A leading theory for the origin of modern humans, the Recent African Origin (RAO) [1], postulates that the ancestors of all modern humans originated in East Africa and that around 100,000 years ago, some modern humans left the African continent and subsequently colonised the entire world, displacing previously established human species such as Neanderthals in Europe [2, 3]. This scenario is supported by the observation that human populations from Africa are genetically the most diverse [2] and that the genetic diversity of non-African populations is negatively correlated with their genetic differentiation towards populations from Africa [3]. Here we add further compelling evidence supporting the RAO model by showing that geographic distance (not genetic distance as in [3]) from East Africa along likely colonisation routes is an excellent predictor for genetic diversity of human populations (R2= 85%). Our results point to a history of colonisation of the world characterised by a very large number of small bottlenecks [4] and limited subsequent gene flow. The pattern of decrease in genetic diversity along colonisation routes is very smooth and does not provide evidence for major genetic discontinuities that could be interpreted as evidence for human “races” [2, 5].
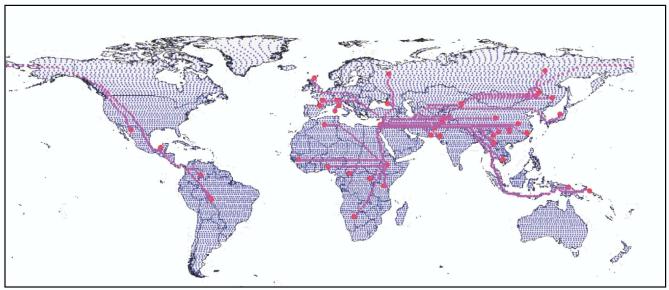
In order to characterise the fine patterns in the decrease of genetic diversity in human populations along likely colonisation routes we analysed a dataset comprising 51 populations distributed worldwide (Figure 1) that have been typed at 377 autosomal microsatellite loci [6] (freely downloadable from: http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm). We first computed geographic distances in a way that is meaningful in the context of modern human settlement history. The point of origin for modern humans was set in Ethiopia, a location where the oldest remains of modern humans have been found [7]. Using graph theory, we then computed the shortest path between this origin and the 51 populations through landmasses wherever possible (see supplementary material for details on the algorithm). This generated migration routes (Figure 1) compatible with the ones proposed by Cavalli-Sforza and colleagues [8], with the exception of the route to America, where the algorithm suggested a scenario with a long stretch at very high latitudes in Asia. To avoid this unrealistic northern migration path, we forced the route to first reach Eastern Asia before veering to high latitudes to move through the Bering straight (Figure 1). In a second step we estimated the genetic diversity for each of the 51 human populations as the unbiased expected heterozygosity HS following Nei (1987) [9].
Figure 1.
Shortest routes (in purple) through landmasses and specified land bridges between the 51 populations analysed (red dots) and a hypothetical East African origin. Geographic distances have been computed as paths connecting vertices (in blue) on land with a newly developed algorithm based on graph theory (see supplementary material).
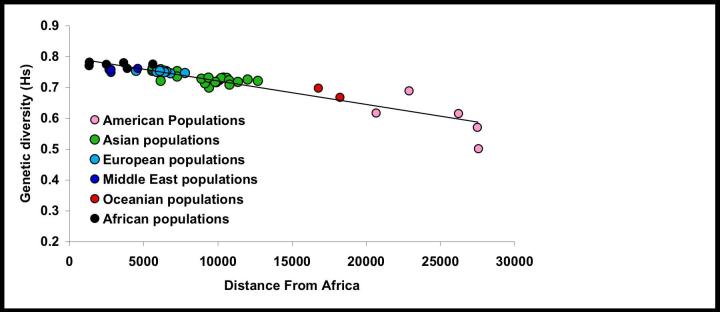
In figure 2 we present the correlation between geographic distance to East Africa and genetic diversity. The negative linear correlation is extremely strong with populations most geographically distant from Ethiopia characterised by the lowest genetic variability. Such a pattern provides extremely strong support for the RAO model of human settlement history since it is only compatible with a colonisation of the world by a single original population of African origin, which subsequently spread throughout the world. The relationship is highly significant (p < 10−4) and geographic distance is an excellent predictor of the neutral genetic diversity of human populations, explaining 85% of the observed variance at a worldwide scale. There is no obvious outlier and we could not detect any effect of the historical census size of the populations or any obvious macro-geographic pattern such as a stepwise decrease in genetic diversity corresponding for instance to a severe bottleneck following the colonisation of a continent (Figure 2). The fact that our analysis points to a monotonous decline in genetic diversity with no obvious genetic discontinuities is interesting in the context of “racial” identifiers as useful proxies for medical applications. While some authors have suggested that human populations are characterised by genetic discontinuities that may prove useful in a clinical context [6, 10], others have argued these are simply artefacts generated by the sampling strategy [2, 5], the latter view is supported by our analysis.
Figure 2.
Relationship between mean genetic diversity of 51 human populations computed over 377 autosomal microsatellite markers and their geographic distances in km from East Africa. The percentage of variance explained by geographic distance is R2 = 85% (p < 10−4). The different colours correspond to the different ethnic groups defined by Rosenberg and colleagues [6]. If those ethnic groups are entered as an additional variable in the model, they do not allow explaining a significantly higher proportion of the variance (p = 0.35). There is also no significant difference between the individual regressions computed for each ethnic group separately (slopes p = 0.12 ; intercepts p = 0.27).
Modelling the underlying parameters most compatible with this strong negative linear correlation will require further investigation. What is clear, however, is that such a pattern of constant loss of genetic diversity along colonisation routes could only have arisen through successive bottlenecks of small amplitude as the range of our species increased, with the populations farthest away from East Africa being furthest from mutation drift-equilibrium [11]. The pattern we observe also suggests that subsequent migration was limited or at least very localised. Independent of the precise causative agents behind that pattern, the quantitative link we provide between geography and neutral genetic diversity should prove very useful to control for human demography when testing whether the geographic distribution of specific human genes have been shaped by natural selection. Our results suggest that information on the geographic coordinates alone is an excellent predictor for the contribution of past demography to the apportionment of genetic diversity in humans.
Supplementary Material
Acknowledgments
We thank Bill Amos, John Archer, Bruno Bucheton, Marie Charpentier, Bénédicte Sanson, Steve Russell and Elizabeth Tyler for comments and insightful discussions. Our research was supported by a Lavoisier Fellowship from the Ministère Français des Affaires Etrangères (FP).
References
- 1.Stringer C, Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988;239:1263–1268. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- 2.Tishkoff S, Kidd K. Implications of biogeography of human populations for ‘race’ and medicine. Nature Genet. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- 3.Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu. Rev. Genomics Hum. Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 4.Excoffier L. Human demographic history: refining the recent African origin model. Curr. Opin. Genet. Dev. 2002;12:675–682. doi: 10.1016/s0959-437x(02)00350-7. [DOI] [PubMed] [Google Scholar]
- 5.Serre D, Pääbo S. Evidence for gradients of human genetic diversity within and among continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 7.White T, Asfaw B, DeGusta D, Gilbert H, Richards G, Suwa G, Howell F. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli-Sforza L, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press; Princeton: 1994. [Google Scholar]
- 9.Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- 10.Bamshad M, Wooding S, Watkins W, Ostler C, Batzer M, Jorde L. Human population genetic structure and inference of group membership. Am. J. Hum. Genet. 2003;72:578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Austerlitz F, JungMuller B, Godelle B, Gouyon P. Evolution of coalescence times, genetic diversity and structure during colonization. Theor. Popul. Biol. 1997;51:148–164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.