, and our understanding of a presumed regulatory network, in which transcription factors and signaling pathways operate, is limited. Here, we show that the expression of the Ebf1 gene involves two promoters that are differentially regulated and generate distinct protein isoforms. We find that interleukin-7 signaling, E2A, and EBF1 activate the distal Ebf1 promoter, whereas Pax5, together with Ets1 and Pu.1, regulates the stronger proximal promoter. In the absence of Pax5, the function of the proximal Ebf1 promoter and accumulation of EBF1 protein are impaired and the replication timing and subcellular localization of the Ebf1 locus are altered. Taken together, these data suggest that the regulation of Ebf1 via distinct promoters allows for the generation of several feedback loops and the coordination of multiple determinants of B lymphopoiesis in a regulatory network.
Early B-cell differentiation is a highly regulated process in which a multipotential progenitor cell is converted into a cell that expresses the B-cell antigen receptor. Several transcription factors and signaling pathways have been implicated in the regulation of this process. In particular, the transcription factors Ikaros and Pu.1 are involved in the regulation of steps preceding the generation of common lymphoid progenitors (CLPs). Pu.1-deficient mice lack B cells, T cells, granulocytes, and monocytes, and they have reduced numbers of multipotential progenitors (44, 45). Pu.1 has been shown to regulate the expression of the Flt3 gene and the interleukin-7 receptor (IL-7R) α gene (7). Both signaling pathways are important for the generation of B cells as mice lacking both Flk2/Flt3 and IL-7R fail to develop B-lineage cells in fetal liver and bone marrow, which are sites for fetal and adult lymphopoiesis, respectively (49). IL-7 signaling results in the activation of the STAT5a/b transcription factors, and consistent with their presumed role in IL-7 signaling, the lymphoid defect in mice carrying a targeted mutation in the IL-7Rα gene can be rescued by expression of a constitutively active form of STAT5 (15). Recent analysis of IL-7-deficient mice showed that the number of CLPs is not significantly changed, but the ability to differentiate into B-lineage cells is greatly diminished in vitro (8).
Several lines of evidence suggest that the specification of the B-cell fate is regulated by the transcription factors early B-cell factor 1 (EBF1) and E2A. First, the targeted inactivations of the Ebf1 and E2a genes result in similar blocks of B-cell differentiation, preceding the onset of rearrangement of the immunoglobulin heavy chain D and JH segments (3, 25, 60). In addition, EBF1 and E2A appear to synergize in the activation of B-lineage gene expression because Ebf1/E2a double-heterozygous mutant mice have a more severe defect in B-cell differentiation than the single-heterozygous mice (35). Finally, forced expression of EBF1 in hematopoietic progenitor cells skews the differentiation along the B-cell pathway (58), and genetic bypass experiments have shown that EBF1 can promote B-cell differentiation in hematopoietic progenitors that are deficient in either Pu.1 or E2A (29, 46).
Another important event in the differentiation of B-lineage cells, the commitment step, is regulated by the transcription factor Pax5 (32). Pax5-deficient mice generate pro-B cells that express early B-cell markers and undergo D-to-JH and proximal VH-to-DJH rearrangements (32). However, Pax5−/− pro-B cells of the bone marrow, which are arrested at that stage, show an unusual lineage plasticity and can differentiate into other cell types upon stimulation with different cytokines (54). Pax5 is also required continuously for the maintenance of the B-cell identity (37, 41). Based on these data, a model for the transcriptional control of B-cell differentiation has been proposed, in which EBF1 and E2A regulate the specification of the B-cell fate, whereas Pax5 is required for the continuous commitment to this lineage.
Initial analysis of the regulatory hierarchy of transcription factors has suggested that E2A activates the expression of Ebf1 and, together, both transcription factors induce the expression of several B-lineage genes, including Pax5 and the λ5, VpreB, and mb1 genes, which encode components of the pre-B-cell receptor (35, 47). However, this simple hierarchical relationship cannot account for various observations. Ectopic expression of E2A induces the expression of EBF1, and the promoter of Ebf1, located 5 kb upstream of the translation initiation codon, contains a binding site for E2A (51). However, the expression of E2A-GFP, in which the Gfp gene has been knocked into the E2a locus, is impaired in EBF1-deficient mice, suggesting that EBF1 may also regulate the expression of E2a (59). Likewise, the hierarchical relationship between EBF1 and Pax5 is not clear. Pax5-deficient bone marrow pre-B cells contain both Ebf1 and E2a transcripts, suggesting that Pax5 acts downstream of EBF1 and E2A (35). Moreover, EBF1 can bind a site in the Pax5 promoter in vitro, and the expression of Pax5 is reduced in Ebf1/E2a double-heterozygous mutant mice (35). In contrast, the forced expression of Pax5 in thymocytes carrying a Pax5 allele in the Ikaros locus results in the activation of a B-cell differentiation program, including the transcriptional activation of Ebf1, suggesting that Pax5 can also act upstream of Ebf1 (13). Based on these data, it has been proposed that the genes involved in the regulation of early B-cell differentiation are linked in a network that may help to stabilize specific developmental decisions (48). However, the insight into the molecular basis of such a network is still limited.
With the aim of gaining more insight into the putative network of transcription factors, we analyzed the regulation of the Ebf1 gene. In addition to the known distal Ebf1 promoter, we identified another Ebf1 promoter, located closer to the initiation codon, and we find that the proximal Ebf1 promoter is more abundantly used and differentially regulated. In addition to the regulation by E2A and EBF1, the distal promoter is also regulated by STAT5, whereas the proximal promoter is regulated by Pax5, Ets1, and Pu.1. Thus, the use of two promoters allows for the generation of distinct regulatory loops that may, in part, account for the postulated network of transcription factors and signaling pathways.
MATERIALS AND METHODS
Cell lines and flow cytometry.
70/Z3, 38B9, PD36, M12, K46, Raji, MEL, and EL4 cells were maintained in RPMI 1640 (Gibco) supplemented with 50 mM β-mercaptoethanol, 1× penicillin-streptomycin-glutamine (PSG; Gibco), and 10% fetal bovine serum (FBS; Perbio). For the maintenance of Ba/F3 cells the RPMI 1640 culture medium was in addition supplemented with 10% conditioned medium from WEHI3 cells as a source of IL-3. HeLa cells were grown in minimal essential medium (Gibco) supplemented with 1× PSG, 1× nonessential amino acids, and 10% FBS. 293, NIH 3T3, and 3T3L1 cells were grown in Dulbecco modified Eagle medium (Gibco) supplemented with 1× PSG and 10% FBS. OP9-MIG stromal cells (43) were cultured in minimal essential medium alpha (Gibco) with 1× PSG and 20% FBS. Bone marrow pro-B cells were grown on a subconfluent OP9-MIG monolayer in RPMI 1640 medium supplemented with 50 mM β-mercaptoethanol, 1× PSG, 10% FBS, and 10% conditioned medium from J558-IL-7 cells as a source of IL-7.
For flow cytometry, cells were obtained from bone marrow of 8- to 12-week-old C57BL/6 mice and from embryos at day E15.5 or E16.5. Isolated cells were incubated with conjugated antibodies, according to standard procedures, and sorted and reanalyzed using a MoFlo cell sorter (Cytomation). For depletion of lineage-positive cells, anti-CD3, -4, -5, -8, -11b, -11c, and -19 and NK1.1 allophycocyanin-conjugated antibodies (BD Pharmingen) were used. Hematopoietic stem cells (HSC; Lin− IL-7R+ c-kithigh Sca-1high), CLPs (Lin− IL-7R+ c-kitlow Sca-1low), pro-B-cell fraction A (Fr. A; B220+ AA4.1+ c-kit+ CD19−) and fractions B and C according to the work of Hardy and Hayakawa (18) (Fr. B, B220+ CD43+ human serum albumin [HSA]+ BP-1−; Fr. C, B220+ CD43+ HSA+ BP-1+), and marginal zone (B220+ CD21high CD23low) and follicular (FO; B220+ CD21low CD23 high) B cells have been sorted by fluorescence-activated cell sorting (FACS) to more than 98% purity.
Mammalian EBF1 expression vectors.
The EBF1β-17 expression vector was generated by fusing the NotI-AatII fragment of pEBF-17 cDNA (16) via a linker to NotI-ApaI into the multiple cloning site of pcDNA3.1-myc-HisA+ (Invitrogen). For the EBF1α expression plasmid the 5′ leader was PCR amplified with the primers 5′-ATA TTA TAG CGG CCG CCA CAG AGG TGG GTG AAT G-3′ (forward) and 5′-CCG GTA GTG GAT CCC ATT ATT-3′ (reverse) and subcloned via NotI-BamHI into pEBF-17 (16); the NotI-PmlI fragment was then introduced into pcDNA3.1-EBF1β-17-myc-HisA+. EBF1β was cloned in a similar manner with the primers 5′-GGC TGC TTC TCA AAG TGA GC-3′ (forward) and 5′-ATA TTA TAG CGG CCG CTG GGG AGA GTC AAA CTG GA-3′ (reverse) and subcloned via NotI-XmaI.
Site-directed mutagenesis.
Mutagenesis was performed by using the QuikChange site-directed mutagenesis kit (Stratagene). Primer sequences to generate point mutations are given in Table S1 in the supplemental material.
Transient transfections and reporter assays.
Ba/F3, 70/Z3, 38B9, PD36, M12, K46, Raji, and EL4 cells were transfected with 30 μg of plasmid DNA by electroporation. 293, NIH 3T3, and 3T3L1 cell transfections were performed using 10 μg of plasmid DNA and the calcium phosphate method (40). The total DNA concentration in each transfection experiment was kept constant by addition of pCI (Promega) expression vector DNA. Cells were harvested in 100 μl reporter lysis buffer 24 to 48 h posttransfection, except for the experiments in which Ba/F3 cells were depleted of IL-3 after transfection. These were incubated for 12 h only. Luciferase assays were conducted according to the manufacturer's instructions (Promega), and β-galactosidase assays were performed as described previously (53).
Reverse transcriptase PCR (RT-PCR).
RNA was purified with Trizol reagent according to the manufacturer's instructions (Invitrogen). Up to 2 μg of total RNA was reverse transcribed using 200 U SuperScriptII (Invitrogen).
Quantitative RT-PCR was performed by using the ABI Prism 7500 sequence detection system (Applied Biosystems) with the standard conditions: 94°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Per PCR 1/20 of the synthesized cDNA was supplemented with 2× SYBR green PCR Master Mix (Applied Biosystems) and the appropriate oligonucleotide primer pair. Primer sequences were designed by Primer Express 2.0 software (Applied Biosystems). Primer sequences and their final concentrations are given in Table S1 in the supplemental material, and Pax5 PCR primers have been reported elsewhere (58).
Immunoblot analysis and generation of anti-EBF1 antibodies.
Fifty micrograms of total protein extracts was separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Immunoblot assays were conducted by the ECL method according to the manufacturer's instructions (Amersham Biosciences). To detect the c-myc-tagged EBF1 isoforms, mouse monoclonal anti-c-myc antibody (1:4,000 dilution; Roche) and the alkaline phosphatase-conjugated goat anti-mouse antibody (1:10,000 dilution; Jackson ImmunoResearch Laboratories, Inc.) were used. For detection of endogenous EBF1 protein, an affinity-purified polyclonal rabbit anti-EBF1 antibody (1:500 dilution, generated against the peptide AHFEKQPPSNLRKSN, amino acids 51 to 66 of EBF1; Quality-Controlled Biochemicals) and the alkaline phosphatase-conjugated goat anti-rabbit antibody (1:10,000 dilution; Jackson ImmunoResearch Laboratories, Inc.) were used. For detection of the EBF1β isoform a rabbit anti-EBF1β antibody (1:3,000 dilution, generated against the peptide Ac-MFGIQESIQRSGSS-KC, amino acids 1 to 24 of EBF1; Biogenes) and the alkaline phosphatase-conjugated goat anti-rabbit antibody (1:10,000 dilution, Jackson ImmunoResearch Laboratories, Inc.) were used. Recombinant EBF1α protein is not detected by the anti-EBF1β antibody. The monoclonal anti-EBF1 antibody was raised in a rat against His-tagged recombinant EBF1 protein comprising amino acids 24 to 422 (21).
Protein purification and electrophoretic mobility shift assay.
Pax5 (residues 1 to 149) and Ets1 (residues 331 to 440) proteins have been expressed and purified as described in the work of Garvie et al. (14). The protocol of the electrophoretic mobility shift assay has been described in the work of Fitzsimmons et al. (12). For the oligonucleotides for the electrophoretic mobility shift assay of Pax5 and Ets1, see Table S1 in the supplemental material.
Primer extension and S1 nuclease protection assay.
For primer extension an antisense oligonucleotide corresponding to nucleotides +88 to +107 of Ebf1β mRNA (EBFr-u4; 5′-CCT GCT GGA TGG AGA TTC TG-3′) was 32P 5′ end labeled, hybridized to 50 μg of total RNA, and extended by 200 U SuperScriptII (Invitrogen) for 1 h at 42°C. Samples were precipitated and separated by denaturing polyacrylamide gel electrophoresis. S1 nuclease protection was performed according to the work of Hagman et al. (16). Ten thousand counts per minute of single-stranded DNA probe was hybridized to 50 μg of total RNA and digested with 50 U of S1 nuclease (Boehringer) for 1 h at 37°C. The single-stranded DNA probe for detection of Ebf1β mRNA 5′ ends was generated by 32P 5′ end labeling an antisense oligonucleotide corresponding to nucleotides +88 to +107 of Ebf1β mRNA (EBFr-u4; 5′-CCT GCT GGA TGG AGA TTC TG-3′).
ChIP.
Primer sequences used for the chromatin immunoprecipitation (ChIP) assay are given in Table S1 in the supplemental material. ChIP was performed according to the work of Fejer et al. (10). Briefly, cells were cross-linked for 9 min in their normal culture medium with 1% formaldehyde at room temperature. The reaction was stopped with 125 mM glycine, and after two washes with ice-cold phosphate-buffered saline (PBS), the cells were lysed in cell lysis buffer (5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, and 0.5% Igepal) and then once in 250 μl nucleus lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS) per 107 cells. Sonication of the nuclear lysates was carried out in a 250-μl volume; the average DNA fragment size was 500 bp, determined for each sample by agarose gel electrophoresis. The lysates were then diluted 1:10 with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.1, 167 mM NaCl). The samples were precleared with blocked protein A-agarose, bovine serum albumin (BSA), and sheared salmon sperm DNA. Immunoprecipitation was carried out with anti-Pax5 antibody (sc-13146; 3 μg; Santa Cruz). Normal mouse immunoglobulin G (IgG) and IgG-agarose conjugate (sc-2027 and sc-2345, respectively) were used as negative controls (Santa Cruz). Immunoprecipitates were collected with protein A agarose (Santa Cruz) and then washed twice with a low-salt buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1), once with high-salt buffer (500 mM NaCl), and twice with a LiCl detergent wash buffer (250 mM LiCl, 1% deoxycholate, 1% Igepal, 1 mM EDTA, 10 mM Tris, pH 8.1). The DNA was then eluted, de-cross-linked, phenol extracted, and finally dissolved in 50 μl water. Ten percent of the input chromatin was used as a control.
Intracellular staining.
To stain EBF1 protein for intracellular FACS analysis, monoclonal rat anti-EBF1 antibodies were used. Typically, 106 cells were resuspended in 50 μl hybridoma supernatant of anti-Fc-block anti-CD16/CD32 and incubated for 10 min on ice. Afterwards, cells were washed with PBS-2% fetal calf serum twice, resuspended in 50 μl of fixation solution A (Caltag; Fix and Perm cell permeabilization kit), and held at room temperature for 15 min. Fixed cells were washed with PBS-2% fetal calf serum twice, resuspended in 25 μl permeabilization solution B (Caltag) and 25 μl of anti-EBF1 hybridoma supernatant, and incubated at room temperature for 15 min. Afterward, the cells were washed with saponin solution (0.5% saponin, 0.5% BSA, 0.02% NaN3 in PBS), and the cell pellet was resuspended in a 1:50 dilution of the secondary antibody anti-rat IgG2a-fluorescein isothiocyanate (FITC; BD Pharmingen) and incubated at room temperature for 15 min. Finally, cells were washed with saponin solution and resuspended in PBS, and FACS analysis was performed.
Replication timing analysis.
The replication timing analysis was performed as described previously (2) with minor modifications. In brief, cells were incubated with 50 μM 5′-bromodeoxyuridine (BrdU; Sigma) for 60 min and stained with 50 μg/ml propidium iodide. Equal numbers of cells were collected for each of the six cell cycle fractions (G1, S1, S2, S3, S4, and G2/M) into lysis buffer, and BrdU-labeled DNA was isolated from each fraction as described previously (17). Equal amounts of BrdU-labeled DNA from Drosophila melanogaster Schneider cells were added to each fraction prior to phenol-chloroform extraction and ethanol precipitation. Isolated DNA was mixed with 0.2 mg of denatured, sheared salmon sperm DNA, sonicated to obtain fragments of approximately 700 bp, immunoprecipitated with 2 μg monoclonal anti-BrdU antibody (Becton Dickinson, Belgium), and incubated with excess secondary antibody (35 μg of rabbit anti-mouse IgG; Sigma). DNA-protein complexes were collected by centrifugation and treated with proteinase K, and BrdU-labeled DNA was recovered using the QIAquick gel extraction kit (QIAGEN). Preliminary observations showed no significant replication timing changes within areas smaller than 150 to 200 kb (data not shown; in concordance with reference 55), and therefore primers for the analysis of the Ebf1 region were designed 200 to 300 kb from each other. Primer sequences are available on request.
FISH.
Bacterial artificial chromosome probes for fluorescence in situ hybridization (FISH) were labeled with digoxigenin using the nick translation kit (Invitrogen). Specific hybridization of bacterial artificial chromosomes was confirmed by metaphase FISH. For two-dimensional (2D) interphase FISH, cells were harvested and hypotonically treated in 75 mM KCl for 5 min at room temperature before being fixed in ice-cold methanol-acetic acid (3:1 ratio). Fixed cells were dropped onto slides, denatured, hybridized, and washed as previously described (57). FISH signals were detected with anti-digoxigenin-FITC (Roche) and amplified with anti-sheep FITC (Vector Laboratories). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). The positions of loci relative to the nuclear periphery were measured as previously described (56).
RESULTS
Identification of multiple and differentially regulated Ebf1 transcription start sites.
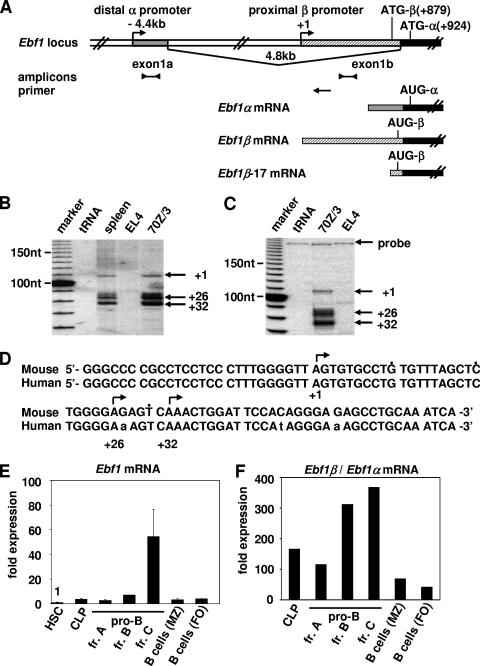
To gain insight into the regulation of the murine Ebf1 gene, we mapped potential promoter regions by determining the 5′ ends of Ebf1 mRNA in pre-B cells. Rapid amplification of cDNA ends with multiple primers, hybridizing with sequences in exon 1 or exon 2 or with genomic sequences upstream of the 5′ end of the longest Ebf1 cDNA, termed Ebf17 (16), indicated that potential 5′ ends are located approximately 879 nucleotides upstream of the 5′-most ATG (data not shown). The nucleotide sequences of the amplification products were found to be colinear with genomic sequences, indicating that they correspond to exon sequences (Fig. 1A). To map the 5′ ends of the corresponding Ebf1 transcripts, we performed primer extensions on RNA from spleen, 70/Z3 pre-B cells, and EL4 T cells, using a 32P-labeled oligonucleotide primer that corresponds to a sequence 107 nucleotides downstream of the putative 5′ ends. In RNA from spleen and 70/Z3 cells, multiple bands corresponding to potential transcription initiation sites were detected within a region of 30 nucleotides (Fig. 1B).
FIG. 1.
The Ebf1 gene is regulated by two promoters, the distal α and proximal β promoters. (A) Schematic overview of the distal Ebf1α and proximal Ebf1β promoters. Two alternative first exons, exon 1a and exon 1b, are indicated. The proximal Ebf1β promoter is located within the first intron of Ebf1α mRNA and 879 nucleotides upstream of the ATG-β, which is spliced out in Ebf1α mRNA. Ebf1α mRNA and Ebf1β mRNA can be distinguished by RT-PCR with primer pairs specific for exon 1a or exon 1b sequences. Indicated are the positions of the PCR amplicons and of the primer used for primer extensions and S1 nuclease protection experiments. (B) Primer extension experiments show multiple bands corresponding to potential 5′ ends of Ebf1β mRNA located 879 nucleotides (nt) upstream of the ATG-β. Potential transcriptional start sites are visible only in 70/Z3 pre-B cells and in spleen, not in Ebf1-negative EL4 T cells. (C) S1 nuclease protection experiments show a similar pattern of protected Ebf1β 5′ ends as in the primer extension experiments. nt, nucleotides. (D) Alignment of the human and mouse Ebf1β promoter transcriptional start sites. (E) Quantitative RT-PCR analysis of EL4 T cells and sorted lymphoid cells shows the upregulation of Ebf1 mRNA during early B-cell development. HSC (Lin− IL-7R+ c-kithigh Sca-1high), CLP (Lin− IL-7R+ c-kitlow Sca-1low), pro-B cell Fr. A (B220+ AA4.1+ c-kit+ CD19−) and Fr. B and Fr. C according to the work of Hardy and Hayakawa (18) (Fr. B, B220+ CD43+ HSA+ BP-1−; Fr. C, B220+ CD43+ HSA+ BP-1+), and marginal zone (MZ; B220+ CD21high CD23low) and follicular (FO; B220+ CD21low CD23high) B cells have been sorted by FACS to more than 98% purity. Ebf1 mRNA levels were normalized to β-actin, and the expression level of HSC was set to 1. (F) Quantitative RT-PCR analysis of endogenous levels of Ebf1α and Ebf1β mRNA. The ratio of Ebf1β to Ebf1α mRNA is displayed. Ebf1β mRNA is predominantly expressed in all analyzed cell types, and the ratio increases during pro-B-cell development, with the highest ratio being in fraction C pre-B cells.
We confirmed these results by an S1 nuclease protection experiment, in which we used a single-stranded DNA probe that had been generated with the same 32P-labeled oligonucleotide as in the primer extension experiment. The pattern of protected products was similar to that of the primer extension analysis, indicating that the protection and extension products correspond to the 5′ ends of Ebf1 transcripts (Fig. 1C). The location of the 5′ ends of these transcripts is approximately 3.9 kb downstream of a previously identified initiation site in the Ebf1 gene (51). To distinguish the two potential promoters of the Ebf1 gene, we refer to the previously described distal promoter as the Ebf1α promoter and the newly identified proximal promoter as the Ebf1β promoter. Based on the splicing of exon 1a in Ebf1α transcripts and the use of an alternative exon 1b in Ebf1β transcripts, the α and β transcripts would contain distinct in-frame AUGs at their 5′ ends (Fig. 1A). The newly identified Ebf1β as well as the Ebf1α promoter is a TATA-less promoter (Fig. 1D) (51). The Ebf1β promoter is highly conserved between mice and humans, whereas the mouse Ebf1α promoter is less conserved (Fig. 1D and data not shown).
Consistent with previous studies (58), the levels of total Ebf1 mRNA, as measured by quantitative RT-PCR with primers lying in common exons, show expression of Ebf1 in CLPs and a marked upregulation in pro-B cells of fraction C according to the nomenclature of Hardy and Hayakawa (Fig. 1E) (18). With the aim of assessing the relative abundance of Ebf1α and Ebf1β transcripts in different tissues and in primary cells representing common lymphoid progenitors or different stages of B-cell differentiation, we performed quantitative RT-PCR using primers that amplify either Ebf1α-specific sequences of exon 1a or Ebf1β-specific sequences of exon 1b. Normalization of the PCR products with the amplification products of β-actin mRNA indicated that the relative abundance of Ebf1β transcripts is significantly higher than that of Ebf1α transcripts, ranging from 41-fold in follicular B cells to 367-fold in fraction C pro-B cells (Fig. 1F). To examine whether the differences in the abundance of α and β transcripts are due to differences in RT-PCR amplifications, we transfected cytomegalovirus (CMV)-based expression plasmids for either Ebf1α or Ebf1β cDNA into 293 cells and found that α and β transcripts accumulate at similar levels (Fig. 3B). In addition, titration of input cDNA showed linear amplification of the primer pairs (data not shown). Taken together, these data suggest that the Ebf1 gene is transcribed by two promoters that are differentially regulated in various cell types.
FIG. 3.
The two isoforms of EBF1 protein, EBF1α and EBF1β, differ in the first 14 amino acids but have the same transactivation potential. (A) Schematic overview of EBF1 protein expression vectors. EBF1α and EBF1β comprise the full 5′ UTR of the Ebf1α and Ebf1β mRNA, respectively, whereas EBF1β-17 differs from EBF1β in its 5′ UTR, which comprises only 45 nucleotides. (B) Real-time RT-PCR of 293 cells transfected with 2 μg of EBF1β-17, EBF1α, or EBF1β together with LEF1 expression vector. Forty-eight hours after transfection Ebf1 and Lef1 mRNA levels were analyzed by RT-PCR and Ebf1 mRNA levels were normalized to Lef1 mRNA. (C) Immunoblot analysis of EBF1 isoforms was performed by transfecting 2 μg of EBF1β-17, EBF1α, EBF1β, or empty vector into 293 cells. After 48 h cells were harvested and total cell extracts were subjected to anti-myc immunoblotting. Cell extracts of EL4 T cells and PD36 B cells were analyzed by anti-EBF1 and anti-EBF1β immunoblotting. (D) Ba/F3 cells were transiently transfected with 2 μg of λ5 luciferase reporter construct, containing nucleotides −299 to +131 of the λ5 gene, together with 1 μg cytomegalovirus β-galactosidase reporter and 0, 0.3, 1, or 3 μg of EBF1β or EBF1α expression plasmids as indicated. (E) Ba/F3 cells were transiently transfected with 2 μg of λ5 luciferase reporter construct together with 1 μg cytomegalovirus β-galactosidase reporter; 3 μg of E47-FD; and 0.3, 1, or 3 μg of EBF1β-17 or EBF1α expression plasmids alone or in pairwise combinations as indicated. (F) Transient transfection of Ba/F3 cells with empty vector only or 3 μg E47-FD in combination with 3 μg EBF1β-17 or EBF1α followed by quantitative RT-PCR of endogenous λ5 mRNA. λ5 mRNA levels were normalized to β-actin mRNA, and the expression level of the empty-vector control was set to 1. Error bars represent the standard deviations of the means of three experiments.
Identification of the Ebf1β promoter and comparison with the Ebf1α promoter.
To examine whether sequences surrounding the presumed 5′ ends of the Ebf1β transcripts have promoter activity, we fused a DNA fragment encompassing approximately 1.7 kb upstream and 0.3 kb downstream of the putative initiation site to the luciferase gene (Fig. 2A). Transfection of this Ebf1 reporter plasmid, termed Ebf1β(−1.7)-luc, into Raji B cells, which show the highest Ebf1β(−1.7)-luc promoter activity, yielded an approximately 30-fold activation relative to the luciferase control plasmid. In addition, a reporter construct in which the Ebf1β promoter fragment has been inverted had the same activity as the empty vector control (Fig. 2A and C). 5′ truncations of the Ebf1β promoter fragment up to position −0.5 kb of the initiation site did not significantly alter the activity of the promoter, suggesting that the included sequences contain the entire Ebf1β promoter (Fig. 2C). To examine the possibility that the distal Ebf1α promoter acts as an enhancer, we performed two sets of experiments. First, we inverted the Ebf1α promoter and found a significant reduction in luciferase activity. Second, we fused the Ebf1α promoter to the Ebf1β promoter in either orientation. In these constructs, the activity of the Ebf1β promoter was similar to that of the construct containing the Ebf1β promoter alone, indicating that the Ebf1α regulatory sequences do not act as an enhancer (Fig. 2C).
FIG. 2.
Cloning of the proximal Ebf1β promoter and comparison with the distal Ebf1α promoter. (A) Schematic overview of the Ebf1α and Ebf1β promoter luciferase reporter constructs. (B) Comparison of the promoter activities of the two Ebf1 promoters in transient-transfection assays of B- and non-B-cell lines. Three micrograms of α(−1.1)-luc, β(−1.7)-luc, or pGl3-luc was transfected together with 1 μg cytomegalovirus β-galactosidase reporter into PD36 pre-B cells, Raji mature B cells, 3T3L1 preadipocytes, EL4 pre-T cells, or NIH 3T3 fibroblasts. The activation (n-fold) compared to that of pGl3-luc is depicted. (C) Transient transfection of Ebf1β promoter constructs into Raji B cells. The empty-vector control pGl3-luc was compared to the Ebf1 promoter constructs α(−1.1)-luc, α(flip)-luc, β(−1.7)-luc, β(flip)-luc, β(−0.5)-luc, α-β-luc, and α(flip)-β-luc. Three micrograms of each luciferase construct was transfected together with 1 μg cytomegalovirus β-galactosidase reporter into Raji B cells. The levels of luciferase activity, normalized to the activity of the cotransfected β-galactosidase reporter, are expressed as activation (n-fold) relative to the luciferase levels of cells transfected with pGl3-luc. All transfections were performed at least three times, and results from representative experiments with standard deviations are shown.
We also compared the relative activities of the Ebf1α and Ebf1β promoters by transfecting the respective luciferase reporter constructs, together with a β-galactosidase reporter plasmid as a transfection control, into cell lines representing different stages of B-cell differentiation or nonlymphoid cells. In the pre-B-cell line PD36, the Ebf1α and Ebf1β promoters augmented the luciferase activity 29- and 7-fold, respectively (Fig. 2B). In Raji cells representing mature B cells, however, the Ebf1α and Ebf1β promoters activated the reporter 15- and 25-fold, respectively. In the T-cell line EL4 and in nonlymphoid cell lines, the activities of the two promoters ranged between three- and ninefold. Thus, both Ebf1 promoters show a limited cell type specificity, whereby the relative activity of the α promoter may be higher in pre-B cells and that of the β promoter higher in B cells.
Generation of distinct but functionally equivalent isoforms of EBF1 by the α and β promoters.
The splicing of the first intron of the Ebf1α pre-mRNA removes the most 5′-located in-frame AUG and generates an mRNA that utilizes an AUG located 42 nucleotides downstream (51). In contrast, the Ebf1β promoter generates an mRNA that includes the first in-frame AUG and could be translated into a longer EBF1 isoform, including an additional 14 residues at the amino terminus (Fig. 1A). Although the most 5′-located AUG does not conform to the Kozak consensus sequence, it is included in peptides that have been identified by mass spectrometry of purified EBF1 protein (T. Grosskopff, G. Mittler, and R. Grosschedl, unpublished data). In addition, EBF1β can be detected with a polyclonal peptide antibody raised against the first 14 amino acids of EBF1β (Fig. 3C). To examine the production of both EBF1α and EBF1β isoforms, we transfected 293 cells with CMV-based expression plasmids containing a Myc epitope tag at the 3′ end of either Ebf1 cDNA (Fig. 3A). The immunoblot analysis indicated that EBF1α and EBF1β proteins differ in their relative abundance and apparent molecular masses (Fig. 3C). The major form of EBF1α migrated faster than the major form of EBF1β, consistent with the lack of 14 amino-terminal residues in EBF1α. The additional slower-migrating forms of EBF1α are most likely generated by protein modifications. The major form of EBF1β accumulated at a level that is approximately 5% of that of EBF1α. To examine whether the long 5′ untranslated region (UTR) of the Ebf1β mRNA results in the lower abundance of EBF1β, we examined the accumulation of protein from the CMV-EBF1β-17 cDNA expression plasmid that lacks most 5′-UTR sequences. In cells transfected with the EBF1β-17 expression plasmid, expression was detected at a level that is similar to that of EBF1α protein (Fig. 3C). Immunoblot analysis to detect endogenous EBF1 in PD36 cell extracts revealed a pattern of bands that resembles the EBF1β protein, which was confirmed by immunoblot analysis with anti-EBF1β antibody (Fig. 3C). As antibodies that detect the EBF1α isoform also recognize the β isoform, we cannot assess the relative abundance of EBF1α in vivo. We also examined whether the RNA steady-state levels of α and β transcripts are comparable. Therefore, we transfected the CMV-based expression plasmids containing either Ebf1α or Ebf1β cDNA together with LEF1 expression vector as a control for transfection efficiency into 293 cells, performed RT-PCR, and found that α and β transcripts accumulate at similar levels (Fig. 3B).
To analyze whether the two isoforms of EBF1 are functionally similar, we transfected IL-3-dependent Ba/F3 cells with a λ5-luciferase reporter construct and the CMV-EBF1α or CMV-EBF1β plasmid. As expected from the lower protein levels observed in the immunoblot of transfected 293 cells (Fig. 3C), EBF1β protein activates the λ5-luciferase reporter to a lesser extent (Fig. 3D). In the following experiments, we used CMV-EBF1α or CMV-EBF1β-17 plasmids, which allow for the accumulation of similar levels of protein. We have previously shown that EBF1, in collaboration with E47, transactivates the λ5 promoter (47). Both EBF1α and EBF1β-17 alone activated the λ5 reporter weakly, but they both synergized with a constitutive dimeric form of E47 to mediate very high levels of λ5 reporter gene expression (Fig. 3E). Moreover, both forms of EBF1 activated an mb1-luciferase reporter construct in combination with E47 (see Fig. S1 in the supplemental material). Based on the ability of EBF1 to act as a “pioneer” transcription factor in the context of higher-order chromatin (26, 27, 38), we compared the potentials of the two EBF1 proteins to activate the expression of the endogenous λ5 gene. In combination with E47, similar activations of λ5 gene expression were observed with EBF1α and EBF1β (Fig. 3F). However, the levels of EBF1- and E2A-induced λ5 expression are still significantly lower than that of the endogenous λ5 gene in pre-B cells. Taken together, these data indicate that the Ebf1α and Ebf1β transcripts generate distinct isoforms of EBF1 that appear to be functionally equivalent. However, we cannot rule out the possibility that the two EBF1 isoforms differ in their potential to regulate other target genes.
Regulation of the Ebf1α promoter by STAT5, E47, and an EBF1 autoregulatory loop.
EBF1 regulates early events in B-cell differentiation, including the specification of the B-cell fate (25, 29, 58). Therefore, the question arises as to the regulation of Ebf1 gene expression by extracellular signaling pathways. Recent experiments have examined the effects of mutations in the IL-7 or IL-7Rα genes on early B-cell differentiation (8, 22). These experiments have shown that the loss of IL-7 signaling results in a very early arrest in B-cell differentiation and loss of Ebf1 gene expression. Importantly, forced expression of a constitutively active form of the nuclear mediator of IL-7 signaling, STAT5-CA, in IL-7Rα−/− pre-pro-B cells resulted in the expression of Ebf1 and a partial rescue of B-cell differentiation (22). To examine whether STAT5-CA is sufficient to activate Ebf1 expression in nonlymphoid cells, we transfected IL-3-dependent Ba/F3 cells with a STAT5-CA expression plasmid and found that the levels of endogenous Ebf1 transcripts were increased ninefold relative to mock-transfected cells (Fig. 4A). Since IL-3 signaling activates STAT5 (30), we also examined the effects of depletion and readdition of IL-3 to Ba/F3 cells that have been transfected with Ebf1 promoter constructs. The readdition of IL-3 increased the activity of the transfected Ebf1α promoter construct by a factor of 66, which is even higher than that observed with the positive control, cyclinD1-luciferase (D1) (28), whereas the Ebf1β promoter was activated 20-fold (Fig. 4B). Finally, we examined the potential activation of Ebf1α- and Ebf1β-luciferase reporter constructs in Ba/F3 cells from which IL-3 had been withdrawn after transfection of increasing amounts of STAT5-CA expression plasmids. STAT5-CA activated the Ebf1α-luciferase reporter in a dose-dependent manner, whereas the extent of the activation of the Ebf1β-luciferase reporter was significantly lower, suggesting that STAT5-CA can induce preferentially the Ebf1α promoter (Fig. 4C). We also examined whether STAT5 is bound to the Ebf1α promoter by performing chromatin immunoprecipitations and amplifications with multiple primer pairs that covered the promoter region between nucleotide positions −1160 and +230 of the Ebf1α promoter. In these experiments, we failed to detect STAT5 binding at the Ebf1α promoter, although we detected STAT5 binding at the cis promoter as a positive control (4), Thus, the regulation of the Ebf1α promoter by STAT5-CA may be indirect. The Ebf1α promoter containing −349 to +245 is activated by STAT5-CA to the same extent as the Ebf1α(−1.1)-luc construct (see Fig. S2 in the supplemental material). By sequence analysis of the mouse Ebf1α promoter we found only one potential STAT5 binding site, at position −104. However, point mutagenesis of this potential binding site did not affect the activation by STAT5-CA (data not shown).
FIG. 4.
Activation of the Ebf1α promoter by STAT5, EBF1, and E47. (A) Transient transfection of Ba/F3 cells with 10 μg STAT5-CA expression plasmid or empty vector. Ebf1 mRNA levels were determined by quantitative RT-PCR analysis and normalized to β-actin mRNA. The expression level of the empty-vector control was set to 1. (B) Transfection of Ba/F3 cells with 3 μg luciferase reporter constructs containing either multimerized STAT-binding sites of the cyclin D1 promoter (D1), Ebf1α(−1.1) promoter sequences (Ebf1α), or the Ebf1β(−1.7) promoter (Ebf1β), together with 1 μg Rous sarcoma virus β-galactosidase reporter as a transfection control. The cells were incubated with or without IL-3 for 12 h prior to the luciferase and β-galactosidase assays. (C) Ba/F3 cells were transiently transfected with 3× D1-SIE1-luc, Ebf1α(−1.1)-luc, or Ebf1β(−1.7)-luc, together with 1 μg Rous sarcoma virus β-galactosidase reporter and increasing amounts of STAT5-CA (0, 3, or 10 μg). IL-3 was withdrawn 12 h prior to the luciferase and β-galactosidase assays. (D) Transient transfection of Ba/F3 cells with empty vector or with 3 μg E47-FD and 3 μg EBF1β-17, followed by quantitative RT-PCR to detect endogenous Ebf1α or Ebf1β mRNA. mRNA levels were normalized to β-actin, and the expression level of the empty-vector control was set to 1. Error bars represent the standard deviations of the means of three experiments. (E) Real-time RT-PCR analysis of fraction B pro-B cells of Ebf1+/− mice show a reduction of Ebf1α, Ebf1β, and total Ebf1 mRNA, relative to wild-type (wt) fraction B pro-B cells (Fr. B; B220+ CD43+ HSA+ BP-1−). Error bars represent the standard deviations of the means of three experiments. (F) Schematic overview of the Ebf1β-luc and Ebf1β(Pu.1mut)-luc promoter luciferase reporter constructs. (G) Transient transfection of HeLa cells with 300 ng of the Ebf1β-luc promoter construct together with 0, 0.3, 1, or 3 μg of Pu.1 and 200 ng of cytomegalovirus β-galactosidase reporter.
The Ebf1α promoter has been previously found to contain binding sites for EBF1 and E47, raising the possibility of an EBF1 autoregulatory loop (51). Therefore, we examined the effects of the ectopic expression of EBF1β-17 and E47 on the expression of both endogenous Ebf1 promoters in Ba/F3 cells. Consistent with the previously observed stimulatory effects of EBF1 and E47 on Ebf1α reporter constructs (51), we observed a 20-fold increase in the accumulation of endogenous Ebf1α transcripts in Ba/F3 cells that have been transfected with EBF1β-17 and E47 expression plasmids (Fig. 4D). However, we did not detect any significant activation of the Ebf1β promoter, suggesting that the EBF1 autoregulatory loop involves specifically the distal Ebf1α promoter. In mice heterozygous for Ebf1, however, a reduction of Ebf1α as well as Ebf1β transcripts is observed, suggesting that the Ebf1β promoter is regulated indirectly by EBF1 (Fig. 4E).
Regulation of the Ebf1β promoter by Pu.1.
Pu.1 has been shown to act upstream of EBF1 in the regulation of B lymphopoiesis (29). In particular, the expression of Ebf1 is abrogated in Pu.1-deficient progenitors and the defect can be rescued by the forced expression of EBF1 (29). Chromatin immunoprecipitation experiments with anti-Pu.1 antibodies also demonstrated the occupancy of a site in the first intron downstream of the Ebf1 exon 1α that is conserved between mice and humans (29). This Pu.1 binding site resides at position +165 of the Ebf1β promoter (Fig. 4F). Transfection of a Pu.1 expression plasmid into HeLa cells augmented the activity of the Ebf1β promoter-luciferase construct in a dose-dependent manner (Fig. 4G). Mutation of the Pu.1 binding site at +165 did not reduce the activation by Pu.1 (data not shown). However, mutation of the 5′-GGAA-3′ motifs at +165 and +303 reduced both basal and Pu.1-activated promoter activity.
Regulation of the Ebf1β promoter by Pax5 and Ets1.
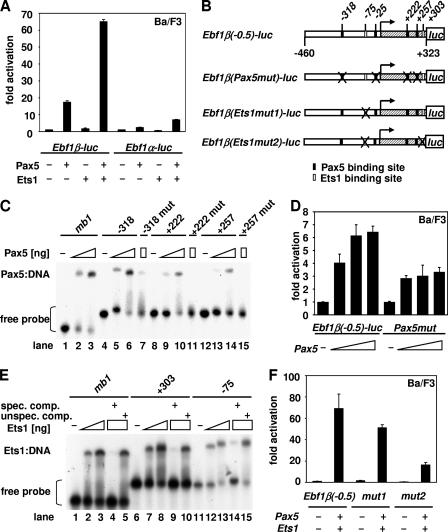
The integration of the Pax5 gene into the Ikaros locus of mice was found to result in the expression of the Ebf1 gene and the generation of B cells in the thymus (13). To examine whether Pax5 may regulate the Ebf1α and/or the Ebf1β promoter, we transfected Ba/F3 cells with a Pax5 expression plasmid, together with a reporter construct in which the 1.3-kb Ebf1α promoter or the 0.9-kb Ebf1β promoter had been linked to the luciferase gene. We detected a marked (17-fold) activation of the Ebf1β promoter but only a weak (twofold) activation of the Ebf1α promoter (Fig. 5A). Based on evidence of the collaboration of Pax5 and Ets1 in the activation of the mb1 promoter (11), we also examined the effect of Pax5 expression in combination with Ets1. The EBF1β promoter was activated even more efficiently than with Pax5 alone.
FIG. 5.
Pax5 and Ets1 collaborate to activate the Ebf1β but not Ebf1α promoter. (A) Ba/F3 cells were transiently transfected with 1 μg cytomegalovirus β-galactosidase reporter and 2 μg of Ebf1α or Ebf1β luciferase reporter construct, together with 3 μg of Pax5 and 3 μg of Ets1 expression plasmids alone or in pairwise combinations as indicated. (B) Overview of the Ebf1β promoter luciferase reporter constructs. Indicated are the Pax5-binding sites at positions −318, −25, +222, and +257 and the Ets1-binding sites at −75 and +303. (C) Electrophoretic mobility shift assays show binding of Pax5 to binding sites at −318, +222, and +257 in the Ebf1β promoter. Recombinant Pax5 protein (0, 50, or 200 ng) was incubated with labeled wild-type or mutant oligonucleotide probes. (D) Mutation of all four Pax5-binding sites in the Ebf1β promoter leads to a twofold reduction of transactivation by Pax5. Ba/F3 cells were transiently transfected with 1 μg cytomegalovirus β-galactosidase reporter and 2 μg of Ebf1β(0.5)-luc or Ebf1β(Pax5mut)-luc reporter construct, together with increasing amounts of Pax5 expression plasmids (0.3, 1, and 3 μg). (E) Electrophoretic mobility shift assays show binding of Ets1 to binding sites at −75 and +303 in the Ebf1β promoter. The indicated amount of recombinant Ets1 protein (0, 100, or 300 ng) was incubated with labeled oligonucleotide probes and assayed in the presence of a 200-fold excess of specific (mb1) or unspecific competitor. (F) Mutation of the Ets1-binding sites at positions −75 and +303 reduces fourfold the potential transactivation by Ets1 and Pax5 on the Ebf1β promoter. Ba/F3 cells were transiently transfected with 1 μg cytomegalovirus β-galactosidase reporter and 2 μg of Ebf1β(0.2)-luc or Ebf1β(Ets1mut)-luc reporter construct, together with 3 μg of Pax5 and 3 μg of Ets1 expression plasmids as indicated.
Inspection of the Ebf1β promoter for potential Pax5-binding sites revealed multiple sequences that could be recognized by Pax5. However, the sequence conservation of Pax5-binding sites is rather low (6), and therefore, we examined 32P-labeled oligonucleotides encompassing potential Pax5-binding sites in electrophoretic mobility shift assays with a recombinant DNA-binding domain of Pax5 (14). Four out of 24 oligonucleotides examined interacted with the DNA-binding domain of Pax5 (Fig. 5B and C and data not shown). Binding of Pax5 to the site at position −318 was as efficient as that to the known Pax5-binding site in the mb1 promoter (Fig. 5C, lanes 1 to 6). Binding to the sites at positions −25, +222, and +257 was approximately fourfold less efficient (Fig. 3C, lanes 8 to 10 and 12 to 14, and data not shown). The specificity of sequence recognition was confirmed by the lower efficiency of binding of mutant oligonucleotides in which conserved nucleotides had been altered (Fig. 5C, lanes, 7, 11, and 15). To examine the effects of the point mutations in the Pax5-binding sites on the Pax5-mediated activation of the Ebf1β promoter, we transfected wild-type and mutated Ebf1β promoter constructs, together with a Pax5 expression plasmid, into Ba/F3 cells. The activation of the mutated Ebf1β promoter by Pax5 was reduced twofold (Fig. 5D). A similar modest effect of mutations in the Pax5-binding sites has been reported for the BLNK promoter (42), suggesting that the specificity of Pax5 function in vivo may depend on its interaction with other DNA-binding proteins.
As Pax5 synergizes with Ets1 in the activation of the Ebf1β promoter, we also examined the promoter sequences for the presence of potential Ets1-binding sites. Two binding sites at positions −75 and +303 were found to interact with the recombinant DNA-binding domain of Ets1 in electrophoretic mobility shift assays (Fig. 5E, lanes 6 to 15). The binding site at position +303 interacted with Ets1 as efficiently as the Ets1-binding site in the mb1 promoter, and the specificity of binding was confirmed by the addition of an excess of unlabeled specific or nonspecific competitor. Based on the formation of a ternary complex of Pax5 and Ets1 on binding sites that are directly juxtaposed in the mb1 promoter (14), we also examined whether a ternary complex is formed on a longer oligonucleotide that encompasses both the Pax5-binding site at +257 and the Ets1-binding site at +303. However, no ternary complex was detected (data not shown), suggesting that Pax5 and Ets1 bind independently of each other to the Ebf1β promoter. To assess the functional role of the Ets1-binding sites, we introduced point mutations in the Ets1-binding sites at −75 and +303. Although Ets1 alone is not able to activate the Ebf1β promoter (Fig. 5A), in cotransfections with Pax5 expression plasmids, mutation of the Ets1-binding sites reduced the activity of the Ebf1β promoter approximately fourfold relative to the wild-type promoter (Fig. 5F). Comparison of the sequence of the Ebf1β promoter revealed a high degree of conservation that includes the Pax5-, Ets1-, and Pu.1-binding sites (see Fig. S3 in the supplemental material).
In vivo occupancy of the Ebf1β promoter by Pax5.
To gain further evidence for the direct binding of Pax5 to the Ebf1β promoter, we examined the in vivo occupancy of the Pax5-binding sites by ChIP experiments with a monoclonal anti-Pax5 antibody. We performed quantitative ChIP experiments on chromatin from cell lines representing mature B cells (K46) or nonlymphoid MEL cells, using PCR primer pairs that flank the Pax5-binding sites in the Ebf1β promoter (Fig. 6A and B). As a positive control, we assayed the binding of Pax5 to the Cd19 promoter (24). In K46 cells, the Ebf1β sequences Ebf1β(−371/−196), Ebf1β(−241/−62), and Ebf1β(+194/+347), including the Pax5 sites at positions −318, +222, and +257, were detected at a level 50% of that at which Cd19 sequences were amplified. In 38B9 cells, Ebf1β(−371/−196) sequences were detected at almost the same level as Cd19 sequences, and the Ebf1β(−241/−62) and Ebf1β(+194/+347) amplicons were detected at lower levels. The Pax5-binding site at −25 [Ebf1β(−89/+24)] was not enriched and amplified. As expected, no binding of Pax5 was detected in MEL cells. We also examined the binding of Pax5 to multiple Ebf1α promoter sequences and failed to detect any significant binding (data not shown).
FIG. 6.
Binding of Pax5 to Ebf1β regulatory sequences in vivo. (A) Schematic representation of the Ebf1β promoter and regions analyzed by ChIP. (B) ChIP was performed in the indicated cell lines with anti-Pax5 and the control IgG antibody. Enrichment (n-fold) comparing immunoprecipitate with the specific anti-Pax5 antibody and the control IgG was determined by real-time PCR; n.d., not detected. (C) Semiquantitative PCR was performed with serial dilutions of template DNA from cultured bone marrow pro-B cells of wild-type (wt) and Pax5−/− mice. Binding can be detected with anti-Pax5 antibodies but not with nonspecific IgG on both the Ebf1β and CD19 promoters. The anti-Pax5 antibodies do not bind to the JunB promoter. (D) Transient transfection of Ba/F3 cells with empty vector only or with 3 μg Pax5 and 3 μg Ets1 followed by quantitative RT-PCR of endogenous Ebf1α, Ebf1β, or total Ebf1 mRNA. mRNA levels were normalized to β-actin, and the expression level of the empty-vector control was set to 1. Error bars represent the standard deviations of the means of three experiments. (E) IL-7-cultured pro-B cells of Pax5-deficient mice have reduced EBF1 protein levels. Intracellular staining of IL-7-cultured pro-B cells was performed with monoclonal anti-EBF1 antibody and secondary anti-rat-FITC antibody. For the control staining only secondary antibody was used. (F) Fraction B pro-B cells of mice deficient for Pax5 show a reduction of Ebf1β mRNA and total Ebf1 mRNA. Real-time RT-PCR analysis was performed with sorted fraction B cells of wild-type (wt) and Pax5−/− mouse fetal liver (B220+ CD43+ HSA+ BP-1−). Ebf1α, Ebf1β, and total Ebf1 mRNAs have been amplified by oligonucleotides specific for exon 1a, exon 1b, or both isoforms. As a control, RT-PCR of B29, mb1, and λ5 was performed. mRNA levels were normalized to β-actin, and the expression level of the wild type was set to 100%. Error bars represent the standard deviations of the means of three experiments.
In IL-7-dependent pro-B-cell cultures from the bone marrow of wild-type mice, sequences that encompass the Pax5 site at −318 could be efficiently immunoprecipitated and amplified using semiquantitative PCR (Fig. 6C). The Pax5-binding sequences in the Cd19 promoter were detected at an approximately twofold-higher efficiency, whereas no binding of Pax5 to junB promoter sequences was detected. The specificity of the immunoprecipitation with anti-Pax5 antibody was further confirmed by the lack of amplification of Ebf1β(−371/196) and Cd19 promoter sequences in assays with Pax5-deficient pro-B cells (Fig. 6C). Quantitative PCRs on three independently immunoprecipitated samples indicated that binding of Pax5 to the −318 and the +222/+257 sites of the Ebf1β promoter in wild-type pro-B cells can be detected at levels that are 70% and 60% of the level observed with the Cd19 promoter, respectively (see Fig. S4 in the supplemental material). Thus, binding of Pax5 to the −318 and +222/+257 regions of the Ebf1β promoter occurs in both pro-B and B cells, although the relative occupancy may differ for these cell types.
Regulation of the endogenous Ebf1 gene by Pax5.
The binding of Pax5 to the Ebf1β promoter in vivo and the transactivation of Ebf1β promoter constructs by Pax5 in transient-transfection experiments strongly suggested a regulation of Ebf1 gene expression by Pax5. To examine the effects of the expression of Pax5 and Ets1 on the generation of Ebf1α and Ebf1β transcripts in their endogenous context, we transfected Pax5 and Ets1 expression plasmids into Ba/F3 cells and determined the accumulation of endogenous Ebf1 transcripts by quantitative RT-PCR (Fig. 6D). As expected, expression of Pax5 and Ets1 resulted in the 12-fold activation of Ebf1β transcripts. Surprisingly, the accumulation of Ebf1α transcripts was even higher (20-fold) despite the lack of Pax5-binding sites in the Ebf1α promoter. This result suggests that Pax5 may activate the endogenous Ebf1α promoter indirectly, possibly via an EBF1 autoregulatory loop (Fig. 4D and E; see below).
To examine whether the generation of Ebf1α and Ebf1β transcripts may be differentially affected in Pax5−/− pro-B cells, we sorted fraction B pro-B cells from the fetal liver of wild-type and Pax5−/− mice. Consistent with previous findings, the numbers of Pax5−/− fraction B pro-B cells was reduced by a factor of 10 and fraction C cells were undetectable (33). Quantitative RT-PCR assays indicated that the overall expression of Ebf1 in Pax5−/− pro-B cells is reduced to approximately 45% of the level found in wild-type pro-B cells, whereby the generation of Ebf1β transcripts is reduced to 30% (Fig. 6F). Only a minor reduction was found for Ebf1α transcripts, indicating that the absence of Pax5 results in a selective decrease in Ebf1β promoter activity. To confirm the identity of the Pax5−/− cells as pro-B cells, we analyzed the expression of B29, mb1, and λ5. As anticipated, expression of B29 was similar in Pax5−/− and wild-type cells, whereas expression of the EBF1 target genes mb1 and λ5 was reduced between 10- and 20-fold (Fig. 6F).
Since Ebf1β mRNA is expressed more efficiently than Ebf1α mRNA, but Ebf1α mRNA is translated more efficiently (see above), we analyzed EBF1 protein levels by intracellular FACS staining. EBF1 protein was detected in wild-type and Pax5−/− IL-7-cultured pro-B cells using monoclonal anti-EBF1 antibody and secondary anti-rat-FITC antibody. For the control staining only secondary antibody was used. IL-7-cultured pro-B cells of Pax5-deficient mice show approximately threefold-reduced EBF1 protein levels, consistent with the reduced Ebf1β promoter activity (Fig. 6E). However, this decrease in EBF1 protein levels may not be sufficient to affect the presumed autoregulatory feedback loop at the Ebf1α promoter, as Ebf1α transcripts can be detected at normal levels (Fig. 6F).
The Ebf1 locus shifts to later replication in the absence of Pax5.
The temporal order of DNA replication is an epigenetic mark reflecting chromatin structure, with early locus replication generally associated with transcription-permissible chromatin (reviewed in reference 9). To investigate the contribution of Pax5 to the regulation of the Ebf1 locus at the chromatin level, the timing of Ebf1 replication was analyzed in wild-type and Pax5-negative pro-B cells. The analysis also included embryonic stem (ES) cells, in which Ebf1 is not transcribed and is known to replicate late in S phase (36). A previously established PCR-based protocol (2) was used, in which nonsynchronized cells were pulse-labeled with BrdU, stained with propidium iodine, and sorted by FACS according to DNA content into six cell cycle fractions, four of which (S1 to S4) corresponded to different stages of S phase. Newly replicated DNA in each fraction was immunoprecipitated with anti-BrdU antibodies and detected by real-time PCR. Controls for the analysis included the α-globin locus replicating early in S phase in all analyzed cell types and a constitutively late-replicating X-chromosome region, X141 (2) (Fig. 7A). Each cell cycle fraction was also “spiked” with BrdU-labeled Drosophila DNA, and Drosophila-specific PCR primers were used to confirm equivalent recovery.
FIG. 7.
Replication timing of the Ebf1 locus is shifted to later replication in Pax5-deficient cells. (A) Replication timing of the early-replicating α-globin gene locus, the late-replicating X141 locus, and the Ebf1 locus in wild-type (wt) and Pax5-deficient pro-B cells is depicted. (B) Schematic overview of the Ebf1 locus. The locations of the primer pairs used for analysis of the replication timing are indicated. (C) In wild-type pro-B cells, Ebf1 replication peaked at the first quarter of S phase (S1). (D) Pax5-negative pro-B cells display later replication (S2) of the Ebf1 locus. (E) An even later replication was observed in ES cells, where Ebf1 is silent. A domain of later replication “centered” at Ebf1 was observed in Pax5-negative and ES cells, which extended to an area of more than 1 Mb, spanning the 37324131I11Rik and EpsinR genes that are active in both pro-B and ES cells.
Ebf1 was found to replicate late in S phase (with a peak in S3) in ES cells, where it is silent (Fig. 7E). In contrast, early Ebf1 replication (in the first quarter of S phase, S1) was observed in wild-type pro-B cells (Fig. 7C). However, a shift to later replication (S2) was observed in Pax5−/− pro-B cells, suggesting that Pax5 contributes to the transcription-permissible chromatin status of the Ebf1 locus (Fig. 7D). The analysis of the region surrounding the Ebf1 locus showed that in wild-type pro-B cells, Ebf1 is located within an approximately 2-Mb-long region of early-replicating DNA. In Pax5-negative pro-B cells as well as ES cells, a domain of later replication timing was observed with a “center” at the Ebf1 locus, spreading to a region of more than 1 Mb. Interestingly, the nearest expressed genes surrounding the Ebf1 locus (Fig. 7B), 3732413I11Rik and EpsinR (located 500 kb upstream and 800 kb downstream, respectively), were active in both ES cells and pro-B cells (data not shown) but replicated significantly earlier in the latter, thus ruling out a direct link between replication timing and transcription.
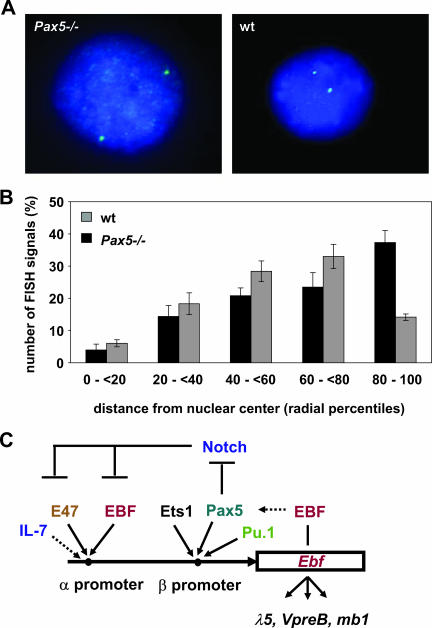
Change in nuclear positioning of the Ebf1 genomic locus in Pax5−/− pro-B cells.
Changes in replication have been shown to correlate with changes in gene chromatin structure, transcriptional activity, large-scale chromatin organization, and subnuclear position (36, 57). We therefore analyzed the large-scale chromatin organization of the Ebf1 locus in Pax5−/− and wild-type cells by 2D-FISH analysis. We observed that the Ebf1 locus was more often positioned towards the periphery of the nucleus in Pax5−/− cells than in wild-type cells (Fig. 8A and B). The nuclear periphery has previously been associated with later replication times and transcriptional inactivity (23, 34, 39, 56, 61). Our observation that in the absence of Pax5, Ebf1 replicates later and locates towards the nuclear periphery is consistent with our finding that Pax5 upregulates transcription from the Ebf1 promoter and suggests that Pax5 regulation of Ebf1 involves large-scale chromatin organization changes at the locus and nuclear repositioning.
FIG. 8.
Fluorescence in situ hybridization shows differential Ebf1 locus positioning in Pax5-deficient compared to wild-type (wt) pro-B cells. (A) Representative images of 2D-FISH analysis of Pax5-deficient and wild-type pro-B cells. (B) Analysis of the nuclear position of the Ebf1 locus shows that the Ebf1 locus is more often detected at the periphery of the nucleus in Pax5-deficient than in wild-type pro-B cells. (C) A complex regulatory network regulates the expression of the two Ebf1 promoters.
DISCUSSION
Ebf1 is a key regulator of early B-cell differentiation that participates in the specification of the cell lineage. Ebf1 is expressed as early as in CLPs, although the functional activity of EBF1 is required for the generation of pro-B cells (fraction B cells) that undergo the rearrangement of immunoglobulin heavy chain genes (25). Moreover, retroviral transduction of Ebf1 in hematopoietic progenitor cells and genetic bypass experiments in Pu.1-deficient progenitors or IL-7-deficient CLPs have shown that EBF1 is sufficient to induce B-cell differentiation, including the activation of E2a and Pax5 (29). These observations have been interpreted to suggest that EBF1 is involved in the initiation of the B-cell differentiation program consistent with the finding that EBF1 acts as a “pioneer” transcription factor in mediating chromatin accessibility and demethylation of DNA (27). However, EBF1 is not sufficient to induce the complete B-cell program in heterologous cell types and requires collaboration with other transcription factors. EBF1 has been found to collaborate with the E2A transcription factors in the activation of the endogenous immunoglobulin surrogate light chain genes in Ba/F3 and HeLa cells. This functional synergy between EBF1 and E2A has also been demonstrated genetically by the B-cell differentiation defect in Ebf1/E2a double-heterozygous mutant mice (35). The combined loss of one Ebf1 and one E2a allele also results in a downregulation of Pax5 expression, suggesting that Pax5 is downstream of EBF1 and E2A (35). Consistent with this hierarchical relationship, an EBF1-binding site has been identified in the Pax5 promoter (35). Moreover, forced expression of EBF1 in E2A- or Pu.1-deficient progenitors results in the activation of Pax5 gene expression (29, 46). However, overexpression of EBF1 and E47 in Ba/F3 cells does not induce the expression of Pax5 (data not shown), suggesting that the activation of Pax5 by EBF1 may require additional transcription factors that are expressed in lymphoid progenitor cells but not in Ba/F3 cells.
In our study, we show that the expression of the Ebf1 gene involves two promoters that are differentially regulated (Fig. 8C). The distal promoter, termed Ebf1α promoter, has been previously shown to be activated by E47 and EBF1 (51). Consistent with these observations, E47 is sufficient to activate the Ebf1α promoter in the context of the endogenous Ebf1 gene in transfected Ba/F3 cells, and this activation is augmented by EBF1 (Fig. 4D and data not shown). The stimulation of the Ebf1α promoter in the endogenous gene context is 20-fold, which is significantly stronger than the previously reported activation of a promoter construct (51). In our experiments, the major effect on the Ebf1α promoter is mediated by E47 and not by EBF1. However, two lines of evidence suggest that the regulation of the Ebf1α promoter by EBF1 generates a positive feedback loop. First, the reduction of the dose of EBF1 in Ebf1 heterozygous mice results in a decrease of Ebf1 gene expression by a factor of 2. Second, the specific activation of the Ebf1β promoter by Pax5 in the endogenous context results in the stimulation of the Ebf1α promoter (Fig. 6D and data not shown). In addition to an EBF1 feedback loop via the Ebf1α promoter, E2A and EBF1 proteins appear to act in a reciprocal feedback loop. The activation of the endogenous Ebf1 gene by E47 in Ba/F3 cells supports the upstream function of E2A proteins in the regulation of Ebf1 gene expression (reference 20 and this study). However, the expression of the E2a gene is also impaired in EBF1-deficient mice (59).
In addition to the regulation of the Ebf1α promoter by E47 and EBF1, we find that this promoter is responsive to IL-7 signaling. Transfection of a constitutively active form of STAT5 (STAT5-CA) into Ba/F3 cells results in a ninefold activation of the endogenous Ebf1 gene (Fig. 4A). In addition, the activity of the isolated Ebf1α but not Ebf1β promoter is augmented by STAT5-CA. However, the effect of STAT5-CA and IL-7 signaling on the Ebf1α promoter may be indirect. The Ebf1α promoter contains a potential STAT5-binding site at position −104, but mutation of this site did not alter the responsiveness of the promoter to STAT5-CA (data not shown). STAT5 has been found to be recruited to promoters via interactions with other DNA-bound transcription factors. For example, the association of STAT5 with the promoters of distal immunoglobulin variable genes has been found to occur in the absence of STAT5-binding sites and has been proposed to be mediated via interaction with DNA-bound Oct proteins (4). We consider this possibility for the regulation of the Ebf1α promoter by STAT5 unlikely because we have not been able to detect binding of STAT5 to the Ebf1α promoter in chromatin immunoprecipitations (data not shown). Therefore, we favor the view that IL-7 signaling and STAT5 regulate the expression of Ebf1 indirectly. The regulation of the Ebf1 gene by IL-7 signaling is also supported by the observations that the accumulation of Ebf1 transcripts is markedly impaired in CLPs from IL-7-deficient and IL-7Rα-deficient mice (8, 22).
The Ebf1β promoter differs in its regulation from the Ebf1α promoter. One of the surprising findings of our study is the regulation of the Ebf1β promoter by Pax5, which binds directly to multiple sites in the Ebf1β promoter and stimulates the activity of the promoter in synergy with Ets1. Although the targeted inactivation of Pax5 results in a block of B-cell differentiation prior to the generation of fraction C pro-B cells and Ebf1 transcripts are detected in Pax5-deficient mice (33), we find that the accumulation of Ebf1β-specific transcripts in preceding fraction B cells is reduced by a factor of 3. We also detected an approximately threefold decrease of intracellular EBF1 by flow cytometry. As the anti-EBF1 antibodies do not discriminate between the α and β isoforms and the anti-EBF1β antibody recognizes a pattern that resembles the pattern of EBF1, we cannot rule out the possibility that EBF1α does not contribute significantly to EBF1 protein expression. In contrast, the abundance of Ebf1α-specific transcripts is not altered. Likewise, an approximately fivefold decrease in total Ebf1 expression was observed in bone marrow pro-B cells of Pax5−/− mice (13). We also observed an activation of the Ebf1β promoter in the context of the endogenous Ebf1 gene in Ba/F3 cells that have been transfected with expression plasmids for Pax5 and Ets1. However, in this experiment we also detected an even higher accumulation of Ebf1α transcripts. As an Ebf1α-luciferase construct is not stimulated by Pax5 and no Pax5 binding could be detected at the Ebf1α promoter in vitro and in vivo (data not shown), the activation of the Ebf1α promoter in the context of the endogenous gene is likely due to a positive feedback loop via EBF1.
Although the effect of Pax5 gene mutation on the expression of Ebf1 is only threefold, a twofold reduction of Ebf1 gene activity has been demonstrated to be significant. Heterozygous mutant Ebf1+/− mice reveal a twofold decrease in the number of pro-B cells and a 10 to 30% reduction in the number of B cells in the spleen (25).
The ectopic expression of Pax5 in thymocytes results in the initiation of a B-cell differentiation program, including the activation of Ebf1 gene expression, whereas the forced expression of Pax5 in E2A-, Pu.1-, or IL-7-deficient CLPs is not sufficient to rescue B-cell development (8, 29, 46). These contrasting observations raise the question of whether Pax5 is directly involved in the establishment of Ebf1 expression. In thymocytes, Notch signaling antagonizes the activity of both E2A proteins and EBF1 (31, 50). Pax5 has been shown to repress the expression of Notch1 (52), and therefore, the forced expression of Pax5 in thymocytes could result in a repression of Notch and activation of E2A protein. According to this view, the expression of Pax5 in thymocytes would lead to the establishment of Ebf1 expression indirectly via activation of E2A proteins, which act on the distal promoter of the Ebf1 gene. Consistent with this scheme, E2A proteins are expressed in CLPs and the expression of Ebf1 is abrogated in the absence of E2A (59).
The Ebf1β promoter is also regulated by Pu.1. Previous experiments have identified a Pu.1-binding site in the first intron of the Ebf1 gene, which is located 165 nucleotides downstream of the start site of the β transcripts and is occupied by Pu.1 in vivo (29). Pu.1 has been found to directly regulate the expression of the IL-7Rα gene (7). Therefore, Pu.1 may act both indirectly via STAT5 in the regulation of the Ebf1α promoter and directly via the Ebf1β promoter. Based on our data, we suggest that the Ebf1α promoter is involved in the initiation of Ebf1 gene expression in CLPs, whereas the proximal Ebf1β promoter is involved in the Pax5-mediated upregulation in fraction B and C pro-B cells and in the maintenance of Ebf1 gene expression.
In support of a role for Pax5 in regulating Ebf1 expression, we show that Pax5 is required for a nuclear-internal location and early replication of the Ebf1 locus in developing B cells. In Pax5-negative pro-B cells, Ebf1 replicates later in S phase and occupies a more peripheral position in the nucleus than in wild-type pro-B cells. However, in the presence of Pax5, Ebf1 replication is advanced and the locus moves away from the nuclear periphery—epigenetic changes that previously had been associated with a transcriptionally permissive state (1, 9). Interestingly, successful immunoglobulin rearrangement was shown to depend on Pax5-mediated relocation of unrearranged immunoglobulin heavy chain alleles from the nuclear periphery to the interior (13, 19). In nonlymphoid cells a similar relationship between developmentally induced gene activation, locus repositioning, and advanced replication timing has been reported (5, 56). The striking temporal difference in the replication of Ebf1 locus in ES cells (late) versus pro-B cells (early) suggests that the epigenetic state of this locus is modified upon hematopoietic commitment. While Pax5 is clearly involved in this process, the observation that the timing of Ebf1 replication is advanced even in pro-B cells that lack Pax5 suggests that other developmentally regulated factors also contribute to the transcriptionally permissive state of the Ebf1 locus. The changes in the replication timing and frequency of subnuclear localization of the Ebf1 locus in Pax5−/− cells cannot be solely accounted for by the regulation of the Ebf1β promoter by Pax5 and raise the interesting possibility that other, yet-unidentified regulatory sequences in the Ebf1 locus may also be regulated by Pax5.
In conclusion, our analysis of the regulation of the Ebf1 gene shows that two distinct promoters integrate regulatory inputs from multiple determinants of B lymphopoiesis, whereby feedback loops, reciprocal activation, and cross-antagonism of individual components generate a complex regulatory network, which may help to stabilize developmental decisions and allow for an accurate differentiation process.
Supplementary Material
Acknowledgments
We are grateful to E. Kremmer (GSF) for the preparation of the monoclonal anti-EBF antibody and Ingrid Falk and Andreas Wurch for FACS sorting. We thank Veronique Azuara for help and advice with the replication timing analyses. Bacterial expression plasmids for the DNA-binding domains of Pax5 and Ets1 proteins have been a kind gift of Cynthia Wolberger. We thank Yuzuru Kanakura for the STAT5A-CA and cyclin D1-luciferase plasmids.
This work was supported by a grant from the German Research Foundation (DFG).
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arney, K. L., and A. G. Fisher. 2004. Epigenetic aspects of differentiation. J. Cell Sci. 117:4355-4363. [DOI] [PubMed] [Google Scholar]
- 2.Azuara, V., K. E. Brown, R. R. Williams, N. Webb, N. Dillon, R. Festenstein, V. Buckle, M. Merkenschlager, and A. G. Fisher. 2003. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 5:668-674. [DOI] [PubMed] [Google Scholar]
- 3.Bain, G., E. C. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 4.Bertolino, E., K. Reddy, K. L. Medina, E. Parganas, J. Ihle, and H. Singh. 2005. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat. Immunol. 6:836-843. [DOI] [PubMed] [Google Scholar]
- 5.Chambeyron, S., N. R. Da Silva, K. A. Lawson, and W. A. Bickmore. 2005. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132:2215-2223. [DOI] [PubMed] [Google Scholar]
- 6.Czerny, T., G. Schaffner, and M. Busslinger. 1993. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 7:2048-2061. [DOI] [PubMed] [Google Scholar]
- 7.DeKoter, R. P., H. J. Lee, and H. Singh. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity 16:297-309. [DOI] [PubMed] [Google Scholar]
- 8.Dias, S., H. Silva, Jr., A. Cumano, and P. Vieira. 2005. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 201:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson, A. D. 2005. Shaping time: chromatin structure and the DNA replication programme. Trends Genet 21:444-449. [DOI] [PubMed] [Google Scholar]
- 10.Fejer, G., M. M. Medveczky, E. Horvath, B. Lane, Y. Chang, and P. G. Medveczky. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts preferentially with the terminal repeats of the genome in vivo and this complex is sufficient for episomal DNA replication. J. Gen. Virol. 84:1451-1462. [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimmons, D., W. Hodsdon, W. Wheat, S. M. Maira, B. Wasylyk, and J. Hagman. 1996. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10:2198-2211. [DOI] [PubMed] [Google Scholar]
- 12.Fitzsimmons, D., R. Lutz, W. Wheat, H. M. Chamberlin, and J. Hagman. 2001. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 29:4154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuxa, M., J. Skok, A. Souabni, G. Salvagiotto, E. Roldan, and M. Busslinger. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvie, C. W., J. Hagman, and C. Wolberger. 2001. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell 8:1267-1276. [DOI] [PubMed] [Google Scholar]
- 15.Goetz, C. A., I. R. Harmon, J. J. O'Neil, M. A. Burchill, and M. A. Farrar. 2004. STAT5 activation underlies IL7 receptor-dependent B cell development. J. Immunol. 172:4770-4778. [DOI] [PubMed] [Google Scholar]
- 16.Hagman, J., C. Belanger, A. Travis, C. W. Turck, and R. Grosschedl. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760-773. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, R. S., T. K. Canfield, M. M. Lamb, S. M. Gartler, and C. D. Laird. 1993. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell 73:1403-1409. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, R. R., and K. Hayakawa. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595-621. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, L. Y., H. E. Liang, K. Johnson, C. Kang, and M. S. Schlissel. 2004. Pax5 activates immunoglobulin heavy chain V to DJ rearrangement in transgenic thymocytes. J. Exp. Med. 199:825-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kee, B. L., and C. Murre. 1998. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 188:699-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieslinger, M., S. Folberth, G. Dobreva, T. Dorn, L. Croci, R. Erben, G. G. Consalez, and R. Grosschedl. 2005. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev. Cell 9:757-767. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi, K., A. Y. Lai, C. L. Hsu, and M. Kondo. 2005. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 201:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosak, S. T., J. A. Skok, K. L. Medina, R. Riblet, M. M. Le Beau, A. G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296:158-162. [DOI] [PubMed] [Google Scholar]
- 24.Kozmik, Z., S. Wang, P. Dorfler, B. Adams, and M. Busslinger. 1992. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol. 12:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, H., and R. Grosschedl. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Lundgren, M., C. M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 27.Maier, H., R. Ostraat, H. Gao, S. Fields, S. A. Shinton, K. L. Medina, T. Ikawa, C. Murre, H. Singh, R. R. Hardy, and J. Hagman. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 5:1069-1077. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura, I., T. Kitamura, H. Wakao, H. Tanaka, K. Hashimoto, C. Albanese, J. Downward, R. G. Pestell, and Y. Kanakura. 1999. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina, K. L., J. M. Pongubala, K. L. Reddy, D. W. Lancki, R. Dekoter, M. Kieslinger, R. Grosschedl, and H. Singh. 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell 7:607-617. [DOI] [PubMed] [Google Scholar]
- 30.Mui, A. L., H. Wakao, A. M. O'Farrell, N. Harada, and A. Miyajima. 1995. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 14:1166-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie, L., M. Xu, A. Vladimirova, and X. H. Sun. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nutt, S. L., B. Heavey, A. G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556-562. [DOI] [PubMed] [Google Scholar]
- 33.Nutt, S. L., P. Urbanek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476-491. [DOI] [PubMed] [Google Scholar]
- 34.O'Keefe, R. T., S. C. Henderson, and D. L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116:1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 36.Perry, P., S. Sauer, N. Billon, W. D. Richardson, M. Spivakov, G. Warnes, F. J. Livesey, M. Merkenschlager, A. G. Fisher, and V. Azuara. 2004. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle 3:1645-1650. [PubMed] [Google Scholar]
- 37.Rolink, A. G., S. L. Nutt, F. Melchers, and M. Busslinger. 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 401:603-606. [DOI] [PubMed] [Google Scholar]
- 38.Sabbattini, P., and N. Dillon. 2005. The lambda5-VpreB1 locus—a model system for studying gene regulation during early B cell development. Semin. Immunol. 17:121-127. [DOI] [PubMed] [Google Scholar]
- 39.Sadoni, N., S. Langer, C. Fauth, G. Bernardi, T. Cremer, B. M. Turner, and D. Zink. 1999. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146:1211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schaniel, C., L. Bruno, F. Melchers, and A. G. Rolink. 2002. Multiple hematopoietic cell lineages develop in vivo from transplanted Pax5-deficient pre-B I-cell clones. Blood 99:472-478. [DOI] [PubMed] [Google Scholar]
- 42.Schebesta, M., P. L. Pfeffer, and M. Busslinger. 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity 17:473-485. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt, T. M., and J. C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17:749-756. [DOI] [PubMed] [Google Scholar]
- 44.Scott, E. W., R. C. Fisher, M. C. Olson, E. W. Kehrli, M. C. Simon, and H. Singh. 1997. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity 6:437-447. [DOI] [PubMed] [Google Scholar]
- 45.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 46.Seet, C. S., R. L. Brumbaugh, and B. L. Kee. 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 199:1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 48.Singh, H., K. L. Medina, and J. M. Pongubala. 2005. Contingent gene regulatory networks and B cell fate specification. Proc. Natl. Acad. Sci. USA 102:4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitnicka, E., C. Brakebusch, I. L. Martensson, M. Svensson, W. W. Agace, M. Sigvardsson, N. Buza-Vidas, D. Bryder, C. M. Cilio, H. Ahlenius, E. Maraskovsky, J. J. Peschon, and S. E. Jacobsen. 2003. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J. Exp. Med. 198:1495-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, E. M., P. Akerblad, T. Kadesch, H. Axelson, and M. Sigvardsson. 2005. Inhibition of EBF function by active Notch signaling reveals a novel regulatory pathway in early B-cell development. Blood 106:1995-2001. [DOI] [PubMed] [Google Scholar]
- 51.Smith, E. M., R. Gisler, and M. Sigvardsson. 2002. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J. Immunol. 169:261-270. [DOI] [PubMed] [Google Scholar]
- 52.Souabni, A., C. Cobaleda, M. Schebesta, and M. Busslinger. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity 17:781-793. [DOI] [PubMed] [Google Scholar]
- 53.Starr, D. B., W. Matsui, J. R. Thomas, and K. R. Yamamoto. 1996. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 10:1271-1283. [DOI] [PubMed] [Google Scholar]
- 54.Urbanek, P., Z. Q. Wang, I. Fetka, E. F. Wagner, and M. Busslinger. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901-912. [DOI] [PubMed] [Google Scholar]
- 55.White, E. J., O. Emanuelsson, D. Scalzo, T. Royce, S. Kosak, E. J. Oakeley, S. Weissman, M. Gerstein, M. Groudine, M. Snyder, and D. Schubeler. 2004. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. USA 101:17771-17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams, R. R., V. Azuara, P. Perry, S. Sauer, M. Dvorkina, H. Jorgensen, J. Roix, P. McQueen, T. Misteli, M. Merkenschlager, and A. G. Fisher. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132-140. [DOI] [PubMed] [Google Scholar]
- 57.Williams, R. R., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163-175. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Z., C. V. Cotta, R. P. Stephan, C. G. deGuzman, and C. A. Klug. 2003. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 22:4759-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang, Y., A. Jackson, L. Pan, K. Shen, and M. Dai. 2004. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 40:1165-1177. [DOI] [PubMed] [Google Scholar]
- 60.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]
- 61.Zink, D., A. H. Fischer, and J. A. Nickerson. 2004. Nuclear structure in cancer cells. Nat. Rev. Cancer 4:677-687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.