Abstract
Heparan sulfate (HS) chains are found in the extracellular matrix, covalently linked to core proteins collectively termed heparan sulfate proteoglycans (HSPGs). A wealth of data has demonstrated roles for HSPGs in the regulation of many cell surface signaling pathways that are crucial during development. Variations in the sulfation pattern along the HS chains influence their ability to interact with molecules such as growth factors, chemokines, morphogens, and adhesion molecules. Sulf1 and Sulf2 are members of a class of recently identified genes that encode heparan sulfate 6-O-endosulfatases (Sulf genes). The removal of 6-O-sulfate from HS via SULF activity influences the function of many factors, including Wnt, fibroblast growth factor, hepatocyte growth factor, heparin-binding epidermal growth factor, and bone morphogenetic protein. Given their possible developmental roles, we have examined Sulf gene expression during mouse embryogenesis. The two Sulf genes are expressed in a broad range of tissues throughout development with largely nonoverlapping expression patterns. Sulf2 transcripts are expressed in the lung, heart, placenta, and ribs. We generated a mouse line possessing a gene trap disruption of the Sulf2 gene. Mice homozygous for the Sulf2 gene trap allele are viable and fertile and have no major developmental defects on several genetic backgrounds. However, we observed strain-specific, nonpenetrant defects affecting viability, lung development, and growth in Sulf2 homozygous animals. These data suggest that Sulf2 may have roles in several tissues but that there is compensation by and/or redundancy with Sulf1.
Heparan sulfate proteoglycans (HSPGs) consist of a protein core possessing several covalently linked heparan sulfate (HS) polysaccharide chains (4). Over 95% of the HS found on mammalian cells is associated with five classes of HSPGs: syndecan, perlecan, glypican, agrin, and collagen XVIII (4). These macromolecules are ubiquitous components of the extracellular matrix and regulate numerous cellular behaviors through their direct interactions with a variety of molecules, including growth factors, chemokines, and adhesion molecules (40).
HS is synthesized as a polysaccharide chain of repeated d-glucuronic acid and d-glucosamine units; however, its fine structure is highly modified by a combination of epimerization, N sulfation, and O sulfation (4, 21). The regulation of HS modifications results in structural heterogeneity that is controlled in a tissue-specific and developmental manner (40). Importantly, many molecules interact preferentially with HS in a manner dependent on its structure, including the degree of sulfation on the N-, 3-O-, and 6-O- positions of glucosamine and the 2-O- position of the uronic acid units (13, 16). Therefore, it is not surprising that the control of HS structure is crucial for proper cellular behavior and development. In Drosophila melanogaster, the regulation of HS structure and HS sulfation has been linked to signal transduction involving the growth factors Wingless (Wg), Hedgehog (Hh), and Fibroblast growth factor (Fgf) (26). In mice, the knockouts of several HS biosynthetic genes have revealed the importance of not only HS but also regulated HS structure in developmental processes such as bone formation, organogenesis, and cancer (6, 15, 35).
Several reports have demonstrated the importance of sulfation at specific positions of HS for normal cellular behavior, most notably the 6-O- position. During heparan sulfate biosynthesis, heparan sulfate 6-O-sulfotransferases (HS6STs) catalyze 6-O sulfation of the N-sulfoglucosamine residue (17). A reduction of HS6ST activity in Drosophila, Caenorhabditis elegans, and zebra fish results in defects associated with reduced fibroblast growth factor (FGF), Slit/Robo, and Wnt signaling, respectively (5, 8, 20).
A novel family of extracellular glucosamine-6-sulfatases (SULF proteins) has been identified in quail, human, mouse, rat, zebra fish, C. elegans, and Drosophila (12, 30-32). The SULF proteins act as endosulfatases and remove the 6-O-sulfate from HS (12). In addition, the SULF proteins are secreted to the cell surface, in contrast to the lysosomal N-acetylglucosamine sulfatase that functions during HS degradation (12, 33, 43). In vitro binding studies have revealed the ability of SULF activity to modulate the binding of several factors to heparin/HS, including vascular endothelial growth factor, FGF-1, and stromal cell-derived factor 1/CXCL12 (41). Similarly, the overexpression of Sulf in cell culture can affect signaling by Wnt, FGF-2, FGF-4, hepatocyte growth factor (HGF), bone morphogenetic protein (BMP), and heparin-binding epidermal growth factor (2, 12, 22, 23, 42, 44). Furthermore, using an ex vivo model of quail development, Dhoot et al. (12) demonstrated the requirement for QSulf-1 in Wnt-dependent gene expression during somite formation.
While significant progress has been made toward identifying factors regulated by SULF activity in vitro, little is known regarding SULF regulation and function in vivo. In this study, we report the embryonic expression pattern of both murine Sulf genes during development. To examine the function of SULF activity in vivo, we generated a mouse strain possessing a gene trap disruption of the Sulf2 gene (previously known as MSulf-2). Mice homozygous for the Sulf2 gene trap allele displayed strain-specific, nonpenetrant defects in viability, growth, and lung development. In inbred genetic backgrounds, Sulf2 homozygous mutant mice did not display any defects in viability and did not have any overt abnormalities.
MATERIALS AND METHODS
Oligonucleotides.
The following oligonucleotides were used for PCR genotyping of Sulf2: oDL180 (Sulf2 exon 6 antisense strand), 5′-TGGGTGCATAGTTGTAACTCG-3′; oDL225 (Sulf2 exon 5 sense strand), 5′-GATTAGCCTGTCTGTCTCGAGC-3′; and oDL299 (pGT1TMpfs sense strand), 5′-GTTGAGATCCAGTTCGATGTAAC-3′.
Generation of Sulf2 mutant mice.
We obtained an embryonic stem (ES) cell line (XST155) containing a gene trap insertion in the sixth intron of the Sulf2 locus (see Fig. 4A) as a gift from William Skarnes (39). The gene trap construct pGT1TMpfs contains a splice acceptor upstream of the transmembrane domain, followed by lacZ (β-galactosidase [β-gal]) and a neomycin resistance fusion gene, TMβgeo (25, 28). The TMβgeo gene is linked via an internal ribosome entry site to the placental alkaline phosphatase gene.
FIG. 4.
Disruption of the murine Sulf2 gene. (A) Schematic representation of the gene trap insertion into the Sulf2 gene between exon 5 and exon 6. (B) TaqMan analysis of Sulf1 and Sulf2 mRNA in total RNA derived from the lungs and kidneys of wild-type (WT), Sulf2+/−, and three independent Sulf2−/− mice. Sulf gene expression was normalized to the expression of hypoxanthine phosphoribosyltransferase and presented as a relative quantitative score relative to Sulf expression in wild-type lungs. (C) RNA in situ analysis of Sulf gene expression in wild-type and Sulf2−/− homozygous E14.5 embryos. Sulf2 message was not detected in Sulf2−/− embryos, and the Sulf1 expression pattern does not appear to change. The signal sequence (S), splice acceptor (SA), transmembrane domain (TM), polyadenylation signal (pA), internal ribosomal entry site (IRES), placental alkaline phosphatase (PLAP), ventricle (V), and ribs (arrowhead) are shown. Bar, 200 μm (C). H&E, hematoxylin and eosin.
XST155 ES cells were injected into C57BL/6 blastocysts (MMRRC at UC Davis), and the resulting chimeric mice were outcrossed to C57BL/6 (Charles River Laboratory, Wilmington, MA) to test for germ line transmission. One chimeric male mouse transmitted the Sulf2 gene trap allele through his germ line and was used in the subsequent studies. The mutant Sulf2 allele and mouse strain were named Sulf2GT(pGT1TMpfs)155Ska (Sulf2 mutant). The wild-type Sulf2 allele (Sulf2+) was genotyped using the oligonucleotides oDL180 and oDL255. The Sulf2GT(pGT1TMpfs)155Ska allele was genotyped using the oligonucleotides oDL180 and oDL225.
The progeny derived from mating male mice chimeric for the XST155 ES cell line to C57BL/6 was considered inbred once (N1). Each successive mating of Sulf2+/− heterozygotes to inbred C57BL/6 was considered one backcross. FVB/N (N1) mice were derived from Sulf2+/− C57BL/6 (N2) parents, resulting in a mixed genetic background (FVB/N, 50%; C57BL/6, 37.5%; FVB/N, 12.5%). The FVB/N mice used in this study were purchased from Charles River Laboratory (Wilmington, MA).
All experiments were performed in accordance with protocols approved by the UCSF Committees on Animal Research (IACUC).
RNA in situ hybridization.
Paraffin sections were placed on acid-etched, 3-aminopropyl-triethoxysilase-treated slides and prepared for in situ hybridization as described previously (3). Plasmids were linearized with the appropriate restriction enzymes to transcribe either sense or antisense 35S-labeled riboprobes. Slides were washed at a final stringency of 65°C in 23× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dipped in emulsion, and exposed for 1 to 2 weeks. DNA was counterstained with Hoechst 33342. The Sulf1 probes include a fragment of the Sulf1 cDNA from nucleotides 1928 to 2613. The Sulf2 probes consist of nucleotides 1043 to 1726 from the Sulf2 cDNA, a region 3′ to the XST155 gene trap insertion site.
TaqMan analysis.
Real-time quantitative PCR was conducted by the Biomolecular Resource Center at the University of California, San Francisco under conditions as previously described (29). RNA was extracted from kidneys and lungs of adult female littermates. Sulf gene expression was normalized to the expression of hypoxanthine phosphoribosyltransferase and presented as a relative quantitative score relative to Sulf expression in wild-type lungs. The following oligonucleotide primers and TaqMan probes were used: Sulf1 forward primer, 5′-TCATTCGTGGTCCAAGCATAGA-3′; Sulf1 reverse primer, 5′-TGGTAGGAGCTAGGTCGATGTTC-3′; Sulf1 probe, 5′-6-carboxyfluorescein- CCAGGGTCGATAGTCCCACAGATTGTTC-butylhydroquinone 1-3′; Sulf2 forward primer, 5′-CTCACGGCTCTTCCCCAAT-3′; Sulf2 reverse primer, 5′-TCTGGGTTGGGTGCATAGTTG-3′; and Sulf2 probe, 5′-6-carboxyfluorescein-CGTCCCAGCACATCACACCGAGTT-butylhydroquinone 1-3′.
β-Galactosidase staining.
Tissue isolation, fixation, and processing for β-galactosidase staining were carried out as described by Hogan (19).
RESULTS
The Sulf genes are broadly expressed during embryogenesis.
We used RNA in situ hybridization to analyze Sulf gene expression between embryonic day 9.5 (E9.5) and E15.5. At E9.5 to E10.5, Sulf1 and Sulf2 mRNA was detected broadly throughout the embryo, with the highest levels in the developing nervous system, including the hindbrain and telencephalic vesicle (Fig. 1). Both Sulf1 and Sulf2 were highly expressed in the developing neural tube, with the Sulf1 mRNA restricted to the floor plate. Sulf2 gene expression was observed in the roof plate, the floor plate, and the ventral half of the neural tube. The expression patterns of Sulf1 and Sulf2 in the E9.5 spinal cord were maintained throughout development to E14.5. The expression patterns of Sulf1 and Sulf2 in the developing nervous system were similar to those seen for the RSulfFP1 and RSulfFP2 rat genes, respectively (31, 32). Sulf2 mRNA was abundant in the dorsal root ganglia and the developing somites, while Sulf1 expression in these tissues was not detected above background. A relatively high level of Sulf1 expression was observed in the hindbrain and the olfactory epithelium lining the olfactory pit.
FIG. 1.
RNA in situ hybridization analysis of Sulf gene expression in E9.5 and E10.5 embryos. Serial sagittal sections of mouse embryos at E9.5 (A) and E10.5 (B and C). The Sulf1 and Sulf2 negative control sense probes resulted in little background. Sulf1 expression was highest in the developing nervous system. Expression of Sulf2 was broad in E9.5 and E10.5 embryos. (C) Enlarged regions boxed in panel B. The olfactory epithelium (arrowhead), dorsal root ganglion (drg), first branchial arch (fb), floor plate (fp), neuroepithelium of the hindbrain (nhb), roof of the hindbrain (rhb), roof plate (rp), somites (sm), and telencephalic vesicle (tv) are shown. Bars, 400 μm (A and B) and 200 μm (C).
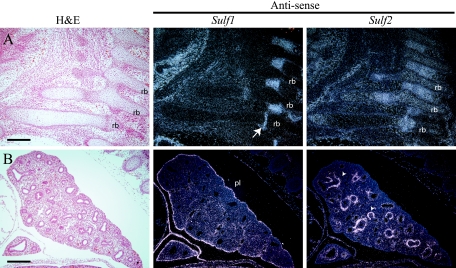
During later stages of development, the Sulf genes maintained broad patterns of expression, with a large degree of overlap in the developing spinal cord and skeleton (Table 1). High levels of Sulf expression were observed in regions of developing cartilage and bone, including the ribs, vertebrae, long bones, and limbs. Although Sulf1 and Sulf2 gene expression overlapped in regions of cartilage development, Sulf2 mRNA was predominant in the ribs and limbs. Sulf2 message was expressed throughout the developing rib, with the highest levels in the proliferating cartilage (Fig. 2A). Within the limbs, Sulf1 message appeared to be restricted to the mesenchyme surrounding developing bones. The liver did not express detectable Sulf message at any embryonic stage examined. Both Sulf genes were expressed in the developing heart; however, only Sulf2 mRNA was detected in the ventricles (see Fig. 4C). During all stages of embryonic development examined, the Sulf genes displayed nonoverlapping patterns of expression in the lung (Fig. 2B). Sulf1 mRNA was seen at low levels in the mesenchyme, with the highest level detected in the pleural lining of the lung. In contrast to that of Sulf1, expression of Sulf2 was restricted to a subset of mesenchymal cells immediately adjacent to the airway epithelium. The expression pattern of Sulf2 was similar to that of smooth muscle alpha actin, suggesting that the Sulf2-expressing cells were likely to be smooth muscle precursors.
TABLE 1.
Embryonic expression of Sulf1 and Sulf2 inferred from RNA in situ hybridizationa
| Testing site | Sulf1 expression | Sulf2 expression | Commentsb |
|---|---|---|---|
| Dorsal root ganglion | − | +++ | A |
| Notochord | − | ++ | A |
| Telencephalic vesicle | +++ | ++ | B |
| Choroid plexus | +++ | ++ | C |
| Hindbrain | +++ | + | D |
| Somites | − | +++ | A |
| Floor plate | +++ | +++ | D |
| Roof plate | − | ++ | A |
| Olfactory epithelium | ++ | − | A |
| Eye | ++ | ++ | B |
| Kidney | + | ++ | A |
| Liver | − | − | A |
| Adrenal gland | + | ++ | D |
| Testes | + | ++ | B |
| Diaphragm | ++ | + | D |
| Heart | + | ++ | A |
| Cartilage | + | +++ | B |
| Lung | + | ++ | A |
| Intestine | + | +++ | D |
| Stomach | ++ | +++ | B |
| Tooth bud | − | − | A |
| Ear | +++ | − | A |
| Skin | ++ | ++ | D |
| Tongue | + | +++ | B |
| First branchial arch | + | ++ | D |
| Placenta (maternal) | − | ++ | A |
| Placenta (embryonic) | − | +++ | A |
| Tip of genital tubercle | +++ | +++ | D |
−, no detectable expression; +, low level of expression; ++, medium level of expression; +++, high level of expression. The level of expression is estimated with respect to the signal intensity of each probe separately.
A, little or no overlapping expression; B, Sulf1 expression restricted to regions of Sulf2 expression; C, Sulf2 expression restricted to regions of Sulf2 expression; D, significant overlap of Sulf1 and Sulf2 expression.
FIG. 2.
Expression of Sulf1 and Sulf2 transcripts in embryonic ribs and lungs. Serial sagittal sections through E14.5 (A) ribs (line) and (B) lungs are shown. (A) Sulf1 transcripts were detected in a restricted region of the developing rib (arrow). Sulf2 message was detected along the length of the rib (rb), with the highest levels in the proliferating cartilage. (B) Sulf1 message was highest in the pleural lining (pl) of the lung, with low levels detected in the mesenchyme. Sulf2 mRNA appeared to be expressed in a subset of mesenchymal cells immediately adjacent to the epithelium (arrowheads). Bars, 100 μm (A) and 200 μm (B). H&E, hematoxylin and eosin.
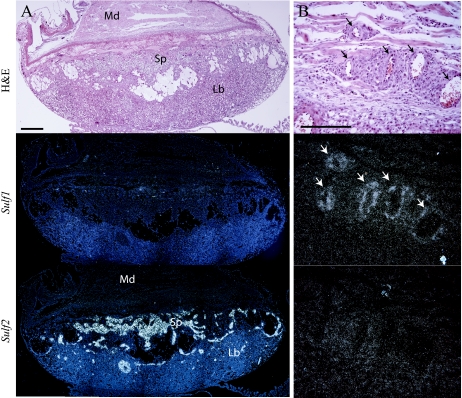
Sulf1 and Sulf2 transcripts were detected in the maternal decidua at 9.5 days postcoitum, displaying reciprocal expression patterns (data not shown). In the conceptus, Sulf2 was expressed in a subset of trophoblast derivatives. At E9.5, Sulf2 was expressed in the ectoplacental cone and trophoblast giant cells, and by E14.5, expression became restricted to the spongiotrophoblast layer (Fig. 3A and data not shown). The embryonic portion of the placenta did not show any detectable Sulf1 transcripts at any time point examined. However, expression of Sulf1 was observed in some vasculature of the maternal decidua and embryo (Fig. 3B and data not shown). Sulf2 transcripts were not detected in placental or embryonic vasculature.
FIG. 3.
Expression of Sulf transcripts in mouse placenta. (A) Serial sections through E14.5 placenta. Little or no Sulf1 expression was detected in the embryonic portion of the E14.5 placenta. Sulf2 message was abundant in the spongiotrophoblast layer (Sp). (B) Sulf1 message was observed in some vasculature (arrows) of the maternal decidua (Md). Sulf2 was not detected in any vasculature examined. Bars, 400 μm (A) and 200 μm (B). Lb, labyrinth; H&E, hematoxylin and eosin.
Generation of Sulf2 mutant mice.
To produce mice deficient in SULF2 activity, we utilized an ES cell line possessing a secretory trap insertional mutation in the Sulf2 gene (XST155, provided by William Skarnes) (25, 28, 37). The XST155 secretory trap insertion resulted in a transcriptional fusion of the first five Sulf2 exons to a reporter gene encoding a type II membrane-spanning domain fused to β-galactosidase/neomycin reporter, β-geo (TMβgeo) (Fig. 4A). The predicted mutant SULF2 protein (SULF2ΤΜβgeo) retained the N-terminal 246 amino acids that included the secretion signal and a truncated sulfatase domain fused to TMβgeo. The resulting SULF2 β-geo fusion protein lacked the C-terminal 629 amino acids of SULF2, including the catalytic cysteine essential for sulfatase activity and a hydrophilic domain necessary for sulfatase activity and HS binding (1). Thus, the SULF2ΤΜβgeo protein was not expected to retain enzymatic activity. The XST155 Sulf2 gene trap ES cells were injected into C57BL/6 recipient blastocysts. Chimeric founder males were then outcrossed to C57BL/6 females to yield mice heterozygous for the Sulf2 gene trap allele. The putative mutant Sulf2 allele was named Sulf2GT(pGT1TMpfs)155Ska. We use a superscript plus sign for the wild-type Sulf2 allele for the remainder of the text.
Mice heterozygous for the Sulf2 gene trap allele (Sulf2+/−) appeared phenotypically normal and were interbred to yield Sulf2 homozygous animals (Sulf2−/−). To confirm that the Sulf2 targeted allele effectively reduced the Sulf2 wild-type transcript, we used real-time quantitative PCR to measure the relative levels of Sulf1 and Sulf2 mRNA in the RNA extracted from the lungs and kidneys of wild-type, Sulf2+/−, and Sulf2−/− littermates (Fig. 4B). Sulf2 transcripts were reduced approximately 50% in Sulf2+/− RNA samples relative to those from wild-type mice. Full-length Sulf2 message was below the level of detection in all of the Sulf2−/− RNA samples examined. RNA in situ hybridization confirmed the absence of detectable wild-type Sulf2 mRNA in E9.5 and E14.5 embryos (Fig. 4C and data not shown). The examination of Sulf1 expression by in situ hybridization did not reveal any detectable change in the Sulf1 expression pattern in either E14.5 or E9.5 embryos (Fig. 4C and data not shown) that would suggest compensation for the lack of Sulf2 mRNA.
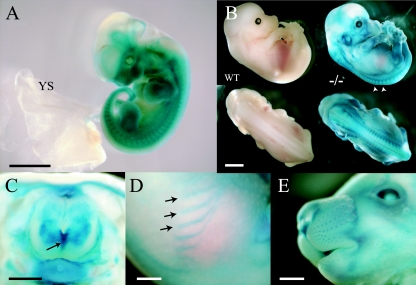
We examined the expression of the TMβgeo reporter in Sulf2+/− embryos at various developmental stages by staining for β-gal activity. At E11.5, TMβgeo was broadly expressed in the embryo, with no detectable expression in the yolk sac (Fig. 5A). Later in development at E13.5 and E15.5, β-gal activity was observed in the dorsal root ganglia, skin, spinal cord, whisker follicles, and cartilage of the ribs and limbs (Fig. 5B to E). The β-gal activity in Sulf2+/− embryos recapitulated endogenous Sulf2 gene expression, as observed by in situ hybridization (see above).
FIG. 5.
Analysis of Sulf2 expression of wild-type and Sulf2+/− heterozygous embryos as revealed by β-galactosidase staining. Whole-mount β-gal staining of mouse embryos at E11.5 (A), E13.5 (B), and E15.5 (C to E). (A) E11.5 Sulf2+/− embryos broadly expressed β-gal, with no staining observed in the yolk sac (YS). (B) Wild-type (WT) E13.5 embryos did not express detectable levels of β-galactosidase. In E13.5 Sulf2+/− embryos, expression was observed in the developing skeleton, dorsal root ganglia (arrowheads), eyes, and vibrissae. (C) Transverse view of E15.5 Sulf2+/−embryo. Strong staining was detected in the floor plate (arrow) and skin. (D and E) β-Galactosidase expression was also detected in the ribs (arrows) and whisker follicles. Bars, 2 mm (A and B), 0.5 mm (C), and 1 mm (D and E).
Sulf2−/− mice display strain-specific lethality.
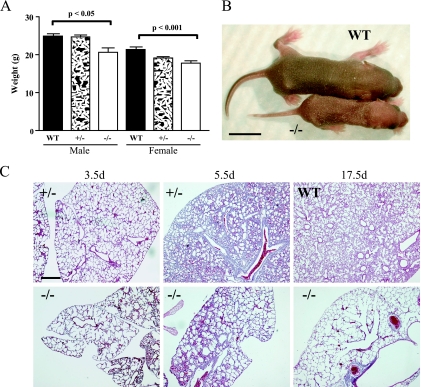
Homozygous Sulf2−/− mutant mice were generated by intercrossing Sulf2+/− heterozygotes on a mixed genetic background (N2; 129P2/Ola × C57BL/6). Genotyping of 3-week-old weanlings revealed significantly fewer Sulf2−/− mice than expected (Table 2). There were 48% fewer Sulf2−/− mice than wild-type littermates and 29% fewer Sulf2−/− mice than predicted for a normal Mendelian distribution. Many of the surviving Sulf2−/− mice at 3 weeks of age were visibly smaller than their wild-type littermates. Weights at 41.5 days of age revealed a significant weight difference between wild-type and homozygous animals (Fig. 6A). Sulf2−/− mice did not differ from their heterozygous littermates in overall health or longevity. Given the high level of Sulf2 expression in the adult ovaries (data not shown), we mated Sulf2−/− females. They were capable of producing viable pups as well as nursing them until weaning.
TABLE 2.
Genotypes of progeny from Sulf2+/− parents
| Strain and generationa | % 129P2 | No. of mice in litter/total no. of mice | No. of mice with indicated genotype
|
P valueb | ||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | ||||
| C57BL/6 | ||||||
| N2 Obs | 25.0 | 20/152 | 52 | 73 | 27 | 0.015* |
| N2 Exp | 38 | 76 | 38 | |||
| N3 Obs | 12.5 | 31/191 | 49 | 102 | 40 | 0.420 |
| N3 Exp | 47.75 | 95.5 | 47.75 | |||
| N7 Obs | 0.8 | 16/109 | 29 | 51 | 29 | 0.800 |
| N7 Exp | 27.25 | 54.5 | 27.25 | |||
| FVB/N | ||||||
| N1 Obs | 12.5 | 10/90 | 22 | 45 | 23 | 1.000 |
| N1 Exp | 22.5 | 45 | 22.5 | |||
| N7 Obs | 0.2 | 9/61 | 16 | 28 | 17 | 0.801 |
| N7 Exp | 15.25 | 30.5 | 15.25 | |||
N indicates the number of successive backcrosses needed to generate parental Sulf2+/−. Obs, observed; Exp, expected.
Probability of a non-Mendelian outcome determined by a χ2 test. *, P < 0.05.
FIG. 6.
Phenotypes associated with Sulf2−/− homozygous mice in a mixed genetic background (N2; 129P2/Ola × C57BL/6). (A) At 41.5 days (d), Sulf2−/− homozygous mice weighed less then their wild-type (WT) littermates. (B) Sulf2−/− animals were occasionally runted from birth. (C) Severely runted Sulf2−/− pups displayed an incompletely penetrant lung phenotype, including enlarged air spaces. Bars, 1 cm (B) and 400 μm (C).
Occasionally, Sulf2+/− intercrosses resulted in severely “runted” pups (Fig. 6B). The runted animals were typically ∼50% of the weight of littermates and were sacrificed at various ages based on their apparent health. Of eight runts observed, six were homozygotes and two were heterozygotes. A portion of the runted Sulf2 homozygotes appeared to be gasping, suggesting that there were lung-related defects. The lungs of three Sulf2−/− runts had enlarged air spaces (Fig. 6C), while the lungs of the remaining five pups were apparently normal at the gross and microscopic levels.
It is possible that the death of runted pups by the time of weaning and genotyping resulted in the non-Mendelian ratio observed with Sulf2+/− matings. However, the frequency of runted pups (∼6% Sulf2−/−) was lower than would account for the reduced number of homozygotes. To investigate whether Sulf2−/− homozygotes die in utero, we examined the embryos from Sulf2+/− heterozygous interbreedings. At E9.5, embryos were collected and genotyped by RNA in situ hybridization. We did not observe a significant deviation from the expected number of Sulf2−/− embryos (Sulf2+/+ and Sulf2+/−, 19; Sulf2−/−, 11). However, by E13.5 and E14.5, there were significantly fewer Sulf2−/− embryos than expected (Sulf2+/+, 13; Sulf2+/−, 16; Sulf2−/−, 2; dead, 3; P < 0.05). We observed dead embryos, which, based on their developmental morphology, appeared to have terminated at various time points ranging from E10.5 to E12.5 (Fig. 7). The dead embryos did not have an identifiable cause of death, and most were not genotyped due to the onset of necrosis and infiltration by maternal inflammatory cells. Sulf2−/− homozygous embryos and their placentas varied in size, with many being smaller than their wild-type littermates. However, no additional defects were observed in the Sulf2−/− embryos or their placentas.
FIG. 7.
Sulf2+/− heterozygous interbreeding in a mixed genetic background (N2; 129P2/Ola × C57BL/6) yielded dead embryos. Embryos were collected at (A to C) E13.5 and (D to F) E14.5. (B, C, and F) Dead embryos ranged in age from E10.5 to E12.5. (E) Sulf2−/− embryos were generally undistinguishable from their wild-type (WT) littermates.
Sulf2 influence on viability is modified by genetic background.
Sulf2−/− homozygous mice generated on a mixed genetic background (N2; 129P2/Ola × C57BL/6) appeared to display numerous phenotypes, including reduced size, lung abnormalities, and possibly reduced embryonic viability. However, none of these phenotypes were completely penetrant, and they all displayed various degrees of expression. In an attempt to exacerbate the Sulf2−/− phenotypes seen in the mixed genetic background, we backcrossed the Sulf2 mutant allele into several inbred backgrounds. The 129P2/Ola strain from which the XST155 ES cell line was derived was not readily available; therefore, we crossed the Sulf2 gene trap allele into the C57BL/6 and FVB/N genetic strains. Sulf2+/− intercrosses at the N3 and N7 generation of backcrossing into the C57BL/6 strain resulted in the expected Mendelian ratio of the Sulf2 mutant allele (Table 2). Inbred Sulf2−/− mice (N7; 129P2/Ola × C57BL/6) did not display any obvious size difference at the time of weaning. However, at 1 year of age, Sulf2−/− mice on the C57BL/6 background weighed ∼25% less than their wild-type littermates (data not shown).
Backcrossing the Sulf2 mutant allele into the FVB/N inbred strain likewise reduced the penetrance of Sulf2 mutant-associated lethality from Sulf2+/− intercrosses (Table 2), and Sulf2−/− mice did not differ from their wild-type littermates in blood cell counts, overall health, or longevity. We tested the fertility of Sulf2−/− homozygous males and females by mating each to wild-type siblings (Table 3). We did not observe any significant reduction in the fertility of Sulf2−/− homozygotes. However, upon Sulf2−/− homozygous intercrossing, we observed reduced litter sizes, suggesting that the absence of both maternal and embryonic Sulf2 can result in embryonic lethality (Table 3). E12.5 embryos collected from Sulf2−/− homozygous intercrosses did not differ in size or general appearance from wild-type embryos (lengths, 8.48 ± 0.13 mm and 8.45 ± 0.1 mm, respectively [means ± standard deviations]). However, the placentas of Sulf2−/− embryos were ∼20% smaller than those of wild-type embryos (cross-sectional areas, 16.30 ± 0.57 mm2 and 20.95 ± 0.61 mm2, respectively; P < 0.01). Other than reduced size, the placentas of Sulf2−/− embryos appeared grossly normal.
TABLE 3.
Litter sizes for Sulf2 breeding pairs
| Male strain | No. of mice in litter after intercrossing of male strain with indicated female straina
|
|
|---|---|---|
| Sulf2+/+ | Sulf2−/− | |
| Sulf2+/+ | 9.33 ± 0.30 (6)c | 8.00 ± 1.17 (4)b |
| Sulf2−/− | 8.78 ± 0.44 (9)b | 3.00 ± 0.35 (9)c |
Data are means ± standard deviations. The value in parentheses represents the number of mice in each breeding pair.
P < 0.0001 by one-way analysis of variance.
P < 0.001 by a comparison test.
Sulf2−/− mice are not sensitive to Ext2 dose.
Two glycosyltransferases, EXT1 and EXT2, are required for HS chain elongation (14). Mice deficient in either Ext1 or Ext2 lack HS and die during early embryogenesis prior to gastrulation (27, 38). The level of HS is important for normal development, as Ext2 heterozygous mice have completely penetrant defects in the developing ribs and long bones and can occasionally develop bony protuberances in their ribs called exostoses (or osteochondromas) (38). We hypothesized that the reduction of total HS in Ext2+/− mice would act as a sensitized background to modify the penetrance and/or expressivity of phenotypes observed in Sulf2−/− mice. We crossed Sulf2−/− mice to Sulf2+/−; Ext2+/− double heterozygotes. The genotyping of the progeny from these crosses revealed 30% fewer Sulf2−/− mice than predicted (n = 83; P < 0.01). This reduction in the observed number of Sulf2−/− homozygotes is consistent with the reduction obtained from Sulf2+/− interbreeding (Table 2). There was no correlation between the Sulf2 and Ext2 genotypes; therefore, eliminating a single Ext2 allele did not modify the penetrance of lethality associated with Sulf2−/− homozygotes.
Although the ribs of Ext2+/− mice are largely normal, they display a 100% penetrant defect consisting of premature hypertrophy in costochondral cartilage. This defect appears as nodules or single misplaced chondrocytes in the rib cartilage. It has been proposed that the defects seen in the ribs of Ext2 heterozygotes are caused by reduced FGF signaling (38). Many reports have demonstrated the requirement of 6-O sulfation for proper FGF signaling and the ability of SULF enzymatic activity to modify FGF/heparan interactions and FGF signaling (22, 41, 44). Therefore, Sulf2−/− mice lacking SULF2 activity in the developing ribs may have altered FGF signaling and thereby modify rib phenotypes observed in Ext2+/− mice. We examined the ribs from Sulf2−/−; Ext2+/− mice but detected no gross effect on the occurrence of Ext2+/−-associated nodules (data not shown). Sulf2−/−; Ext2+/− mice were also susceptible to the formation of rib exostoses. Overall, the analysis of skeletons from wild-type Sulf2 homozygotes and Sulf2−/−; Ext2+/− mice did not reveal any bone defects associated with a lack of SULF2 activity or any genetic interaction between Sulf2 and Ext2.
DISCUSSION
The ability of the SULF proteins to act as HS 6-O-endosulfatases and their localization to the cell surface make them good candidates for enzymes capable of regulating numerous signaling events during development. As an initial investigation into the regulation and function of SULF activity in vivo, we have characterized the expression of both Sulf1 and Sulf2 during mouse embryonic development. Sulf1 and Sulf2 expression is detected broadly throughout development, and many organs displayed a complementary expression pattern. Interestingly, the Sulf genes displayed overlapping expression patterns in tissues associated with complex assortments of instructive bioactive molecules, such as the developing limb, neural tube, and choroid plexus. The widespread expression of the Sulf mRNA during development suggests that the Sulf genes may play multiple roles in regulating signaling factors in many tissues; however, it is possible that the Sulf genes are subject to posttranscriptional regulation and act in a more restricted pattern during development. Analysis of SULF protein expression will be informative and may provide more clues regarding the possibility of redundancy.
Overexpression analyses of SULF protein in cell lines have clearly demonstrated the ability of SULF activity to positively or negatively affect signaling by factors important in development, including Wnt, FGF, heparin-binding epidermal growth factor, HGF, and BMP (2, 12, 22, 23, 42). Less well characterized are the requirements for heparan 6-O sulfation and SULF activity in vivo. To define the role of SULF activity in vivo, we generated a mouse strain deficient for Sulf2. A gene trap insertion of β-geo into the Sulf2 gene resulted in the disruption of the sulfatase domain and deletion of the C-terminal 629 amino acids. Both quantitative reverse transcriptase PCR and RNA in situ analysis revealed the absence of detectable full-length Sulf2 transcript in Sulf2−/− mice.
In a mixed genetic background, Sulf2−/− homozygotes were occasionally runted with lung abnormalities. Nonrunted Sulf2−/− homozygotes were significantly smaller than their wild-type littermates but were otherwise fertile and healthy. Furthermore, we observed a non-Mendelian ratio with Sulf2 heterozygous interbreedings, suggesting nonpenetrant lethality associated with Sulf2−/− homozygotes. The lethality associated with Sulf2−/− mice appears to occur over a range of time points, from E9.5 to E13.5. Although growth retardation and embryonic lethality can be caused by a multitude of perturbations, these phenotypes are often associated with defects during implantation, cardiovascular development, or a failure to make the transition from maternal to fetal hematopoiesis (9, 10). It is intriguing that although Sulf2 is not detected in the developing vasculature, it is the primary source of Sulf in both the placenta and the embryonic heart. Furthermore, inbred Sulf2−/− females mated to Sulf2−/− males yielded smaller litter sizes and produced embryos with smaller placentas than wild-type mice. However, we did not observe any gross histological defects in either the embryos or the placentas. Since Sulf2 mutant heterozygous breeding did not result in significant lethality associated with Sulf2−/− embryos, maternal SULF2 must be sufficient to rescue the lethality observed in Sulf2−/− crosses. In addition, Sulf2−/− females are capable of producing normal litter sizes when crossed to wild-type males; therefore, embryonic SULF2 is sufficient to rescue an absence of a maternal contribution. We believe that there is both an embryonic and a maternal Sulf2 contribution to placental function. Mouse placental development requires the activity of several signaling pathways that are modulated by HSPGs (Wnt, platelet-derived growth factor, BMP, FGF-4/FGF receptor 2, and HGF/Met) (11, 18). It is possible that in the absence of SULF2, signaling through one or more pathways is altered, resulting in a compromised placenta unable to support normal embryonic development.
We tested the possibility that an overall reduction in heparan sulfate may sensitize developing embryos to a reduction in SULF2 activity. Mice are sensitive to Ext2 dose since Ext2+/− mice have penetrant defects in rib development. However, Sulf2−/−; Ext2+/− double mutant mice were viable and did not have any gross embryonic defects. It is worth noting that as young adults, 50% of Sulf2−/−; Ext2+/− males developed genital abnormalities not seen in the single mutant animals. The underlying defect associated with this phenotype will require further investigation.
Sulf2−/− mice generated by Sulf2+/− intercrosses displayed low penetrance phenotypes affecting viability, lung development, and postnatal growth. These defects were not observed upon backcrossing of the Sulf2 allele into C57BL/6 and FVB/N strains, suggesting that there are modifier loci capable of influencing the Sulf2 phenotypes. However, on the FVB/N background, Sulf2− homozygous intercrosses did result in reduced embryonic viability. A recent study by Lamanna et al. reports an independent Sulf2 knockout showing no obvious phenotypes (24). However, those authors report anecdotal data that confirm our observations of nonpenetrant lethality of Sulf2 homozygotes as well as reduced body weight. It should be noted that many of the heparan-modifying enzymes knocked out in the mouse display nonpenetrant phenotypes and/or various expressivities. Mice deficient in either heparan sulfate 2-sulfotransferase or glucosaminyl N-deacetylase/N-sulfotransferase-1 exhibit 100% lethality but have nonpenetrant defects associated with skeletal and eye development (7, 34). Furthermore, gene knockout of heparan sulfate 3-O-sulfotransferase-1 results in a strain-specific lethality (36). The phenotypes seen in Sulf2−/− mice suggest a function for Sulf2 in several tissues. However, to study the role of Sulf2 during embryonic development, it will be necessary to identify a genetic background sensitive to Sulf2 levels. The phenotypes associated with the Sulf2−/− animals appeared to be correlated with the proportion of the 129P2/Ola genetic background. Backcrossing the Sulf2 mutant allele into the 129P2/Ola genetic background may yield a strain with more-penetrant phenotypes and facilitate the analysis of Sulf2 function during embryogenesis.
Given the requirement for proper regulation of heparan modifications and, specifically, heparan 6-O sulfation during development, it was interesting to find no gross embryonic abnormalities in Sulf2-null animals when backcrossed into several inbred strains. Two plausible explanations can be offered for our observations. First, unlike activity for enzymes such as HS6STs, which are required for heparan 6-O sulfation, Sulf activity likely acts to fine tune the levels of heparan 6-O sulfation. Therefore, it is not surprising that a lack of Sulf activity results in more-subtle effects than a reduction in HS6ST levels. Second, the expression patterns of Sulf1 and Sulf2 display overlap in many tissues, and therefore, redundancy may explain the lack of a phenotype. The analysis of Sulf1 and Sulf2 double mutant mice will address the issue of redundancy and may be a necessary step for understanding the role of Sulf function during embryonic development.
In summary, Sulf2 may have multiple roles during murine embryonic development as demonstrated by strain-specific phenotypes, but Sulf2 activity is not essential for development or postnatal viability. The function of SULF activity may become more apparent during stress response or disease. Several studies have associated the misregulation of Sulf expression with several forms of cancer, including human ovarian cancer, pancreatic cancer, and hepatocellular carcinoma (22, 23). Furthermore, we have previously described the upregulation of Sulf2 during human breast cancer and in two mouse models of mammary carcinoma, MMTV-Wnt1 and MMTV-Neu (29). The examination of these tumor models and other mouse models of disease in a Sulf2-null background may be an important step in furthering our understanding of Sulf function.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA057621 [Z.W.], HL075602 [Z.W.], ES012801[Z.W.], and GM57411 [S.D.R.]) and a fellowship from the U.S. Department of Defense Breast Cancer Program (DAMD17-03-1-0485 to D.H.L.).
We thank William Skarnes for the Sulf2 XST155 gene trap ES cell line, Dominique Stickens for the use of Ext2 mice, Ying Yu and Bernard Thompson for excellent animal care, and Helen Capili for sectioning. We also thank Angela A. Andersen and Ian D. Chin-Sang for critically reading the manuscript.
Footnotes
Published ahead of print on 20 November 2006.
REFERENCES
- 1.Ai, X., A. T. Do, M. Kusche-Gullberg, U. Lindahl, K. Lu, and C. P. Emerson, Jr. 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 281:4969-4976. [DOI] [PubMed] [Google Scholar]
- 2.Ai, X., A. T. Do, O. Lozynska, M. Kusche-Gullberg, U. Lindahl, and C. P. Emerson, Jr. 2003. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 162:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht, U., G. Eichele, J. A. Helms, and H. Lu. 1997. Visualization of gene expression patterns by in situ hybridization, p. 23-48. In G. Daston (ed.), Molecular and cellular methods in developmental toxicology. CRC Press, Boca Raton, FL.
- 4.Bernfield, M., M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729-777. [DOI] [PubMed] [Google Scholar]
- 5.Bink, R. J., H. Habuchi, Z. Lele, E. Dolk, J. Joore, G. J. Rauch, R. Geisler, S. W. Wilson, J. den Hertog, K. Kimata, and D. Zivkovic. 2003. Heparan sulfate 6-O-sulfotransferase is essential for muscle development in zebrafish. J. Biol. Chem. 278:31118-31127. [DOI] [PubMed] [Google Scholar]
- 6.Blackhall, F. H., C. L. Merry, E. J. Davies, and G. C. Jayson. 2001. Heparan sulfate proteoglycans and cancer. Br. J. Cancer 85:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock, S. L., J. M. Fletcher, R. S. Beddington, and V. A. Wilson. 1998. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 12:1894-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bülow, H. E., and O. Hobert. 2004. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron 41:723-736. [DOI] [PubMed] [Google Scholar]
- 9.Conway, S. J., A. Kruzynska-Frejtag, P. L. Kneer, M. Machnicki, and S. V. Koushik. 2003. What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis 35:1-21. [DOI] [PubMed] [Google Scholar]
- 10.Copp, A. J. 1995. Death before birth: clues from gene knockouts and mutations. Trends Genet. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 11.Cross, J. C. 2005. How to make a placenta: mechanisms of trophoblast cell differentiation in mice—a review. Placenta 26(Suppl. A):S3-S9. [DOI] [PubMed] [Google Scholar]
- 12.Dhoot, G. K., M. K. Gustafsson, X. Ai, W. Sun, D. M. Standiford, and C. P. Emerson, Jr. 2001. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293:1663-1666. [DOI] [PubMed] [Google Scholar]
- 13.Esko, J. D., and U. Lindahl. 2001. Molecular diversity of heparan sulfate. J. Clin. Investig. 108:169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esko, J. D., and S. B. Selleck. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71:435-471. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, E., and L. Kjellen. 2001. Heparan sulfate: lessons from knockout mice. J. Clin. Investig. 108:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habuchi, H., O. Habuchi, and K. Kimata. 2004. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj. J. 21:47-52. [DOI] [PubMed] [Google Scholar]
- 17.Habuchi, O. 2000. Diversity and functions of glycosaminoglycan sulfotransferases. Biochim. Biophys. Acta 1474:115-127. [DOI] [PubMed] [Google Scholar]
- 18.Hemberger, M., and J. C. Cross. 2001. Genes governing placental development. Trends Endocrinol. Metab. 12:162-168. [DOI] [PubMed] [Google Scholar]
- 19.Hogan, B. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 20.Kamimura, K., M. Fujise, F. Villa, S. Izumi, H. Habuchi, K. Kimata, and H. Nakato. 2001. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene. Structure, expression, and function in the formation of the tracheal system. J. Biol. Chem. 276:17014-17021. [DOI] [PubMed] [Google Scholar]
- 21.Kramer, K. L., and H. J. Yost. 2003. Heparan sulfate core proteins in cell-cell signaling. Annu. Rev. Genet. 37:461-484. [DOI] [PubMed] [Google Scholar]
- 22.Lai, J., J. Chien, J. Staub, R. Avula, E. L. Greene, T. A. Matthews, D. I. Smith, S. H. Kaufmann, L. R. Roberts, and V. Shridhar. 2003. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J. Biol. Chem. 278:23107-23117. [DOI] [PubMed] [Google Scholar]
- 23.Lai, J. P., J. R. Chien, D. R. Moser, J. K. Staub, I. Aderca, D. P. Montoya, T. A. Matthews, D. M. Nagorney, J. M. Cunningham, D. I. Smith, E. L. Greene, V. Shridhar, and L. R. Roberts. 2004. hSulf1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology 126:231-248. [DOI] [PubMed] [Google Scholar]
- 24.Lamanna, W. C., R. J. Baldwin, M. Padva, I. Kalus, G. Ten Dam, T. H. van Kuppevelt, J. T. Gallagher, K. von Figura, T. Dierks, and C. L. R. Merry. 2006. Heparan sulfate 6-O-endosulfatases: discrete in vivo activities and functional cooperativity. Biochem. J. 400:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leighton, P. A., K. J. Mitchell, L. V. Goodrich, X. Lu, K. Pinson, P. Scherz, W. C. Skarnes, and M. Tessier-Lavigne. 2001. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410:174-179. [DOI] [PubMed] [Google Scholar]
- 26.Lin, X., and N. Perrimon. 2000. Role of heparan sulfate proteoglycans in cell-cell signaling in Drosophila. Matrix Biol. 19:303-307. [DOI] [PubMed] [Google Scholar]
- 27.Lin, X., G. Wei, Z. Shi, L. Dryer, J. D. Esko, D. E. Wells, and M. M. Matzuk. 2000. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev. Biol. 224:299-311. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell, K. J., K. I. Pinson, O. G. Kelly, J. Brennan, J. Zupicich, P. Scherz, P. A. Leighton, L. V. Goodrich, X. Lu, B. J. Avery, P. Tate, K. Dill, E. Pangilinan, P. Wakenight, M. Tessier-Lavigne, and W. C. Skarnes. 2001. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 28:241-249. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto-Tomita, M., K. Uchimura, A. Bistrup, D. H. Lum, M. Egeblad, N. Boudreau, Z. Werb, and S. D. Rosen. 2005. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia 7:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morimoto-Tomita, M., K. Uchimura, Z. Werb, S. Hemmerich, and S. D. Rosen. 2002. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 277:49175-49185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagamine, S., S. Koike, K. Keino-Masu, and M. Masu. 2005. Expression of a heparan sulfate remodeling enzyme, heparan sulfate 6-O-endosulfatase sulfatase FP2, in the rat nervous system. Brain Res. Dev. Brain Res. 159:135-143. [DOI] [PubMed] [Google Scholar]
- 32.Ohto, T., H. Uchida, H. Yamazaki, K. Keino-Masu, A. Matsui, and M. Masu. 2002. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes Cells 7:173-185. [DOI] [PubMed] [Google Scholar]
- 33.Parenti, G., G. Meroni, and A. Ballabio. 1997. The sulfatase gene family. Curr. Opin. Genet. Dev. 7:386-391. [DOI] [PubMed] [Google Scholar]
- 34.Ringvall, M., J. Ledin, K. Holmborn, T. van Kuppevelt, F. Ellin, I. Eriksson, A. M. Olofsson, L. Kjellen, and E. Forsberg. 2000. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J. Biol. Chem. 275:25926-25930. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson, R. D. 2001. Heparan sulfate proteoglycans in invasion and metastasis. Semin. Cell Dev. Biol. 12:89-98. [DOI] [PubMed] [Google Scholar]
- 36.Shworak, N. W., S. HajMohammadi, A. I. de Agostini, and R. D. Rosenberg. 2002. Mice deficient in heparan sulfate 3-O-sulfotransferase-1: normal hemostasis with unexpected perinatal phenotypes. Glycoconj. J. 19:355-361. [DOI] [PubMed] [Google Scholar]
- 37.Skarnes, W. C., J. E. Moss, S. M. Hurtley, and R. S. Beddington. 1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stickens, D., B. M. Zak, N. Rougier, J. D. Esko, and Z. Werb. 2005. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development 132:5055-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stryke, D., M. Kawamoto, C. C. Huang, S. J. Johns, L. A. King, C. A. Harper, E. C. Meng, R. E. Lee, A. Yee, L. L'Italien, P. T. Chuang, S. G. Young, W. C. Skarnes, P. C. Babbitt, and T. E. Ferrin. 2003. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 31:278-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tumova, S., A. Woods, and J. R. Couchman. 2000. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int. J. Biochem. Cell Biol. 32:269-288. [DOI] [PubMed] [Google Scholar]
- 41.Uchimura, K., M. Morimoto-Tomita, A. Bistrup, J. Li, M. Lyon, J. Gallagher, Z. Werb, and S. D. Rosen. 2006. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viviano, B. L., S. Paine-Saunders, N. Gasiunas, J. Gallagher, and S. Saunders. 2004. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 279:5604-5611. [DOI] [PubMed] [Google Scholar]
- 43.von Figura, K., B. Schmidt, T. Selmer, and T. Dierks. 1998. A novel protein modification generating an aldehyde group in sulfatases: its role in catalysis and disease. Bioessays 20:505-510. [DOI] [PubMed] [Google Scholar]
- 44.Wang, S., X. Ai, S. D. Freeman, M. E. Pownall, Q. Lu, D. S. Kessler, and C. P. Emerson, Jr. 2004. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. USA 101:4833-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]