The availability of a complete and accurate set of aminoacylated tRNAs is fundamental to protein biosynthesis in all three kingdoms of life [1]. Despite this rigid requirement, many bacteria and archaea lack asparaginyl- and/or glutaminyl-tRNA synthetase (AsnRS and/or GlnRS, respectively), the canonical enzymes that catalyze the direct biosynthesis of Asn-tRNAAsn and Gln-tRNAGln [1]. In organisms lacking either or both of these enzymes, the requisite aminoacyl-tRNAs are biosynthesized indirectly via a two-step process (Fig. 1). First, a non-discriminating aspartyl- or glutamyl-tRNA synthetase (ND-AspRS or ND-GluRS) generates Asp-tRNAAsn or Glu-tRNAGln respectively. Second, the bi-functional Asp-tRNAAsn/Glu-tRNAGln amidotransferase (Asp/Glu-Adt or GatCAB) catalyzes a transamidation reaction to repair both of these misacylated tRNAs. A Glu-tRNAGln-specific amidotransferase (Glu-Adt or GatDE) is also used in some species instead of, or in addition to, Asp/Glu-Adt. A recent review has been published describing these amidotransferases [2]. Asp/Glu-Adt is typically assayed by thin-layer chromatography (TLC) to separate the different amino acid products [3]. This assay is timeconsuming and cannot distinguish between Asp and Gln or between Asn and Glu. Here we report an improved assay for Asp/Glu-Adt that uses thin-layer electrophoresis (TLE) [4, 5] for amino acid separation instead of TLC. The electrophoretic step is rapid and leads to improved resolution.
Figure 1.
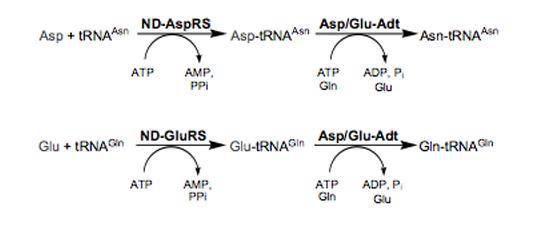
Biosynthesis of Asn-tRNAAsn and Gln-tRNAGln in the absence of AsnRS and GlnRS. Top panel: An ND-AspRS catalyzes the biosynthesis of Asp-tRNAAsn. Next, Asp/Glu-Adt converts Asp-tRNAAsn into Asn-tRNAAsn in an ATP-dependent reaction that uses glutamine as a nitrogen donor. Bottom panel: Gln-tRNAGln is biosynthesized in the same two-step process, beginning with an ND-GluRS (or GluRS2 [11, 12], not shown). In both cases, misacylation is shown in grey.
Although the overall reaction catalyzed by Asp/Glu-Adt is the simple conversion of an amino acid carboxylate (in either Asp or Glu) into a carboxyamide (in Asn or Gln), this transformation takes place while the amino acid is attached to a tRNA, making product detection complicated because of the large size of the aminoacyl-tRNA substrate compared to the small chemical change introduced by the enzyme. A typical assay for Asp/Glu-Adt begins with either [14C]-Asp-tRNAAsn or [14C]-Glu-tRNAGln [6]; these radiolabeled aminoacyl-tRNAs are generated using the corresponding ND-AspRS or ND-GluRS. Each of these tRNAs is incubated with Asp/Glu-Adt, ATP, and non-radiolabeled glutamine as the ammonia donor. Time points are removed and quenched in 3 M sodium acetate (pH 5), followed by phenol extraction. Each quenched aliquot, containing a mixture of unreacted [14C]-Asp-tRNAAsn or [14C]-Glu-tRNAGln and of enzymegenerated [14C]-Asn-tRNAAsn or [14C]-Gln-tRNAGln, is deacylated by mild alkaline treatment and the released amino acids are subsequently separated by TLC and visualized by phosphorimaging of the 14C-labeled amino acids. Asp/Glu-Adt has also been assayed by HPLC, after amino acid derivatization with a fluorescent label to facilitate detection; this method is not as sensitive as the standard TLC/radioactivity method described above, but it has the advantage that it does not require radioactivity [7].
Because of the polar nature of free amino acids, the TLC plates are developed in highly aqueous solvents (Solvent A: isopropanol: formic acid: acetic acid: water in an 8:1:1:4 ratio, Fig. 2A) and take 1-2 hours for complete elution. As can be seen in the TLC plate shown in Fig. 2A, these conditions effectively resolve Asp (Rf = 0.63) from Asn (Rf = 0.70) and Glu (Rf = 0.70) from Gln (Rf = 0.60). The drawbacks to this method are the time it takes to develop the TLC plate, the fact that Asn and Glu (lanes 2 and 4) and Asp and Gln (lanes 1 and 5) have identical and nearly identical retention factors respectively, and occasional streaking. Because Asp/Glu-Adt is confronted with both Asp-tRNAAsn and Glu-tRNAGln in vivo in many organisms [8-10], an assay that allows for the resolution of all four relevant amino acids would be highly useful for experiments designed to mimic in vivo conditions.
Figure 2.

TLE as a method to assay Asp/Glu-Adt. A. Phosphorimage of a TLC plate developed in Solvent A. The origin is not shown and is approximately 2.25 inches below the lowest spot. B. TLE plate electroeluted for 40 minutes at 100 volts in Solvent B. The solid line denotes the site of sample loading (origin); the directions of the cathode (+) and the anode (-) are also labeled. In both A and B, each lane was spotted at the origin with 50 nCi of the relevant 14C-labelled amino acid, as labeled. Lane 1-Asp; Lane 2-Asn; Lane 3-1:1 mixture of Asp:Asn; Lane 4-Glu; Lane 5-Gln; Lane 6-1:1 ratio of Glu:Gln. C. An assay of Asp/Glu-Adt using TLE. Asp/Glu-Adt was assayed as described in the text. Lanes 1-3 represent amino acid controls (Lane 1-Asp, Lane 2-Asn, Lane 3-1:1 ratio of Asp:Asn). Lane 4 shows the results from the no enzyme control where the Asp-tRNAAsn remains unreacted. Lane 5 shows the results from the Asp/Glu-Adt assay where approximately 50% of the Asp-tRNAAsn has been converted to Asn-tRNAAsn.
We decided to evaluate TLE as a technique for better amino acid resolution in Asp/Glu-Adt assays. In TLE, compounds are eluted electrophoretically and consequently the charged states of the different amino acids contribute to their mobility across the stationary phase. We first evaluated TLE separation of Asp, Asn, Glu, and Gln in the absence of Asp/Glu-Adt (Figure 2B) using standard TLC plates (Precoated silicagel 60F254 plate, Fisher Scientific). Square plates were cut to dimensions suitable for elution in a standard submerged horizontal gel electrophoresis chamber (e. g. a 15 cm wide × 7 cm long TLC plate in a Bio-Rad mini-sub gel GT). Each amino acid was separately soaked along a line at the center of the plate (the origin, Figure 2B) and the plate was gently loaded with running buffer (Solvent B: 0.1 M NaH2PO4, pH 7). The plate was placed in the electroelution chamber and the anode and cathode chambers were filled with running buffer. Filter paper was soaked in running buffer and used to bridge the buffer beds at each electrode with the TLC plate. The plate was subjected to 100 volts for 40 minutes and then developed by exposure to an enhanced phosphorimager screen (Amersham Biosciences) and the plate was imaged on a Molecular Dynamics Storm 840 Phosphorimager. As can be seen in Figure 2B, TLE led to clear resolution of Asp (Rf = +0.20) and Asn (Rf = -0.34) and of Glu (Rf = +0.14) and Gln (Rf = -0.43), with better separation and less streaking than that obtained by standard TLC. TLE also led to the resolution of Asn and Gln (compare lanes 2 and 5, Fig. 2B, and Rf values given above) and partial resolution of Asp and Glu (compare lanes 1 and 4, Fig. 2B, and Rf values given above) and is therefore superior to TLC for its ability to resolve all four amino acids.
Next, we verified TLE as a method to assay Asp/Glu-Adt (Figure 2C). Helicobacter pylori Asp/Glu-Adt (310 nM) was incubated with [14C]-Asp-tRNAAsn (2.5 μmoles, 25 μM, 500 μCi), 4 mM ATP, 1 mM Gln, 4 mM MgCl2, 1.25 mM KCl, in 10 mM Hepes, pH 7.5, for 10 minutes. A parallel no enzyme control experiment was also conducted. The reactions were quenched and precipitated. The resultant pellets were resuspended in 50 μL 25 mM KOH and deacylated at 65 °C for 10-15 minutes. Following deacylation, mixtures were neutralized with 12.5 μL 0.1 M HCl and pellets were dried in a Savant SC110A Speed Vac Plus. Pellets were resuspended in 7 μL H2O and analyzed by TLE (3 μL spots) as described above. As can be clearly seen in Figure 2C (compare lanes 4 and 5), Asp/Glu-Adt converted approximately 50% of the Asp-tRNAAsn into Asn-tRNAAsn and the radiolabelled amino acids from this reaction were successfully resolved by TLE. Furthermore, TLC analyses are hindered by occasional diffusion and streaking of the amino acid; these problems were not observed in any of our TLE experiments.
In summary, we report here the development of a TLE assay for Asp/Glu-Adt. This assay is facile, reproducible and faster than the standard TLC assay typically used with this enzyme.
Acknowledgments
Acknowledgements: Financial support from Johns Hopkins University and the National Institutes of Health (GM071480) is gratefully acknowledged. PC is supported by a DPST Fellowship from the Royal Thai Government. The authors thank Dr. Charles Merryman for an introduction to TLE and Professor Marc Greenberg for use of his Phosphorimager.
Glossary
Abbreviations used:
- AsnRS
asparaginyl-tRNA synthetase
- Asp/Glu-Adt
Asp-tRNAAsn/Glu-tRNAGln amidotransferase
- GlnRS
glutaminyl-tRNA synthetase
- ND-AspRS
non-discriminating aspartyl-tRNA synthetase
- ND-GluRS
non-discriminating glutamyl-tRNA synthetase
- TLC
thin-layer chromatography
- TLE
thin-layer electrophoresis
References
- [1].Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- [2].Feng L, Tumbula-Hansen D, Min B, Namgoong S, Salazar JC, Orellana O, Soll D. Transfer RNA-Dependent Amidotransferases: Key Enzymes for Asn-tRNA and Gln-tRNA Synthesis in Nature (2005) in The Aminoacyl-tRNA Synthetases. In: Ibba M, Francklyn C, Cusack S, editors. Landes Bioscience; Georgetown, Texas: pp. 314–319. [Google Scholar]
- [3].Schon A, Kannangara CG, Gough S, Soll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–90. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- [4].Fisher TL, Reingold ID, Fitzsimmons TL. Thin-Layer Electrophoresis. Journal of Chemical Education. 2001;78:1241–1243. [Google Scholar]
- [5].Merryman C, Green R. Transformation of aminoacyl tRNAs for the in vitro selection of “drug-like” molecules. Chem Biol. 2004;11:575–82. doi: 10.1016/j.chembiol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- [6].Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A. 1997;94:11819–26. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001;40:6450–7. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- [8].Becker HD, Min B, Jacobi C, Raczniak G, Pelaschier J, Roy H, Klein S, Kern D, Soll D. The heterotrimeric Thermus thermophilus Asp-tRNA(Asn) amidotransferase can also generate Gln-tRNA(Gln) FEBS Lett. 2000;476:140–4. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- [9].Raczniak G, Becker HD, Min B, Soll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J Biol Chem. 2001;276:45862–7. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- [10].Salazar JC, Zuniga R, Raczniak G, Becker H, Soll D, Orellana O. A dual-specific Glu-tRNA(Gln) and Asp-tRNA(Asn) amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500:129–31. doi: 10.1016/s0014-5793(01)02600-x. [DOI] [PubMed] [Google Scholar]
- [11].Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Soll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci U S A. 2003;100:13863–8. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc Natl Acad Sci U S A. 2003;100:11297–302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]


