Abstract
Background
The programmed cell death 2 (Pdcd2) gene on mouse chromosome 17 was evaluated as a member of a highly conserved synteny, a candidate for an imprinted locus, and a candidate for the Hybrid sterility 1 (Hst1) gene.
Results
New mouse transcripts were identified at this locus: an alternative Pdcd2 mRNA skipping the last two coding exons and two classes of antisense RNAs. One class of the antisense RNA overlaps the alternative exon and the other the entire Pdcd2 gene. The antisense RNAs are alternative transcripts of the neighboring TATA-binding protein gene (Tbp) that are located mainly in the cell nucleus. Analogous alternative PDCD2 forms truncating the C-terminal domain were also detected in human and chicken. Alternative transcripts of the chicken PDCD2 and TBP genes also overlap. No correlation in the transcription of the alternative and overlapping mRNAs was detected. Allelic sequencing and transcription studies did not reveal any support for the candidacy of Pdcd2 for Hst1. No correlated expression of Pdcd2 with the other two genes of the highly conserved synteny was observed. Pdcd2, Chd1, and four other genes from this region were not imprinted in the embryo.
Conclusion
The conservation of alternative transcription of the Pdcd2 gene in mouse, human and chicken suggests the biological importance of such truncated protein. The biological function of the alternative PDCD2 is likely to be opposite to that of the constitutive form. The ratio of the constitutive and alternative Pdcd2 mRNAs differs in the tissues, suggesting a developmental role.
The identified Tbp-alternative Pdcd2-antisense transcripts may interfere with the transcription of the Pdcd2 gene, as they are transcribed at a comparable level. The conservation of the Pdcd2/Tbp sense-antisense overlap in the mouse and chicken points out its biological relevance. Our results also suggest that some cDNAs in databases labeled as noncoding are incomplete alternative cDNAs of neighboring protein-coding genes.
Background
The mouse Pdcd2 gene has been mapped to mouse chromosome (Chr) 17, tail-to-tail to the gene for TATA-binding protein (Tbp), which makes it a candidate for the mouse Hst1 gene [1,2]. The distance between the stop codons of Tbp and Pdcd2 is 4.8 kb. The arrangement of these two genes is also conserved in human [1] and at least five nonmammalian vertebrates [3]. The proteasome subunit C5 gene (Psmb1) is syntenic with these two genes in the mouse, being located head-to-head with Tbp, and in all vertebrates investigated thus far [3]. These genes are nonrandomly distributed also in nonvertebrate genomes [1,4]. The coordinated expression of these three genes could be one of the possible reasons for this observation.
In the rat, transcription of the Pdcd2 (also called Rp8) gene was induced when thymocytes underwent programmed cell death caused by applying radiation or dexamethasone [5] and in fibroblasts after exposure to neutron radiation [6]. The amount of the rat protein increased in nuclei of kidney fibroblasts after prolonged hypoxia [7]. The mRNA of the human orthologous gene, PDCD2, was upregulated in peripheral blood mononuclear cells from patients with chronic fatigue syndrome [8]. However, the upregulation of the mouse and human orthologous genes has not been universally associated with apoptosis [9-11].
The product of the Pdcd2 gene and its orthologs consists of two domains: a MYND zinc-finger DNA-binding domain found in transcription regulators and a highly conserved C-terminal (CT) domain of unknown function. The human PDCD2 gene has two forms. The alternative, less abundant PDCD2 mRNA is formed due to a polyadenylation (polyA) signal in the third intron. The resulting product thus carries the MYND, but not the CT domain [GenBank: NM_144781]. PDCD2 is negatively regulated by the B-cell lymphoma 6 (BCL6) gene in lymph node germinal centers of human tonsils [12], but no such repression was found in immortalized primary B cells [13]. PDCD2 is also a regulator of the host cell factor C1 (HCFC1) gene; mutations in HCFC1 arrest cells in the middle of the G1 phase of the cell cycle [14]. PDCD2 is a candidate for a tumor-suppressor gene on human Chr 6q27 [12-15]. PDCD2 and TBP are candidate type 1 diabetes susceptibility loci [16,17]. A possible haploinsufficiency effect at the 6q27 tumor-suppressor locus has been suggested [15]. A haploinsufficiency gene (Thl1) is also located on mouse Chr 17, in a region that includes Pdcd2 [18-20]. The mouse Pdcd2 gene has been considered a candidate imprinted gene, as it is differentially expressed in 9.5-day-post-coitum (E9.5) parthenogenetic versus androgenetic mouse embryos of the C57BL/6J (henceforth B6) strain [21].
The aim of this study was to characterize in detail transcription within the Pdcd2 locus and its vicinity, as well as its imprinting status and candidacy for the Hst1 locus. To elucidate the reasons for the conservation of the synteny surrounding Pdcd2, we also analyzed the transcription and imprinting of the neighboring genes.
Results
Alternative transcript of the mouse Pdcd2 gene
To look for isoforms of the mouse Pdcd2 mRNA, we used the BLAST program to identify ESTs matching the Pdcd2 genomic sequence. The presence of a putative alternative Pdcd2 exon, which starts 1217 bp from the 3' end of the constitutive Pdcd2 transcript, was detected [GenBank: AA199080, clone 657319 from E12.5 embryo]. The EST clone was then sequenced from its 3' end and a polyA tail was found. The 3' end was preceded by a polyA signal, AATAAA. No A-rich sequence that could cause a spurious priming of the oligo-dT primer during cDNA preparation was found in the genomic DNA. The alternative transcript of Pdcd2 was confirmed by RT-PCR of testicular RNA (data not shown). The alternative mRNA contained a new exon on its 3' end, which replaced the fifth and sixth constitutive Pdcd2 exons and thus also changed the open reading frame (ORF). The alternative transcript thus does not contain the highly conserved CT domain, as with the human PDCD2 isoform 2 (Fig. 1). However, unlike the Pdcd2 ORF, the human ORF is truncated by the end of exon 3 [GenBank: NM_144781]. The 3' RACE method confirmed the 3' end of the alternative Pdcd2 transcript also in testicular RNA. The sequenced RACE product was the same as deduced from the EST clone 657319. The length of the Pdcd2 alternative exon was 1147 bp. The 3' end of the alternative exon matched another EST sequence from E12 embryo (BB097798), but the alternative exon was 1156 bp in length according to this EST [GenBank: DQ906042].
Figure 1.
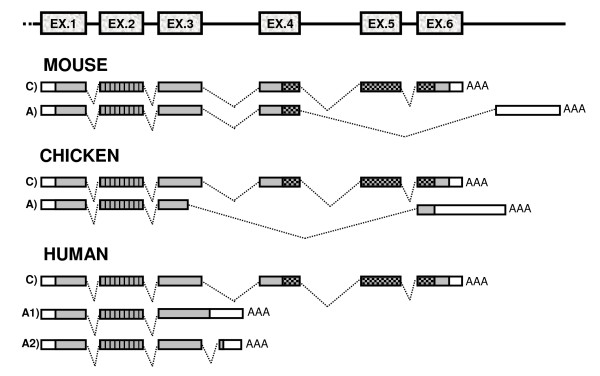
Schematic representation of the constitutive (C) and alternative (A) transcripts encoding PDCD2 in the mouse, chicken and human. Empty and grey rectangles represent the untranslated regions and ORFs, respectively. Vertically hatched rectangles represent the MYND zinc-finger domain in exon 2, and crosshatched rectangles represent the conserved domain of unknown function (CT) coded by exons 4, 5, and 6. EX., exon; AAA, polyA tail.
RNAs transcribed from the opposite strand of the Pdcd2 locus and its vicinity
The BLAST output of the genomic region covered by the alternative Pdcd2 transcript also contained five other ESTs (AA792289 from skin, CA887845 from differentiated neural stem cells, AA792289 from skin, BY723771 from hypothalamus, and CX219724 from neurosphere). Due to their orientation, these ESTs could represent an antisense transcript of the alternative exon of Pdcd2. The opposite end of the AA792289 EST clone was therefore sequenced. The antisense orientation was confirmed by the presence of polyA tail preceded by a polyA signal (AATAAA). There was no A-rich sequence in the genomic DNA that would cause a mispriming of the oligo-dT primer during cDNA preparation. The 3' RACE method and sequencing were then used to identify the antisense transcript in mouse testis as well. The EST and RACE sequences were identical and matched the genomic Pdcd2 sequence in the entire alternative exon and a part of the alternative intron, 382 bp apart from the constitutive polyA site of Pdcd2. The sequences did not contain the sequence of the Pdcd2 constitutive exons (Fig. 2). By RT-PCR and sequencing, it was confirmed that this transcript (termed Pdcd2as1) is a novel alternatively polyadenylated form of the neighboring Tbp gene.
Figure 2.
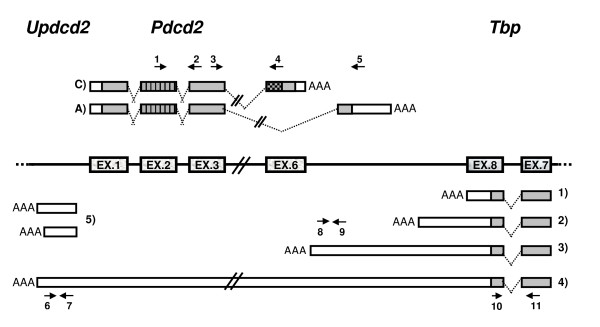
Schematic representation of Pdcd2 and Tbp transcripts in mouse testes. 1) to 4): forms of Tbp transcripts; 1) constitutive Tbp transcript, 2) alternative Tbp transcript not reaching Pdcd2, 3) Pdcd2as1 transcript overlapping the entire alternative Pdcd2 exon, 4) Pdcd2as2 transcript, the longest Tbp alternative, overlapping the entire Pdcd2 gene, 5)Updcd2 – two putative transcripts upstream of Pdcd2 using the same promoter as Pdcd2. Numbered arrows represent the primers used for real-time PCR. Primer pair 1–2 detects both Pdcd2 transcripts, 3–4 just constitutive Pdcd2, 3–5 just alternative Pdcd2, 6–7 Pdcd2as2, 8–9 both antisense mRNAs (Pdcd2as1 and Pdcd2as2), 10–11 all forms of Tbp. For other legend see Fig. 1.
Four other ESTs, two from mammary tumor (AW211687 and AW212179) and two from a forelimb of E13 embryo (CJ049248 and CR514396) were identified upstream of the 5' end of Pdcd2, suggesting there may be a new gene transcribed from the same promoter as Pdcd2, but in the opposite orientation. This 5' region is not conserved in human. The transcription status of this mouse region was investigated by RT-PCR and RACE methods. A transcript encompassing this sequence upstream of the Pdcd2 gene was found in mouse testes and other tissues. At least two alternative forms of this transcript (termed Updcd2) formed by different polyadenylation were identified by 3' RACE and RT-PCR, one of them having the same 3'-end as the EST CR514396 (Fig. 2). 5' RACE detected the 5' end consistent with EST CJ049248 [GenBank: DQ906044]. The bi-directional function of the Pdcd2 promoter is also supported by our finding of two tags in the CAGE (Cap Analysis of Gene Expression) database [22]. The CAGE method measures the expression levels of transcription starting sites by sequencing 5' ends of transcripts prepared through modifying their caps. The CAGE tags T17F00DAE0F6 and T17F00DAE115 suggest that transcription also occurs in the Updcd2 direction. However, there are about ten times more CAGE tags in the Pdcd2 direction, suggesting the promoter activity is much stronger in the direction of Pdcd2 than in that of Updcd2.
On the other hand, these identified RNAs could also represent a novel alternative transcript of the Tbp gene, an antisense read-through of the entire Pdcd2 gene. Therefore, RT-PCR with a Updcd2-specific primer used for reverse transcription and PCR with primer pairs along the Pdcd2 and Tbp genes were used to identify this read-through transcript (henceforth Pdcd2as2). As the PCR products along the entire Pdcd2 gene, the Pdcd2-Tbp intergenic region and the last exons of Tbp genes were successfully amplified and sequenced, the Pdcd2as2 transcript appears to be another new alternative form of Tbp originating by alternative polyadenylation and encompassing the entire Pdcd2 gene in the antisense orientation [GenBank: DQ906043]. Thus, our data indicate that there is both a new alternative Tbp mRNA and a new gene sharing the Pdcd2 promoter.
Furthermore, the BLAST search of the Tbp-Pdcd2 intergenic region revealed nine ESTs, which apparently represent another alternative transcript(s) of the Tbp gene. Three 3' EST sequences (AU020825, AU019062, and the polyA-containing BG069472, all from the embryo) extend the previously characterized 3'UTR of Tbp by 416 bp. Six EST sequences (BQ934602 otocysts, BM945950 E18.5 brain, BG975205 mammary tumor, CK634594 E9.5-10.5 head, CB841791 E15.5 eye and BB068539 from E15 testes) overlap the former three ESTs and each other and extend by 1660 bp in the 3'direction. A 3'RACE product was obtained from a testicular RNA and sequenced. It contains the polyadenylation signal ATTAAA and carries the same end as EST BB068539. No A-rich sequence that could cause an oligo-dT mispriming during cDNA preparation was found in the genomic DNA.
Taken together, besides the Updcd2 transcripts, there are three to four alternative Tbp forms transcribed from the opposite DNA strand in the Pdcd2 locus and its vinicity, including Pdcd2as1, Pdcd2as2, 3'RACE-confirmed mRNA, and the transcript suggested by the three embryonic ESTs incl. BG069472. The alternative mRNAs of the Tbp gene arise by alternative polyadenylation and differ in overlaps with the Pdcd2 gene (Fig. 2).
Transcription analyses of mouse Pdcd2 and Tbp loci
The expression of all identified Pdcd2 and Tbp transcripts was detected by RT-PCR in all eight tissues examined, as well as in different developmental stages of the testis (data not shown). To find whether the antisense transcripts could participate in the regulation of the ratio between the mRNA isoforms [23,24], the level of expression of Pdcd2 and Tbp alternative transcripts was more accurately determined by quantitative real-time RT-PCR (QRT-PCR) in seven mouse tissues (Fig. 3), as well as in developmental stages of the testis (Fig. 4) using primers detecting the Pdcd2 constitutive, alternative, Pdcd2as1 and Pdcd2as2 transcripts (primers Fig. 2, Additional file 1).
Figure 3.
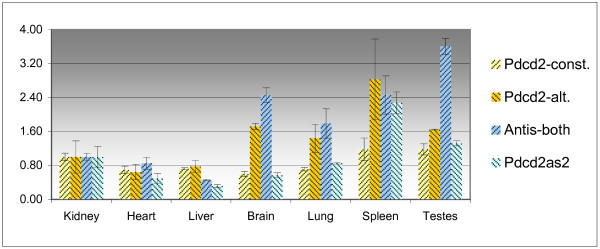
Quantification of the expression of Pdcd2 and Tbp transcripts in seven mouse tissues. For better visualization of the results, the relative quantity in the kidney was taken as one; const., constitutive transcript; alt., alternative transcript, Antis-both, Pdcd2as1 and Pdcd2as2 transcripts.
Figure 4.
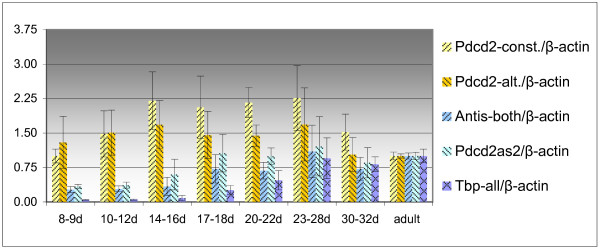
Relative quantification of the expression of Pdcd2 and Tbp transcripts in the testes of mice of different age; normalized by the expression of β-actin. Adult mice were 2 to 2.5 months old. For better visualization of the results, the relative quantity in the adult mouse testes was taken as one; for other legend see Fig. 3.
In tissues, relatively higher levels of the alternative Pdcd2 transcript correlated with upregulation of the Pdcd2-antisense alternative Tbp transcripts. However, no such relationship was detected in the developmental stages of testes (Fig. 4).
The levels of all examined transcripts were determined using the absolute quantification method in seven mouse tissues (Fig. 5, Table 1). The levels of the alternative Pdcd2 transcripts are on average 40 times lower compared to the level of the constitutive Pdcd2 transcript in the analyzed tissues. The Pdcd2as2 transcript was expressed in tissues on average 5 times less than the constitutive Pdcd2. The levels of the Pdcd2as1 transcript in tissues were comparable to that of the constitutive Pdcd2 transcript. The level of all Tbp forms (detected by primers from the TBP-coding region) in the testes was nearly 30 times higher than the level of Pdcd2-antisense alternative Tbp transcripts, in contrast to only 3-fold difference in other tissues (Table 1).
Figure 5.
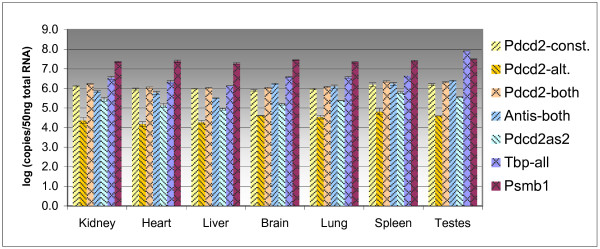
Absolute quantification of the expression of Pdcd2, Tbp and Psmb1 transcripts in seven mouse tissues. The values represent the common logarithm of the number of copies of the particular transcript in the sample corresponding to the amount of 50 ng of total RNA.
Table 1.
Absolute quantification of the expression of Pdcd2 and Tbp transcripts in seven mouse tissues. The values represent the number of copies of the particular transcript in the sample corresponding to the amount of 5 pg of total RNA.
| Tissue/Transcript | Pdcd2-const. | Pdcd2-alt. | Pdcd2-both | Antis-both | Pdcd2as2 | Tbp-all |
| Kidney | 134 | 2 | 169 | 70 | 28 | 342 |
| Heart | 93 | 2 | 110 | 59 | 14 | 218 |
| Liver | 97 | 2 | 104 | 32 | 9 | 136 |
| Brain | 80 | 4 | 108 | 171 | 16 | 382 |
| Lung | 95 | 3 | 117 | 125 | 24 | 362 |
| Spleen | 158 | 7 | 227 | 171 | 63 | 448 |
| Testes | 158 | 4 | 203 | 251 | 36 | 8140 |
const., constitutive transcript; alt., alternative transcript
The variations of the expression of the sense/antisense (SA) transcripts in different cell types could make the SA relationship in developmental stages of the testis undetectable. To identify the Pdcd2/Tbp SA transcript ratios in homogenous cell populations, pure flow sorted populations of spermatogenic cells were isolated and the mRNA levels of Pdcd2 and antisense Tbp transcripts were assessed by QRT-PCR (Fig. 6). The alternative Pdcd2 transcript was detected just in the population of leptotene spermatocytes. The expression of the antisense transcripts exhibited neither positive nor negative correlation with this restricted alternative Pdcd2 expression, suggesting that there is no regulation of the Pdcd2 alternatives by the antisense RNAs in these cells (Fig. 6). Also, the higher quantity of total Tbp transcripts was detected in pachytene spermatocytes and spermatids.
Figure 6.
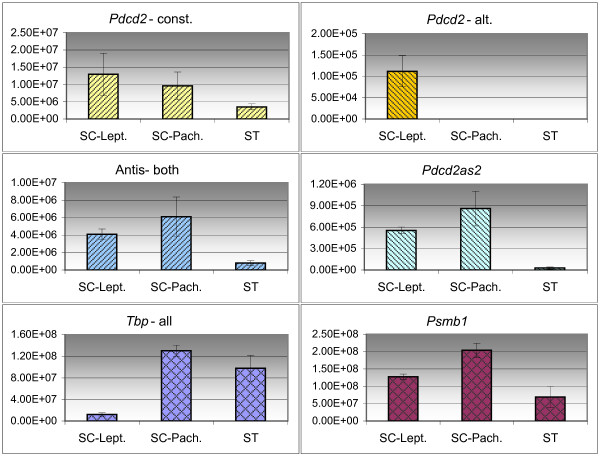
Absolute quantification of the expression of Pdcd2 and Tbp transcripts in flow-sorted spermatogenic cell populations. The values on axis x represent the number of copies of the particular transcript in the sample corresponding to the amount of 50 ng of total RNA. SC-Lept, leptotene+zygotene spermatocytes; SC-Pach., pachytene spermatocytes; ST, spermatids; const., constitutive transcript; alt., alternative transcript; Antis-both, Pdcd2as1 and Pdcd2as2 transcripts.
Long perfect dsRNA duplexes, resulting from the antisense transcription, can be substrates for adenosine deaminase (ADAR) enzymes that catalyze the hydrolytic deamination of adenosines to inosines. Massive A/I editing can cause the subsequent retention of the edited RNAs in the nucleus [25,26]. To test whether the Pdcd2as1 or Pdcd2as2 transcripts can drive the expression of the sense Pdcd2 transcripts by such a mechanism, the mRNA levels of Pdcd2 and both their antisense RNAs were measured in cytoplasmic and nuclear fractions by QRT-PCR. The nuclear/cytoplasmic ratios of the tested genes disclose the nuclear localization of Pdcd2as1 and Pdcd2as2 transcripts, but cytoplasmic localization of the Pdcd2 mRNAs (Fig. 7).
Figure 7.
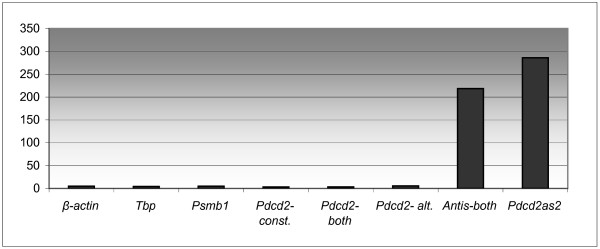
The ratio of transcripts in nuclear versus cytoplasmic RNA fractions in adult B6 testes. const., constitutive transcript; alt., alternative transcript, Antis-both, Pdcd2as1 and Pdcd2as2 transcripts.
Alternative and antisense transcription at the chicken, rat, and human PDCD2 loci
Recently, the chicken region containing the PDCD2, TBP, and PSMB1 genes was sequenced and major transcripts identified by RT-PCR ([3], [GenBank: AY376311]). The genes are in the same order and orientation as their mouse and human orthologs. The organization of the coding exons of the chicken PDCD2 gene (also called Q6JLB0_CHICK) is the same as in the mouse and human. Provided that the alternative and antisense transcripts are important for the function of the Pdcd2 orthologous genes, these features should also be conserved in the chicken genome. Conserved overlapping transcription could also shed light on the synteny conservation. We therefore looked for alternative and antisense transcription at the chicken PDCD2 locus by EST analysis, RACE and RT-PCR. A putative alternative PDCD2 transcript that suggests skipping exons 5 and 4 and a part of exon 3 was detected in the dbEST database (BU309667 from the heart, CO773817 from the testes). The presence of the alternative transcript in chicken was confirmed by RT-PCR and sequencing of testicular RNA. This alternative mRNA does not contain the CT domain, as with the mouse and human alternative transcripts. However, unlike mouse or human, the chicken ORF is truncated before the end of exon 3 (Fig. 1, [GenBank: DQ906045]).
The overlapping transcription of chicken PDCD2 and TBP was suggested by ESTs (BU272933 from cerebrum, BU462305 and BU476463 from the ovary) and it was confirmed by RT-PCR of testicular RNA using primers from the intergenic region in combination with primers from the exons of TBP and PDCD2, respectively. The sequence of the PCR products indicated an overlap of at least 80 bp between alternative PDCD2 and TBP transcripts [GenBank: DQ906046, DQ906047]. The SA overlap is thus conserved in chicken. Our RT-PCR data also indicate that the chicken Tbp gene has another alternatively polyadenylated form even longer than the one published by Yamauchi et al. [27].
The human PDCD2 has an alternative transcript formed by polyadenylation in the third intron [GenBank: NM_144781]. However, EST BQ187150 from fetal eye and eight others suggest alternative splicing rather than alternative polyadenylation (Fig. 1, human transcript 2A). No overlapping transcription in the human is indicated by the ESTs, although there are 18 ESTs (including polyadenylated AW207335) in the PDCD2-TBP intergenic region, which could represent an alternative human PDCD2 isoform(s), generated by alternative polyadenylation. However, the analysis of this region is hindered by an A-rich sequence.
There is also a putative alternatively polyadenylated transcript of Rp8, the rat homolog of the mouse Pdcd2 gene (polyadenylated rat EST AI704628 from E16.5 ventricle). Two ESTs matching the Rp8-Tbp intergenic region (AI412868 from the brain and CO571404 from the testes) suggest overlapping transcription in rat, because they are in the opposite orientation. There are also two ESTs in the third Rp8 intron (CB730473 and CB727152, hypothalamus) in the opposite direction than the Rp8 gene. The alternative form of the Rp8 gene lacking the CT domain has not been suggested by any EST yet.
Analysis of coordinated expression of three genes of a highly conserved synteny
The orthologs of Pdcd2 and its neighboring genes Tbp and Psmb1 are nonrandomly distributed in eukaryotic genomes [1,3], but the reason for this observation is unknown. As one of the possible reasons could be the coordinated expression of these three genes, we have tested this hypothesis by QRT-PCR in mouse tissues. The three genes are housekeeping, but the expression of Tbp has been reported to be highly elevated in the testis [28], while the other genes were not tested in this organ. As shown in Fig. 8, Tbp but not Pdcd2 or Psmb1 were overexpressed in the testis. Thus, there are some regulatory elements of Tbp not accessible by the other two genes. Indeed, analyses of a much broader panel of tissues and stages by microarrays suggested a different pattern of expression for these three genes [29]. It is therefore improbable that the reason for the synteny conservation is the co-expression of these genes.
Figure 8.
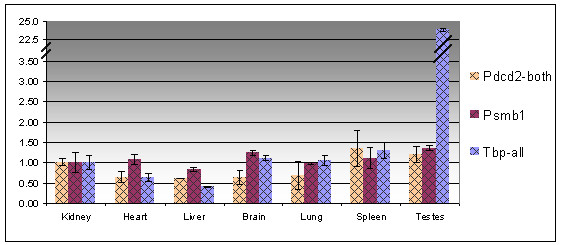
Quantification of the expression of Pdcd2, Tbp and Psmb1 transcripts in different mouse tissues. For better visualization of the results, the relative quantity in kidney was taken as one.
Evaluation of Pdcd2 as a candidate for Hst1
To evaluate the Pdcd2 gene as a candidate for the Hst1, its testicular transcript was isolated by PCR from the C3H/J mouse testicular library and sequenced. The C3H/J strain carries the Hst1f allele, but it has the same predicted protein product as B6 (Hst1S allele). No polymorphism between B6 and C3H/J strains was detected in 249 bp of the promoter region of Pdcd2 (data not shown).
In 15-day-old mice, significant differences between the frequency of pachytene spermatocytes in the testis of prospectively fertile and sterile hybrids can be detected (10% versus 2% [30]), suggesting that Hst1 could be differentially expressed at this stage. The amount of the Pdcd2 transcripts was therefore compared in fertile and sterile hybrid mouse testis by Northern blotting with the β-actin probe used as a standard (data not shown) and by QRT-PCR using the Gapdh gene as a standard (Fig. 9). The alternative Pdcd2, Pdcd2as1 and Pdcd2as2 were also tested by QRT-PCR. There were no significant differences in the expression of these four transcripts between the prospectively fertile and sterile 15-day-old hybrids. Therefore, Pdcd2 is unlikely to be the Hst1 gene. A significant upregulation of the Tbp, Pdcd2as1 and Pdcd2as2 transcripts was detected in fertile compared to sterile adult hybrids (Fig. 9). This difference is probably caused by distinct cellular composition of adult fertile and sterile testes that differ in the number of pachytene spermatocytes and spermatids [30]. These cell types vary in the pattern of expression of Tbp transcripts (Fig. 6).
Figure 9.
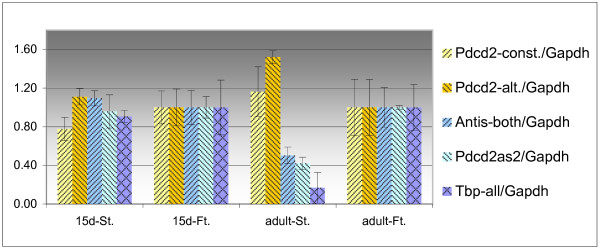
Relative quantification of the expression of Pdcd2 and Tbp transcripts in the testes of fertile (Ft.) and sterile (St.) hybrids at the age of 15 days (15d) and adult. The values were normalized by the expression of Gapdh and compared relatively to the expression of the particular transcript in the testes of a fertile hybrid 15 days old and of an adult, respectively.
Analysis of genomic imprinting of mouse Pdcd2 and neighboring genes
Differential expression of the mouse Pdcd2 gene was reported in E9.5 parthenogenetic versus androgenetic mouse embryonic tissues of the B6 strain, suggesting it may be paternally imprinted [21]. Nikaido and colleagues have also suggested that the chromodomain 1 (Chd1) gene located 180 kilobases from Pdcd2 is maternally imprinted. The imprinting of Pdcd2 and neighboring loci could explain the conservation of the synteny. To determine whether the Pdcd2 gene and its neighbors are imprinted, polymorphisms in their transcripts were searched comparing the sequences of the mouse strains B6 and PWD/Ph (henceforth PWD) [31]. One single-nucleotide polymorphism (SNP) was detected in the coding region of the constitutive Pdcd2 transcript. This synonymous G/A transition is located in exon 5 and thus not present in the alternative Pdcd2 transcript (Additional file 2). Primers used to amplify the constitutive mRNA were from exons 2 and 5 and did not detect the Pdcd2as2 transcript. The SNP was analyzed in Pdcd2 transcripts from RNAs of E9.5 embryos and placentas obtained from reciprocal backcrosses (B6 × (PWD × B6)) and ((B6 × PWD) × B6). Backcross animals were used to control the contamination of placentas by maternal tissues. The DNAs of the embryos were genotyped for Chr 17 and the RNAs from placentas of B6/B6 embryos were tested by RT-PCR for a B6/PWD polymorphic marker in Tbp (PWD-specific 6-bp insertion). We found that the placentas were strongly contaminated by maternal tissue and thus were not useful for testing the imprinting status (data not shown). Therefore, the imprinting status of the Pdcd2 gene was determined only in the embryos and yolk sacs from reciprocal F1 crosses and backcrosses of B6 and PWD. As both B6 and PWD alleles were present in B6/PWD heterozygous embryos, the Pdcd2 constitutive transcript was imprinted neither in mouse E9.5 embryo nor in yolk sac. Polymorphisms were also detected in Pdcd2as1 and Pdcd2as2 mRNAs in the region not overlapping with the Pdcd2 transcripts as well as in five other genes from this region, including Chd1, Tbp, Psmb1, D17Ph4e, and Dll1 (Additional file 2). All these genes were tested for imprinting in the same way (data not shown). None of these genes was imprinted.
Discussion
We have analyzed in detail the transcription pattern of the putative apoptotic gene Pdcd2 [9] and the tightly linked Tbp gene, a key factor in transcription initiation of all eukaryotes [32].
One new alternative Pdcd2 transcript, arising by alternative splicing, and at least three new transcripts of Tbp originated by alternative polyadenylation have been identified in the mouse. The presence of the alternative Pdcd2 mRNA was also found in human [GenBank: NM_144781] and chicken. Although the mechanism and structure of these transcripts is different, in all three species the alternative Pdcd2 transcript encodes the MYND zinc-finger domain but not the highly conserved CT domain, suggesting the biological importance of such truncated protein. The function of the alternative Pdcd2 product can be deduced from the reported study of cell-cycle regulation [14]. The cell line used has a mutation in the HCFC1 gene, causing an arrest of the cell growth at the non-permissive temperature. Transfection of a construct carrying the functional HCFC1 gene rescued the arrest. Cotransfection of the complete PDCD2 greatly decreased the colony formation, while cotransfection with a truncated PDCD2, consisting just of the MYND domain (as in the alternative transcripts), increased it [14]. The function of the alternative PDCD2 is therefore likely to be opposite to that of the constitutive form. The ratio of the constitutive and alternative Pdcd2 mRNAs differs in the tissues, suggesting a developmental role.
We identified four alternatively polyadenylated alternative forms of Tbp in the mouse. The regulation of the stability of Tbp mRNAs could be a potential role for all identified alternative forms of Tbp, as the 3' UTR determines the life-time of mRNA [34]. Moreover, two of four newly identified transcripts of the Tbp gene overlap with Pdcd2 transcripts forming the SA pairs. The overlapping transcription of some alternative Pdcd2 and Tbp transcripts was also confirmed in the chicken. The conservation of SA transcription could explain the conserved synteny of these two genes, although no correlated expression of Tbp, Pdcd2 and Psmb1, three genes of conserved synteny [3], was observed. The mouse Pdcd2as1 transcript overlaps just the alternative Pdcd2 transcript, while Pdcd2as2 overlaps the entire Pdcd2 gene (both Pdcd2 forms). We determined that their level of transcription represents the minority (about 30% in six mouse tissues, 3% in the testes) of total Tbp transcription and that they are mostly localized in the cell nucleus. Both these facts suggest that they play some other regulatory role in gene expression than the protein coding one. Moreover, they are transcribed at a level sufficient to interfere with the transcription of the neighboring Pdcd2 gene. A possible mechanism of function of the Pdcd2as2 transcript could be the downregulation of Pdcd2 expression at the level of transcription via antisense-induced methylation of the CpG island of Pdcd2 or by post-transcriptional degradation of mRNA after the formation of dsRNA [35]. Antisense transcripts can participate in the regulation of the ratio between mRNA isoforms [23,35-37]. Correlated expression of the Pdcd2 transcripts and overlapping antisense RNAs was suggested by our quantitative expression studies in mouse tissues, but it was not confirmed by the quantification of these transcripts in various developmental stages of the testis nor in flow-sorted populations of spermatogenic cells. The pattern of the expression of Pdcd2, Tbp, and Psmb1 genes in sorted populations of spermatogenic cells is in agreement with the Affymetrix microarray data of Namekawa et al. [38], measuring gene expression in STAPUT-sorted spermatogenic cell populations. Schmidt and Schibler [28,39] found that the Tbp gene is upregulated in the testis. We have confirmed their suggestion that the increased transcription of Tbp begins already in pachytene spermatocytes (Fig. 6). On the other hand, mRNAs of both the Pdcd2-antisense Tbp alternative transcripts are present at a higher level in spermatocytes and downregulated in spermatids, suggesting a distinct role for these mRNAs and the constitutive Tbp transcript in these cells.
There are several possible reasons why we could not detect any SA relationship. First, the antisense transcript could influence the expression of its sense partner at the level of mRNA translation. This regulation is typical for micro-RNAs and small RNAs, which inhibit the target mRNAs without destroying the template [40,41]. However, this mechanism is improbable, as micro-RNAs do not show perfect complementarity with their target sequence in the majority of eukaryotes [42,43]. To test this hypothesis, the protein level of alternative and constitutive Pdcd2 needs to be determined. Second, the antisense regulation could be very dynamic or specific; the rapid change of SA ratio could be detectable just after specific stimuli in particular tissues or cells [42].
We excluded that the antisense transcripts could drive the expression of the sense RNA via mechanisms connected with massive A/I editing of long perfect dsRNA duplexes, resulting from the antisense transcription and causing subsequent retention of the edited transcripts in the nucleus [25,26], as we did not detect any difference in the sequence of cDNA of the alternative Pdcd2 exon prepared from the nuclear RNA fraction. Moreover, we did not find any dramatic difference in the nuclear vs. cytoplasmic ratio of Pdcd2 transcripts in comparison to control genes. Our results also suggest that the heterogeneity of the cell types in the testes was not the reason of the absence of any SA relationship through the measurement of Pdcd2-Tbp SA expression in the homogenous testicular cell populations. The proper biological function of Pdcd2as1 and Pdcd2as2 transcripts thus remains to be determined.
A large number of putative antisense transcripts have been found recently by mapping ESTs and/or cDNAs in the mouse genome [44-47]. However, only a relatively small number of SA loci have been experimentally characterized by expression analysis [42,48]. Here we present the identification and detailed characterization of a specific SA locus. The recent large-scale studies [47,49] suggested that 63 to 72% of all genome-mapped transcription units in the mouse overlap with antisense cDNA or EST and multiple-sized transcripts are often generated from the SA loci [48]. In general, over 65% of transcriptional units produce multiple splice variants, and transcript diversity also arises through alternative promoter usage and alternative polyadenylation [49]. The significance of gene overlap has been shown [50] by an evolutionary approach, as genes overlapping in mammals are more likely to have the same organization in the pufferfish. The Pdcd2/Tbp SA overlap is conserved in the mouse and chicken, suggesting its biological relevance.
We did not find any support for the candidacy of Pdcd2 for Hst1 by allelic sequencing and transcription analysis. Our data indicate that Pdcd2, Chd1, and four other genes from this region are not imprinted in E9.5 embryos as was suggested by Nikaido et al. An explanation for these conflicting results could be that Pdcd2 and Chd1 are non-imprinted, but regulated by imprinted genes. Alternatively, the conflict may be due to the use of placentas contaminated by maternal tissues by Nikaido et al. [21]. It has been shown recently that the expression profiling of uniparental mouse embryos is highly inefficient in identifying novel imprinted genes [33].
Conclusion
The alternative Pdcd2 transcripts, which encode the DNA-binding MYND zinc-finger domain but not the CT domain, were identified in the mouse, human and chicken, suggesting the biological importance of such putative truncated protein. The alternative PDCD2 protein lacking the CT domain could interfere with the regulatory effect of the complete PDCD2 protein on gene expression of specific genes. The ratio of the constitutive and alternative Pdcd2 mRNAs differs in various tissues, suggesting a developmental role.
The identified Tbp-alternative Pdcd2-antisense transcripts seem to play some regulatory role in gene expression, compared to the protein-coding function of the constitutive Tbp mRNA. They are mostly localized in the cell nucleus and transcribed at a level sufficient to interfere with the transcription of the Pdcd2 gene. The conservation of Pdcd2/Tbp SA overlap in the mouse and chicken also point out their biological relevance. Furthermore, our data suggest that at least some of the cDNAs identified in the large-scale sequencing projects labeled as noncoding RNAs are in fact incomplete alternative cDNAs of neighboring protein-coding genes.
Methods
RNA preparation, reverse transcription, real-time PCR
Total RNA was prepared using Trizol (Invitrogen). Reverse transcription (RT) was performed with MuMLV or SuperScript II (Invitrogen). Quantification of mRNAs was performed using a real time PCR system (LightCycler, Roche). An aliquot corresponding to 50 ng of total RNA was added to the reactions with FastStart DNA Master SYBR Green I kit and cycled in the LightCycler. As controls, RT reactions without reverse transcriptase were utilized. The assays were done several times with independently collected samples. The data were analyzed using LightCycler Software version 3.5.3 (Roche).
Polymerase chain reaction (PCR) and sequencing
Each template or control was added to 50 μl reactions prepared as master mixes containing final concentration 0.15 nM of each of the four dNTPs, 50 mM KCl, 10 mM Tris-HCl, pH 8.8, 1.6 mM MgCl2, 0.08% Nonindet P40, 1.5 U of Taq polymerase (MBI Fermentas), and 15 pmol of each primer (see Additional file 1 for all used primers). Additives betaine (1 M, Sigma) and/or DMSO (7%, Sigma) were used for GC-rich templates. The amplification products were denatured in a thermal cycler (Applied Biosystems) at 94°C for 2 min, then cycled 25 to 38 times at 94°C for 30 s, at the corresponding primer annealing temperature for 30 s, and at 68°C for 2.5 min, and, finally, incubated at 70°C for 5 min. The PCR products were resolved on agarose gels with ethidium bromide along with a size marker and photographed under UV illumination to check their sizes. The sequencing reactions were performed with the Big Dye kit according to the manufacturer's instructions (Applied Biosystems) and run in a sequencing capillary machine ABI310 (Applied Biosystems).
Isolation of the 3' end of Pdcd2 alternative and antisense transcript by 3' RACE
3' RACE experiments were performed with 0.5 μg of total 28-day-old mouse testes RNA and 10 pmol of the poly-T primer 3'CDS according to the manufacturer's instructions (SMART RACE Kit, Clontech). The first-strand cDNA synthesis was performed at 50°C for 30 min in 540 mM Trehalose, 1× RACE first strand buffer (Clontech), 10 mM DTT, 1 mM dNTPs, and 100 units SuperScript reverse transcriptase (Invitrogen). After incubation, first-strand cDNA reaction was diluted 1:5 with water and incubated at 75°C for 10 min to inactivate the reverse transcriptase.
Two-and-half microliters were used for PCR reactions in a total reaction volume of 50 μl and subjected to 35 cycles of amplification. Each cycle consisted of a 94°C denaturation (30 s), 56°C annealing (30 s), and 68°C extension (3 min) step. The first round of PCR was performed with 0.5 μM of the gene-specific primer and the Universal primer mix (UPM-Clontech). Reactions with only one primer added served as negative controls.
Ten percent of the PCR reactions were analyzed on agarose gels. PCR products were diluted 1:20 and 1 μl served as a template to a second round of PCR using 0.5 μM nested gene-specific primer, and Nested Universal Primer (NUP, Clontech). Amplifications were performed for 20 cycles using standard PCR conditions. PCR products were visualized on agarose gels, isolated from the gel and sequenced.
5'RACE Invitrogen GeneRacer kit
The 5' end of the Updcd2 transcript was cloned and identified with GeneRacer kit (Invitrogen) using gene-specific Updcd2 Racer primers (Additional file 1). This method is designed to capture only the intact full-length transcripts while eliminating any truncated mRNA. Total RNA from mouse testes was first treated with calf intestinal phosphatase to dephosphorylate the 5'-ends of any truncated mRNA. Following decapping the intact mRNAs with tobacco acid pyrophosphatase, GeneRacer RNA oligo was ligated specifically to the 5'-ends of the decapped mRNAs. After the synthesis of the first strand cDNA using the oligo- [dT] primer with Superscript II RNase H- reverse transcriptase, the 5'-end of the Pdcd2 mRNA was amplified by PCR followed by nested PCR according to the procedure described in the GeneRacer kit using Pdcd2 gene-specific primers, and the 5' primer and nested 5' primer supplied by the kit. The PCR products were isolated, cloned and sequenced.
GenBank accession numbers
DQ906042- Mus musculus Pdcd2 alternative transcript mRNA, complete cds,
DQ906043- Mus musculus Pdcd2-antisense 2 (Pdcd2as2) transcript, Tbp mRNA with alternative 3'UTR, partial cds,
DQ906044- Mus musculus upstream Pdcd2 (Updcd2) noncoding mRNAs sharing the Pdcd2 promoter, partial sequence,
DQ906045- Gallus gallus PDCD2 alternative transcript mRNA, complete cds,
DQ906046- Gallus gallus PDCD2 transcript with alternative 3'UTR (a3PDCD2) mRNA, partial sequence,
DQ906047- Gallus gallus TBP alternative transcript 2 (cTBP2), TBP mRNA with alternative 3'UTR, partial sequence.
Preparation of testicular single-cell suspension and FACS
Cells were isolated from B6 two-month-old male mice by using two-step enzymatic digestion to remove interstitial cells [51], using the EKRB medium (pH 7.2) supplemented with collagenase (50 mg/ml) and DNase (1 mg/ml). The single-cell suspensions were pelleted and resuspended in appropriate buffer.
Two million cells were diluted in 1.5 ml EKRB + 1% fetal calf serum (FCS) with DNase (1 mg/ml) and stained with Hoechst (1 mg/ml) for 1 h at 32°C. Just before analysis, PI (propididium iodide, 1 mg/ml) was added to exclude dead cells. Analysis and cell sorting were performed in FACS (Fluorescence Activated Cell Sorter) Vantage (Becton, Dickinson and Company). One hundred thousands of leptotene-zygotene spermatocytes, pachytene spermatocytes and spermatids for RNA isolation were sorted directly to RLT solution with mercaptoethanol (QIAGEN RNeasy micro kit). Twenty thousand cells of each fraction were sorted to EKRB medium with 1% FCS and Hoechst (1 mg/ml) to check the purity of the spermatocyte fractions after the immunostaining using an antibody against the synaptonemal complex.
Isolation of the cytoplasmic/nuclear RNA fraction
The testicular single-cell suspensions (5 × 107 cells) were pelleted and resuspended in 300 μl RLN buffer (50 mM Tris-Cl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) supplemented with 1 mM DTT and RNAsin (1000 U/ml). The mixture was incubated on ice for 5 min and spun in a microcentrifuge at 14 000 rpm for 5 min. The supernatant containing the cytoplasmic fraction was separated from the pellet (nuclear fraction). The RNAs were prepared using Trizol (Invitrogen).
Authors' contributions
OM carried out experimental work described in the paper, participated in designing the study and drafted the manuscript. ZT conceived the study, participated in its design, and together with JF and OM participated in the writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Primers used for RT-PCR, RACE and real-time PCR. The primer name, 5'-> 3' sequence and usage are noted. In QRT-PCR primer pairs, conditions of the reaction using the real-time PCR system (LightCycler, Roche) are noted in the brackets. Primer names beginning with "Ch" amplify chicken Pdcd2, Tbp genes. F, forward primer; R, reverse primer; alt., alternative.
Testing the imprinting status of the transcripts in the Hst1 region. B6 × PWD polymorphisms used to test the imprinting status of the transcripts in the Hst1 region are noted in the table. The example of the imprinting analysis (for Pdcd2 constitutive transcript) are depicted below.
Acknowledgments
Acknowledgements
We thank David Homolka for help with FACS of testicular cells, Dr. Petr Jansa for chicken RNA, Dr. Heinz Himmelbauer for EST clones, V. Fotopulosova and M. Landikova for technical assistance, and Dr. P. Jansa, three anonymous reviewers, and S. Takacova for comments on the manuscript. This work was supported by grants Nos. 204/01/0997 and 301/05/0738 from the Grant Agency of the Czech Republic, No. A5052406 from the Grant Agency of the Academy of Sciences of the Czech Republic, and Nos. 1M6837805002 (Center for Applied Genomics) and AV0Z50520514 from the Ministry of Education, Youth and Sports of the Czech Republic. J.F. is an International Research Scholar of the Howard Hughes Medical Institute.
Contributor Information
Ondrej Mihola, Email: mihola@biomed.cas.cz.
Jiri Forejt, Email: jforejt@biomed.cas.cz.
Zdenek Trachtulec, Email: trachtul@biomed.cas.cz.
References
- Trachtulec Z, Forejt J. Synteny of orthologous genes conserved in mammals, snake, fly, nematode, and fission yeast. Mamm Genome. 2001;12:227–231. doi: 10.1007/s003350010259. [DOI] [PubMed] [Google Scholar]
- Trachtulec Z, Mihola O, Vlcek C, Himmelbauer H, Paces V, Forejt J. Positional cloning of the Hybrid sterility 1 gene: fine genetic mapping and evaluation of two candidate genes. Biol J Linn Soc. 2005;84:637–641. doi: 10.1111/j.1095-8312.2005.00460.x. [DOI] [Google Scholar]
- Trachtulec Z, Vlcek C, Mihola O, Forejt J. Comparative analysis of the PDCD2-TBP-PSMB1 region in vertebrates. Gene. 2004;335:151–157. doi: 10.1016/j.gene.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Trachtulec Z, Hamvas RM, Forejt J, Lehrach HR, Vincek V, Klein J. Linkage of TATA-binding protein and proteasome subunit C5 genes in mice and humans reveals synteny conserved between mammals and invertebrates. Genomics. 1997;44:1–7. doi: 10.1006/geno.1997.4839. [DOI] [PubMed] [Google Scholar]
- Owens GP, Hahn WE, Cohen JJ. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol Cell Biol. 1991;11:4177–4188. doi: 10.1128/mcb.11.8.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloschak GE, Chang-Liu CM, Panozzo J, Libertin CR. Low doses of neutrons induce changes in gene expression. Radiat Res. 1994;138:S56–59. doi: 10.2307/3578762. [DOI] [PubMed] [Google Scholar]
- Shakib K, Norman JT, Fine LG, Brown LR, Godovac-Zimmermann J. Proteomics profiling of nuclear proteins for kidney fibroblasts suggests hypoxia, meiosis, and cancer may meet in the nucleus. Proteomics. 2005;5:2819–2838. doi: 10.1002/pmic.200401108. [DOI] [PubMed] [Google Scholar]
- Kaushik N, Fear D, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, et al. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol. 2005;58:826–832. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Hacker G. Cloning of mouse RP-8 cDNA and its expression during apoptosis of lymphoid and myeloid cells. DNA Cell Biol. 1995;14:189–193. doi: 10.1089/dna.1995.14.189. [DOI] [PubMed] [Google Scholar]
- D'Mello SR, Galli C. SGP2, ubiquitin, 14 K lectin and RP8 mRNAs are not induced in neuronal apoptosis. Neuroreport. 1993;4:355–358. doi: 10.1097/00001756-199304000-00003. [DOI] [PubMed] [Google Scholar]
- Chen Q, Qian K, Yan C. Cloning of cDNAs with PDCD2(C) domain and their expressions during apoptosis of HEK293T cells. Mol Cell Biochem. 2005;280:185–191. doi: 10.1007/s11010-005-8910-z. [DOI] [PubMed] [Google Scholar]
- Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu A, et al. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci USA. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusam S, Vasanwala FH, Dent AL. Transcriptional repressor BCL-6 immortalizes germinal center-like B cells in the absence of p53 function. Oncogene. 2004;23:839–844. doi: 10.1038/sj.onc.1207065. [DOI] [PubMed] [Google Scholar]
- Scarr RB, Sharp PA. PDCD2 is a negative regulator of HCF-1 (C1) Oncogene. 2002;21:5245–5254. doi: 10.1038/sj.onc.1205647. [DOI] [PubMed] [Google Scholar]
- Steinemann D, Gesk S, Zhang Y, Harder L, Pilarsky C, Hinzmann B, Martin-Subero JI, Calasanz MJ, Mungall A, Rosenthal A, et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer. 2003;37:421–426. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Seryogin YA, Turakulov RI, Savost'anov KV, Titovich EV, Zilberman LI, Kuraeva TL, Dedov II, Nosikov VV. Evaluation of IDDM8 susceptibility locus in a Russian simplex family data set. J Autoimmun. 2005;24:243–250. doi: 10.1016/j.jaut.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Owerbach D, Pina L, Gabbay KH. Association of a CAG/CAA repeat sequence in the TBP gene with type I diabetes. Biochem Biophys Res Commun. 2004;323:865–869. doi: 10.1016/j.bbrc.2004.08.159. [DOI] [PubMed] [Google Scholar]
- Browning VL, Bergstrom RA, Daigle S, Schimenti JC. A haplolethal locus uncovered by deletions in the mouse T complex. Genetics. 2002;160:675–682. doi: 10.1093/genetics/160.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DE, Bergstrom RA, Munroe RJ, Lee BK, Browning VL, You Y, Eicher EM, Schimenti JC. Overlapping deletions spanning the proximal two-thirds of the mouse t complex. Mamm Genome. 2003;14:817–829. doi: 10.1007/s00335-003-2298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell GR, Munroe RJ, Schimenti JC. Transgenic rescue of the mouse t complex haplolethal locus Thl1. Mamm Genome. 2005;16:838–846. doi: 10.1007/s00335-005-0045-8. [DOI] [PubMed] [Google Scholar]
- Nikaido I, Saito C, Mizuno Y, Meguro M, Bono H, Kadomura M, Kono T, Morris GA, Lyons PA, Oshimura M, et al. Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res. 2003;13:1402–1409. doi: 10.1101/gr.1055303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- Murphy PR, Knee RS. Identification and characterization of an antisense RNA transcript (gfg) from the human basic fibroblast growth factor gene. Mol Endocrinol. 1994;8:852–859. doi: 10.1210/me.8.7.852. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Carmichael GG. Antisense starts making more sense. Nat Biotechnol. 2003;21:371–372. doi: 10.1038/nbt0403-371. [DOI] [PubMed] [Google Scholar]
- Yamauchi J, Sugita A, Fujiwara M, Suzuki K, Matsumoto H, Yamazaki T, Ninomiya Y, Ono T, Hasegawa T, Masushige S, et al. Two forms of avian(chicken) TATA-binding protein mRNA generated by alternative polyadenylation. Biochem Biophys Res Commun. 1997;234:406–411. doi: 10.1006/bbrc.1997.6653. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility gene located in the T/t- H-2 supergene on chromosome 17. In: Reisfeld S, Ferone RA, editor. Current trends in histocompatibility. New York: Plenum Press; 1981. pp. 103–131. [Google Scholar]
- Gregorova S, Forejt J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies-a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha) 2000;46:31–41. [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Ruf N, Dunzinger U, Brinckmann A, Haaf T, Nurnberg P, Zechner U. Expression profiling of uniparental mouse embryos is inefficient in identifying novel imprinted genes. Genomics. 2006;87:509–519. doi: 10.1016/j.ygeno.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/S0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–193. doi: 10.1210/er.14.2.184. [DOI] [PubMed] [Google Scholar]
- Knee R, Murphy PR. Regulation of gene expression by natural antisense RNA transcripts. Neurochem Int. 1997;31:379–392. doi: 10.1016/S0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. Developmental testis-specific regulation of mRNA levels and mRNA translational efficiencies for TATA-binding protein mRNA isoforms. Dev Biol. 1997;184:138–149. doi: 10.1006/dbio.1997.8514. [DOI] [PubMed] [Google Scholar]
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Good L. Translation repression by antisense sequences. Cell Mol Life Sci. 2003;60:854–861. doi: 10.1007/s00018-003-3045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A. Natural antisense transcripts. RNA Biol. 2005;2:53–62. doi: 10.4161/rna.2.2.1852. [DOI] [PubMed] [Google Scholar]
- Werner A, Berdal A. Natural antisense transcripts: sound or silence? Physiol Genomics. 2005;23:125–131. doi: 10.1152/physiolgenomics.00124.2005. [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R. Naturally occurring antisense: transcriptional leakage or real overlap? Genome Res. 2005;15:364–368. doi: 10.1101/gr.3308405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for RT-PCR, RACE and real-time PCR. The primer name, 5'-> 3' sequence and usage are noted. In QRT-PCR primer pairs, conditions of the reaction using the real-time PCR system (LightCycler, Roche) are noted in the brackets. Primer names beginning with "Ch" amplify chicken Pdcd2, Tbp genes. F, forward primer; R, reverse primer; alt., alternative.
Testing the imprinting status of the transcripts in the Hst1 region. B6 × PWD polymorphisms used to test the imprinting status of the transcripts in the Hst1 region are noted in the table. The example of the imprinting analysis (for Pdcd2 constitutive transcript) are depicted below.


