Summary
Objectives
As part of an investigation of connecting health professionals and the lay public to both disease and genomic information, we assessed the availability and nature of the data from the Human Genome Project relating to human genetic diseases.
Methods
We focused on a set of single gene diseases selected from main topics in MEDLINEplus, the NLM’s principal resource focused on consumers. We used publicly available websites to investigate specific questions about the genes and gene products associated with the diseases. We also investigated questions of knowledge and data representation for the information resources and navigational issues.
Results
Many online resources are available but they are complex and technical. The major challenges encountered when navigating from phenotype to genotype were (1) complexity of the data, (2) dynamic nature of the data, (3) diversity of foci and number of information resources, and (4) lack of use of standard data and knowledge representation methods.
Conclusions
Three major informatics issues arise from the navigational challenges. First, the official gene names are insufficient for navigation of these web resources. Second, navigational inconsistencies arise from difficulties in determining the number and function of alternate forms of the gene or gene product and maintaining currency with this information. Third, synonymy and polysemy cause much confusion. These are severe obstacles to computational navigation from phenotype to genotype, especially for individuals who are novices in the underlying science. Tools and standards to facilitate this navigation are sorely needed.
Keywords: Phenotype, genotype, databases (genetic), computational navigation, information systems, interoperability
Introduction
The human genome sequence contains the genetic code that forms the basis of each human being. The human DNA sequence, determined by the massive Human Genome Project (HGP) that spans multiple laboratories, countries and continents, was completed in draft form in the spring of 2001 [1]. The subsequent activities have concentrated on progressing from draft form to completed form, switching to a focus on the determination of what are the functions of the genes, and what variations exist in the genome. The information arising from the HGP promises to alter our perceptions of disease and health and to change the way medicine is practiced. The nature of our genes and how they give rise to various illnesses (both the common diseases of middle/advanced age and the rarer single gene disorders) are being explored in depth with the potential of improved health and longevity for the public.
The publicity associated with the HGP has led many people to ask questions about the health implications and specifically, “What data are coming from the Human Genome Project that relate to my disease and my risks for disease?” An exploration of the databases that hold the information about connections between specific genes and diseases reveals 1400 human genes that have been proven to cause at least one disease and where the DNA sequence and molecular function are determined.1 This extensive information is available over publicly accessible internet sites, but not easily accessible to the public because of its technical nature.
This project was undertaken to assess what data from the HGP are available on common inherited diseases and how accessible the data are to the lay public. The work focused on the major information resources containing consumer health information, genome and proteome knowledge and the methods to navigate among them. This paper will focus on informatics issues that arise when navigating the information systems with the HGP data. The project provides the foundation for creating an integrated information system to connect the public to the health implications of the HGP data.
Background
Over 300 information resources are publicly available over the internet and have data associated with various aspects of genes, gene function, and diseases across multiple species [2]. Table 1 lists some of the major information resources containing data that relate directly to human genetic diseases or to data from the Human Genome Project that relate directly to human genes that cause disease. The basic data of the HGP (and all other species) resides in synchronized sequence databases held by the National Library of Medicine’s (NLM) GenBank [3], the DNA Data Bank of Japan (DDBJ) [4] and the European Molecular Biology Laboratory’s (EMBL) Nucleotide Sequence Database [5]. The data pertaining to the gene products arising from these sequences are contained in databases held by the same three groups in the LocusLink [6], SWISS-PROT/TrEMBL [7], and KEGG [8] systems, but also in various other public resources. The major resource for human genetic diseases is the Online Mendelian Inheritance in Man (OMIM) [9] catalog of human genes and human genetic diseases, closely linked to other NLM resources. The scientific literature references for all of this work are held in MEDLINE [6] by the NLM. Most of the other gene resources listed in Table 1 are derived from these basic sources and add value to them in numerous ways.
Table 1.
Some major information resources pertaining to human genes and genetic diseases
| Primary Focus: | |
| Disease information | |
| • MEDLINEplus | medlineplus.gov/ |
| • ClinicalTrials.gov | clinicaltrials.gov/ |
| • Gene Reviews | genetests.org/ |
| • Genetic and Rare Diseases Information Center | genome.gov/HEALTH |
| • Online Mendelian Inheritance in Man (OMIM) | ncbi.nlm.nih.gov/OMIM |
| • Genes and Disease | ncbi.nlm.nih.gov/disease/ |
| • Atlas of Genetics and Cytogenetics in Oncology and Hematology | infobiogen.fr/services/chromcancer/ |
| • Cancer.gov | cancer.gov/ |
| • Unified Medical Language System (UMLS) | umlsinfo.nlm.nih.gov |
| Genetic disease testing | |
| • GeneTests | genetests.org/ |
| Gene, gene product, gene function information | |
| • LocusLink | ncbi.nlm.nih.gov/LocusLink/ |
| • SwissProt/TrEMBL | expasy.org/sprot/ |
| • Human Gene Mutation Database (HGMD) | archive.uwcm.ac.uk/uivcm/mg/hgmd0.html |
| • GeneCards | bioinformatics.weizmann.ac.il/cards/ |
| • Genome Data Bank (GDB) | gdb.org/ |
| • ENZYME Nomenclature Database (Enzyme Commission) | expasy.org/enzyme/ |
| • Kyoto Encyclopedia of Genes and Genomes (KEGG) | genome.ad.jp/kegg/ |
| • Gene Ontology Consortium (GO) | geneontology.org/ |
| • Database of Interacting Proteins (DIP) | dip.doe-mbi.ucla.edu/dip/Main.cgi |
| • Protein Information Resource (PIR) | pir.georgetown.edu/ |
| • International Protein Index (IPI) | ensembl.org/IPI/ |
| Gene names | |
| • Genew- Human Genome Nomenclature Committee (Human Genome Organization) | gene.ucl.ac.uk/nomenclature/ |
| Chromosome location and maps | |
| • Map Viewer | ncbi.nlm.nih.gov/mapview |
| DNA sequences | |
| • GenBank | ncbi.nlm.nih.gov/Genbank/ |
| • EMBL nucleotide sequence database | ebi.ac.uk/emb/ |
| • DNA Data Bank of Japan (DDBJ) | ddbj.nig.ac.jp |
| • Single nucleotide polymorphisms (dbSNP) | ncbi.nlm.nih.gov/SNP |
| • Reference Sequences (RefSeq) | ncbi.nlm.nih.gov/LocusLink/refseq.html |
| • Unigene | www.ncbi.nlm.nih.gov/UniGene/ |
| Homologies among species | |
| • HomoloGene | ncbi.nlm.nih.gov/HomoloGene/ |
| Journal literature | |
| • MEDLINE | pubmed.gov |
Some consumer-focused information resources have information on genetic diseases, but the consumer resources are not usually closely interlinked with the molecular biology databases.The NLM has two such resources: MEDLINEplus [10] with over 100 genetic diseases in its main topics and subtopics, and ClinicalTrials.gov [11] with information on clinical trials for many genetic diseases. The GeneTests [12] system of the University of Washington is focused on health professionals rather than the public. It lists commercially available laboratory tests for the genetic diseases; the related Gene Reviews presents clinical summaries done by genetics experts.
Methods
An analysis in June 2002 of MEDLINE-plus, the NLM’s principal resource focused on consumers, revealed that several specific inherited diseases were main topics or subtopics in MEDLINEplus and also fulfilled four other criteria: 1) entries in OMIM for a specific disease, 2) entries in LocusLink for specific gene products for the OMIM disease, 3) disease summary in GeneReviews; and 4) at least three commercial laboratories doing DNA tests as listed in GeneTests. Thirteen of these diseases were examined in detail to investigate the potential navigation from phenotype (disease) to genotype and the current systems that contained the data of interest. The thirteen diseases studied were
achondroplasia
Canavan disease
cystic fibrosis
Duchenne muscular dystrophy
Gaucher disease
Huntington disease
limb girdle muscular dystrophy
Marfan syndrome
myotonic dystrophy
neurofibromatosis
phenylketonuria
polycystic kidney disease
tuberous sclerosis.
The selection as a topic or subtopic in MEDLINEplus guarantees a range of materials on patient education2, family support3, glossaries of genetics concepts4 and explanations of genetic testing5.
The specific questions of this study related to where genetic data that would potentially be of interest to patients and their families as well as health care professionals could be found. The questions arose from experience with genetic counseling sessions where these were the questions often asked by patients or considered important by genetic counselors before giving an informed prognosis and risk estimates to the patients.
What gene(s) causes the disease?
On which chromosomes are the genes located?
What are the normal functions of the gene product(s)
What mutations have been found in the genes?
What are the functions of the mutated gene product(s)?
Which laboratories are performing DNA tests for the mutations?
Are there gene therapies or clinical trials for the disease?
Do the genes cause any other diseases besides the target disease?
What names are used to refer to the genes and the diseases in these resources?
The study considered where the information resided to answer these questions and the navigation issues encountered. We also investigated whether the genes, gene products and diseases were included in the Unified Medical Language System® version 2002AA (UMLS®)6 in order to determine if these specific concepts were included in the vocabularies covered by that system. We considered questions of knowledge and data representation for the information resources and navigational questions among systems. The systems where these questions were investigated were the publicly available resources listed in Table 1 that also lists the URLs for all of the systems described in this paper. The search strategies were executed and the data evaluated by the first author who is trained in both medical genetics and informatics.
Results
The results will be presented in two examples, one for the disease achondroplasia and one for the disease Marfan syndrome. Table 2 presents the answers to the nine questions for the disease achondroplasia. This example serves to demonstrate the type and complexity of the data. The system navigation from a consumer system (MED-LINEplus) to the bioinformatics resources for the genotype information is given by the example of the disease Marfan syndrome. Figure 1 illustrates the path through the information resources used to answer the questions about Marfan syndrome; the traversal was from the consumer health resources through disease-related resources and then to the bioinformatics resources. Table 3 shows the information resources that in general hold answers to the questions of this study.
Table 2.
The answers to the questions for the disease achondroplasia.
Seventeen separate information resources listed in Table 1 (MEDLINE, OMIM, LocusLink, GeneReviews, GeneTests, Gene Ontology, HGMD, HGNC, GeneCards, PIR, ENZYME, DIP, Atlas of Cytogenetics in Oncology and Hematology, Cancer.gov, RefSeq, ClinicalTrials.gov, UMLS) contained pieces of this data.
|
Fig. 1.
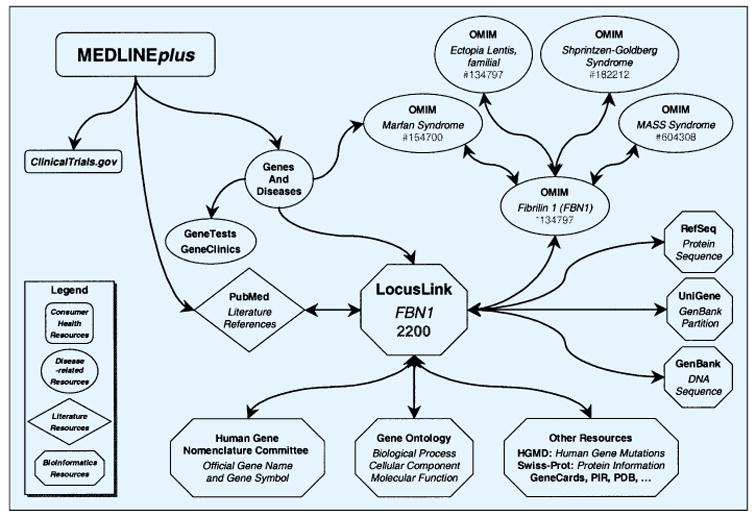
Navigation path from phenotype to genotype through the information resources used to answer the questions of the study as they pertain to Marfan syndrome
Table 3.
Information resources that hold answers to the questions of the study.
The complete set of data to answer the questions only comes after traversing all resources. Note that while a consumer audience can use OMIM to find information on almost all of these questions, it is generally considered dense reading and not readily understandable.
|
All of the data sought for the set of thirteen diseases was found in the systems navigated, although the full description of normal gene function was not always satisfactory without reading the primary literature. It was easier to navigate completely online from phenotype to genotype with some of the diseases investigated than with others. An example comes from the Marfan Syndrome and is shown in Figure 1. Four of the thirteen diseases can be traversed in an analogous manner (Gaucher disease, Huntington disease, Marfan Syndrome, myotonic dystrophy (for one of the two causative genes)). Genes and Disease serves as the only linking system between the consumer-health oriented MEDLINE-plus system and the knowledge bases of molecular biology. Without a linking system, the phenotype-genotype connections are much more difficult when starting from MEDLINEplus and rely entirely on the prior knowledge of the navigator. Clinical trial information would only come through a link from the MEDLINEplus page to ClinicalTrials.gov since none of the other systems link their users directly to the clinical trials for the diseases in question.
For these thirteen target diseases, there were 31 genes, 189 gene names, 59 gene products (including isoforms), 56 associated diseases, and 240 disease names. The list of synonyms for the gene and gene product names is undoubtedly incomplete because there is no general agreement on what names to use and because there is no single source to collect all of them. All of the thirteen target diseases were represented in the UMLS and the Medical Subject Headings (MeSH)7 used to index the literature; in some cases the specific disease name or subtype name was not found in MeSH as a main heading but rather was an entry term for a more general disease category (e.g., MeSH includes muscular dystrophies as the official MeSH Heading but includes the multiple types of muscular dystrophy as entry terms). The full list of diseases caused by the genes was not always found in the UMLS (including MeSH). 28 of the 31 genes were found in both UMLS and MeSH; two gene products were found in a general category but not the specific gene; one gene (FKRP) was not found in 2002 UMLS and is still not in the 2003AA UMLS. In general, the phenotype to genotype knowledge is most sketchy as it relates to a description of normal function of the genes.
Four major challenges were encountered when navigating from phenotype to genotype:
-
Complexity of data: The sheer complexity and volume of the data emanating from the HGP is daunting even to the scientific community. The gene names and gene product names are especially complex. The number of synonyms and the non-intuitive nature of the synonyms for various diseases, genes, and gene symbols make it difficult to find comprehensive information. The nomenclature committee of the Human Genome Organization (HUGO) decides upon an “official” gene name and gene symbol and keeps an online database with the official name and symbol. However, the official gene names are the result of scientific compromise and debate, and are frequently unwieldy and difficult to remember or use computationally, inviting the use of more synonyms. For example, the official name for the gene that when mutated causes Duchenne and Becker Muscular Dystrophy and X-linked cardiomyopathy is dystrophin (muscular dystrophy, Duchenne and Becker type). Most people abbreviate this to dystrophin, even though this makes the name identical to the name for the gene product in the rat (Dystrophin) (note upper case D) and the fruit fly (dystrophin) (note lower case d) and similar to that of the mouse (dystrophin, muscular dystrophy). Furthermore, many knowledge bases throughout the world do not uniformly use the official gene names and gene symbols. For example, for the gene that when mutated causes myotonic dystrophy type 1, the official gene product name is dystrophia myotonica-protein kinase and the official symbol is DMPK. But SWISS-PROT uses the entry name DMK_HUMAN and the protein name myotonin-protein kinase. Neither the SWISS-PROT entry name or protein name is included in the synonym list of Locus-Link or the HGNC database. However, the only entry terms into the Gene Ontology database are the SWISS-PROT ID or entry names.
The official gene names often include metadata that link to one or more diseases, although not necessarily the complete list of diseases. The fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) gene gives metadata about two of the eleven disorders caused by mutations in the gene. Besides disease name, other metadata are often included in the gene names or symbols, including the species (e.g., FBN1_HUMAN), the disease inheritance pattern (e.g., polycystic kidney disease 1 (autosomal dominant)), and the biochemical pathway (e.g., polycystin precursor). While it is not exceptional in biomedical terminology to have metadata as part of the terms, the extent to which this occurs in the human genes names is exceptional.
Dynamic nature of the data: The existing scientific and clinical systems are in a constant state of flux because of the rapidity of developments coming from a global research effort. The situation is unlikely to settle down within the foreseeable future. From the time a journal article appears with a new disease-gene connection, it takes almost six months for the knowledge to cascade through all of the interconnected bioinformatics systems. The nature and number of gene products is still scientifically labile. More examples of genes with multiple gene products (isoforms) arise daily, some of which are active in specific tissues or at a specific developmental stage. For example, the human dystrophin gene produces eighteen known isoforms from the use of alternate promoters or alternate exons. Knowledge about these situations is still emerging.
-
Diverse foci and number of data/knowledge base systems: Table 1 only lists a handful of the information resources available with data from the HGP. With over 300 resources, it is a daunting task to gather all of the information and navigate the systems. Further, most of the existing systems with information related to the data from the HGP are focused on the scientific or subspecialty medicine communities and presuppose a working knowledge of the science behind the databases and the tools.
Most systems focused on consumer access to health information, such as MEDLINEplus, do not generally link to the genomics knowledge bases because of the lack of an obvious way to connect the two worlds. Furthermore, the scientific and clinical databases are difficult for the general public to comprehend, and the consumer systems do not link to them largely for this reason.
Data and knowledge representation: The lack of standard methods for representing the data makes it a challenge to navigate manually or to manipulate those data computationally. The data fields often include information that does not strictly adhere to the definition of the field’s contents. For example, in the list of synonyms for a gene in the Genome DataBase (GDB) there is the Unigene number, a number from the NCBI system that relates a GenBank partition that serves as the reference standard gene so that all researchers will use the same amino acid sequence as a reference. In the Gene Ontology database, the IPI (International Protein Index) number is listed in the synonym field. The IPI number references a set of database records and amino acid sequences of the same protein in major protein databases. The SWISS-PROT lists the Enzyme Commission number as a synonym although it is more like a functional category.
Many of the genome knowledge bases have information on multiple species to allow for a cross-species comparison of gene function. Whereas this is helpful for scientific research, choosing the correct database entry is difficult since the gene symbols and gene names can be very similar across species. Here, once again unlike most biomedical terms, the use of upper and lower case often denotes meaning. For example, the gene symbol for the human fibrillin 1 gene is FBN1 while the gene symbol for the fibrillin 1 gene in the mouse is Fbn1. Further ambiguity arises from the use of MFS1 as an alternate gene symbol for fibrillin 1 since MFS1 is also used as an abbreviation for the disease Marfan Syndrome caused by a mutation in the gene. Overall, it is fairly common for a disease name to be used as a synonym for a gene name.
The lack of consistent use of terms leads to more difficulties with automated processing of the data. An example comes from the Gene Ontology [15], the most widely used ontology in molecular biology. It represents knowledge in three domains: molecular function, biological process and cellular component. It uses terms as part of the ontology that are well known through the biomedical literature, such as “protein tyrosine kinase”, but uses these terms in an unusual sense. This entity can be found in many of the protein databases and is generally understood to represent the biological molecule. But in the Gene Ontology, it is defined as the enzyme reaction that it performs: “catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyro-sine phosphate”. Thus the term becomes shorthand for several entities that come together to produce a chemical reaction, in addition to the connotation of being a specific physical entity.
Discussion
Three major informatics issues of navigation and data complexity arose during the course of this study:
First, the official gene names are insufficient for navigation of these web resources. Navigation is accomplished primarily by hotlinks. No single resource has all of the known symbols and synonyms, although the HGNC provides the best list available. No universal identifying number exists for genes and gene products, although there are attempts at such; e.g., the International Protein Index (IPI) lists cross-reference numbers to identify entries representing the same human protein across the SwissProt/Trembl, RefSeq, and Ensembl resources. The parenthetical remarks, incomplete metadata, and lack of universal identifying numbers make the gene names very difficult for computational navigation [13]. It is equally difficult to navigate in the other direction: from specific genes to consumer-oriented disease descriptions [14].
Second, navigational inconsistencies arise from difficulties in determining the number and function of alternate forms of the gene or gene product and maintaining currency with the information. The various resources do not agree on this information, a result of the different updating cycles of each independent resource and the rapid pace of research.
Third, the issues involved with synonymy (multiple terms with the same meaning) and polysemy (multiple meanings for the same term) cause much confusion. This is compounded by the use of the synonym data fields to maintain cross-references to other systems. The use of the same term for multiple meanings causes difficulties in determining whether the appropriate information is retrieved and with frequent confusion of data categories, another navigational hazard. All of these practices cause confusion in understanding, terminological systems and navigation.
However, there are some knowledge representation tools and systems [15–20] being developed, a testament to the recognized need for interoperability and traversal of heterogeneous resources including such important considerations as links to the literature. The Gene Ontology Consortium [21] has played a valuable role in the development of a controlled vocabulary across species, although the genes themselves are not directly addressed by this group. These tools can be augmented to improve the linkages between the health-related and bioinformatics resources. An active research community holds promise that the situation will improve.
Conclusion
There is a tremendous amount of data arising from the results of the Human Genome Project. This study investigated the methods and resources to find associated data focused on the genes and gene products related to a set of human disease genes. Specific informatics issues causing hazards for computational navigation are (1) terminology inconsistencies especially with gene names with parenthetical remarks and incomplete metadata; (2) navigational inconsistencies; (3) inconsistent use of terms and methods of representing information.
Overall, such disparity exists between the focus of consumer health systems and bioinformatics systems that uninterrupted manual or computational navigation from one type to another is usually not successful. These are severe obstacles; tools and standards to facilitate this navigation are sorely needed. The difficulties in finding the data are further compounded by challenges of presenting it to a consumer audience. The answers to the public queries of “What data is coming from the Human Genome Project that relates to my disease and my risks of disease?” are increasingly available but not easily retrievable. Of course, even the best Internet information system will not be able to replace diagnosis and guidance from qualified health professionals.
Footnotes
Conventions used in this paper: Gene names, gene products and gene symbols are in italics.
Results from a search on 12-19-2002 of Locus-Link with the query has_seq AND disease_ known AND organism = human.
JAMA patient page: www.ama-assn.org/public/journals/patient/archive/pat1114.htm
Genetic Alliance: www.geneticalliance.org/
Genetics Education Center: www.kumc.edu/gec/
Nemours Foundation: http://kidshealth.org/parent/system/medical/genetics.html
Unified Medical Language System: http://umlsinfo.nlm.nih.gov/
Medical Subject Headings (MeSH): www.nlm.nih.gov/mesh/
References
- 1.The Human Genome. Nature. 2001;409 (6822):813–958. doi: 10.1038/35057264. [DOI] [PubMed] [Google Scholar]
- 2.Baxevanis AD. The Molecular Biology Database Collection: 2002 update. Nucleic Acids Res. 2002;30 (1):1–12. doi: 10.1093/nar/30.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2002;30 (1):17–20. doi: 10.1093/nar/30.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tateno Y, Imanishi T, Miyazaki S, Fukami-Kobayashi K, Saitou N, Sugawara H, et al. DNA Data Bank of Japan (DDBJ) for genome scale research in life science. Nucleic Acids Res. 2002;30 (1):27–30. doi: 10.1093/nar/30.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoesser G, Baker W, van den Broek A, Camon E, Garcia-Pastor M, Kanz C, et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2002;30 (1):21–6. doi: 10.1093/nar/30.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, et al. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 2002;30 (1):13–6. doi: 10.1093/nar/30.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 2002;3 (3):275–84. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30 (1):42–6. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30 (1):52–5. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller N, Lacroix EM, Backus JE. MEDLINE-plus: building and maintaining the National Library of Medicine’s consumer health Web service. Bull Med Libr Assoc. 2000;88 (1):11–7. [PMC free article] [PubMed] [Google Scholar]
- 11.McCray AT. Better access to information about clinical trials. Ann Intern Med. 2000;133 (8):609–14. doi: 10.7326/0003-4819-133-8-200010170-00013. [DOI] [PubMed] [Google Scholar]
- 12.Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002;19 (5):501–9. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]
- 13.Bodenreider O, Mitchell JA, McCray AT. Evaluation of the UMLS as a Terminology and Knowledge Resource for Biomedical Informatics. Proc AMIA Symp. 2002:61–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan P, Mitchell JA, Bodenreider O, Pant G, Menczer F. Proceedings of the International Workshop on Bioinformatics and Multi-Agent Systems (BIXMAS 2002) Bologna; Italy: July 15, 2002. Web crawling agents for retrieving biomedical information. 2002. [Google Scholar]
- 15.Stevens R, Goble CA, Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinform. 2000;1 (4):398–414. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]
- 16.Oliver DE, Rubin DL, Stuart JM, Hewett M, Klein TE, Altman RB. Ontology development for a pharmacogenetics knowledge base. Pac Symp Biocomput. 2002:65–76. [PubMed] [Google Scholar]
- 17.Chen RO, Felciano R, Altman RB. RIBOWEB: linking structural computations to a knowledge base of published experimental data. Proc Int Conf Intell Syst Mol Biol. 1997;5:84–7. [PubMed] [Google Scholar]
- 18.Kazic T. Semiotes: a semantics for sharing. Bioinformatics. 2000;16 (12):1129–44. doi: 10.1093/bioinformatics/16.12.1129. [DOI] [PubMed] [Google Scholar]
- 19.Wroe C, Stevens R, Goble C, Ashburner M. An evolutionary methodology to migrate the gene ontology to a description logic environment using DAML+OIL. Pac Symp Biocomput. 2003 doi: 10.1142/9789812776303_0058. in press. [DOI] [PubMed] [Google Scholar]
- 20.Stevens R, Baker P, Bechhofer S, Ng G, Jacoby A, Paton NW, et al. TAMBIS: transparent access to multiple bioinformatics information sources. Bioinformatics. 2000;16 (2):184–5. doi: 10.1093/bioinformatics/16.2.184. [DOI] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11 (8):1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]


