Abstract
Inbred mouse strains have been maintained for more than 100 years, and they are thought to be a mixture of four different mouse subspecies. Although genealogies have been established, female inbred mouse phylogenies remain unexplored. By a phylogenetic analysis of newly generated complete mitochondrial DNA sequence data in 16 strains, we show here that all common inbred strains descend from the same Mus musculus domesticus female wild ancestor, and suggest that they present a different mitochondrial evolutionary process than their wild relatives with a faster accumulation of replacement substitutions. Our data complement forthcoming results on resequencing of a group of priority strains, and they follow recent efforts of the Mouse Phenome Project to collect and make publicly available information on various strains.
Long before the beginning of mouse genetics, humans in Eastern Asia were already breeding mice in order to obtain animals with different coat colors. Modern mouse genetics, however, did not start until the early 20th century with William Castle’s studies on inheritance. Most of his mice derived from collections of mice fanciers, and they were the ancestors of many inbred strains that are still used today (Rader 2004).
Mouse strains are known to have mixtures of various ancestral genomes from different Mus musculus (house mouse) subspecies (for review, see Yoshiki and Moriwaki 2006). Different molecular markers suggest that the main contributors are M. musculus musculus (Bishop et al. 1985), M. musculus domesticus (Yonekawa et al. 1982; Sakai et al. 2005), and, to a lesser extent, M. musculus castaneus (Sakai et al. 2005). One other subspecies that is usually considered to have contributed is M. musculus molossinus (Sakai et al. 2005), although this itself is supposed to be a hybrid between M. m. musculus and M. m. castaneus (Lundrigan et al. 2002; Wade et al. 2002).
More than 450 mouse inbred strains have been established since the first—DBA/2 (dilute, brown, non-agouti)—was developed by Castle’s student Clarence Cook Little in what would become The Jackson Laboratory (Beck et al. 2000; Rader 2004). Furthermore, in different laboratories worldwide, many substrains of each strain have also been maintained.
A mouse strain is defined as inbred if the animals have been crossed brother × sister for ≥20 consecutive generations and individuals of the strain can be traced back to a single ancestor pair at the 20th or subsequent generation (Eppig et al. 2005). Theoretical studies indicate that, at this time, ≥98.6% of loci should be homozygous, but many strains have been bred for >150 generations, which makes them homozygous at virtually every locus (Beck et al. 2000).
Beck et al. (2000) extensively documented inbred mice genealogies, suggesting that independent inbreeding processes occurred in at least three regions of the globe: (1) Castle’s mice (Group B) and C57-related strains (Group E) originated from Abbie Lathrop’s stocks in the United States; (2) Swiss mice (Group A) derived from mice from Switzerland; and (3) strains derived from colonies from China and Japan (Group C). Little is known, however, of mitochondrial DNA (maternal) phylogenies of these strains. So far, there has been only one study on complete mtDNA sequences from different inbred strains, with the purpose of revising the complete mouse mtDNA reference sequence (Bayona-Bafaluy et al. 2003). The study of mtDNA phylogenies has the potential to elucidate the matrilineal lineage of common inbred strains and its relationship to the main subspecific lineages.
Mammalian mitochondrial DNA (mtDNA) is a circular double-stranded molecule that encodes 13 genes of the respiratory chain. Defects in these molecules have been associated with a variety of disorders that may affect different tissues in different ways (for review, see Wallace 1999). A number of mouse models have recently been developed that have clarified how mutations in mtDNA are transmitted. Examples include transmitochondrial mice carrying heteroplasmic point mutations (Sligh et al. 2000), heteroplasmic mice with mtDNAs from variants characteristic of two different strains (Battersby and Shoubridge 2001), and mice with homoplasmic replacement of endogenous mtDNA (McKenzie et al. 2004). With this recent increasing interest in mtDNA from mice models, it becomes important to know the mtDNA sequence in each of the different inbred strains.
The analysis of complete mtDNA sequences of inbred mice is also useful for the establishment of mutation/substitution rates. Given that, in principle, these restricted animal populations have a reasonably well documented history, with no inclusion of foreign DNA, variation that occurs can be generated only by mutation. Moreover, study of mice mitochondrial phylogenies may become helpful in clarifying differences that have been reported between mutation rates estimated from pedigrees and substitution rates calculated from phylogenies (Howell et al. 2003; Ho et al. 2005; Ho and Larson 2006).
Two main issues were addressed while performing this work: (1) validation of published genealogies and clarification of the matrilineal origin of common inbred strains and (2) evolutionary analysis of inbred strains. It was developed by focusing on 16 selected strains that are part of the Mouse Phenome Project (Bogue 2003), which aims to enhance the resources available for laboratory mice by collecting phenotypic and genotypic characteristics of these animals and making them publicly available through a Web-accessible database. These data may ultimately help researchers track down the genes involved in particular phenotypes, by allowing association of phenotypes with genotypes for each strain. A set of 40 priority strains has been established, and for 15 of them the complete genome is now being resequenced (Pearson 2004). By performing the mtDNA characterization of these 15 strains (plus C57BL/6), we are also contributing to this project.
Results
mtDNA diversity
We have sequenced the complete mtDNA of 32 mice belonging to 16 inbred strains (two of each strain): 12 common inbred (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+tf/J, C3H/HeJ, C57BL/6J, DBA/2J, FVB/NJ, KK/HlJ, NOD/LtJ, NZW/LacJ) and four wild-derived inbred strains, each representing a different subspecies (CAST/EiJ, MOLF/EiJ, PWD/PhJ, and WSB/EiJ). Out of the 16 strains sequenced we obtained 12 different haplotypes. For all inbred strains, including the wild-derived, sequencing of the second animal of the same strain always confirmed the first sequence (haplotype) obtained.
We combined our data with 13 complete sequences available online, giving a total of 20 different haplotypes consisting of (1) 11 common inbred strains plus (2) two more divergent (NZB and MilP) common inbred haplotypes, (3) two cell lines, and (4) five wild-derived inbred strains’ haplotypes. The mtDNAs of all 11 common inbred strains were very similar. Only 15 substitutions were detected in the 16,299 mtDNA nucleotides, 14 inside genes and one in a ribosomal RNA (Supplemental Table 1). No substitutions were observed in the control region. Insertions in transfer RNA genes were also observed in sites previously reported as highly polymorphic (Bayona-Bafaluy et al. 2003; Supplemental Table 1). This similarity between common inbred strains has already been reported for some of these strains (Bayona-Bafaluy et al. 2003). However, we have extended the number of strains and included mice from different groups (Beck et al. 2000): A (Swiss mice), B (Castle’s mice), C (strains derived from China and Japan), and E (C57-related strains).
For strains C3H and C57BL/6, we obtained different haplotypes from the published ones. C3H/HeJ differs from C3H/He (Kiebish and Seyfried 2005) at position 8889, and the two C57BL/6 sequences differ at position 11,780. Based on the data of Kiebish and Seyfried (2005) and on a personal communication with J.A. Enríquez (Universidad de Zaragoza, Spain), we concluded that the substitution at 11,780 was probably an artifact, and will therefore be ignored, while that at 8889 was likely real. All other common inbred strains that we could compare with published data matched the reported sequences.
The four strains derived from wild mice that we sequenced are highly divergent from each other, and three of them also diverge from the reference C57BL/6 sequence: CAST (M. m. castaneus) differs from the reference in 377 positions, PWD (M. m. musculus) and MOLF differ (M. m. molossinus) in 386 positions. The MOLO strain (M. m. molossinus) mtDNA that was already reported (Akimoto et al. 2005) differed from MOLF in 15 sites. WSB, the M. m. domesticus representative, shows only 10 differences from the reference.
Phylogenetic analyses
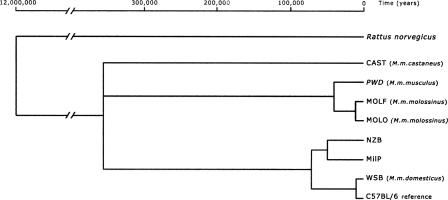
We evaluated the possible branching order through a preliminary network analysis (data not shown) and tried to assess which was the first split within the wild-derived mice by maximum likelihood analysis. Since the closest outgroup is the rather divergent Rattus norvegicus, we did not include the control region in this first tree. The maximum likelihood values were not different enough to determine which of the three most divergent groups separated first, and therefore we left this deepest slip unresolved with the following three branches diverging (Fig. 1): (1) Mus musculus castaneus-derived strain, (2) M. m. musculus and M. m. molossinus representatives, and (3) a group that includes the M. m. domesticus representative and all the common inbred strains, here represented by the C57BL/6 branch. Apart from NZB and MilP, all common inbred strains clustered together with the M. m. domesticus representative.
Figure 1.
Maximum likelihood phylogenetic tree of five wild-derived and three common inbred strains. Rattus norvegicus was used as an outgroup.
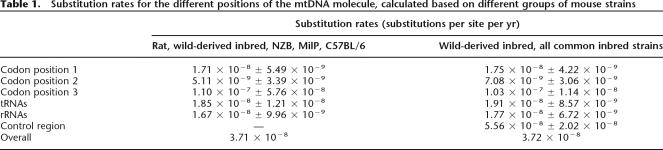
We estimated coalescence times for these branches assuming a divergence time between Mus and Rattus of 12 million yr (paleontological data from Jacobs et al. 1989; Jaeger et al. 1986 in Michaux et al. 2002; Suzuki et al. 2004). The results date the split between the three main groups at 371,000 ± 91,300 yr ago, consistent with published mtDNA data that place the divergence between Mus musculus subspecies between 0.1 Mya (based on mitochondrial cytochrome B) and 0.5 Mya (based on mitochondrial and nuclear genes combined) (Suzuki et al. 2004).
Based on the same assumption, we obtained an overall substitution rate for the mouse coding mtDNA of 3.7 × 10−8 substitutions per site per yr. This value is rather similar to the 1.26 × 10−8 substitutions per site per yr obtained by Mishmar et al. (2003) for the human mtDNA coding region. A higher substitution rate of 1.1 × 10−7 substitutions per site per yr is observed for third codon positions, while the second codon positions present the lowest substitution rate of 5.1 × 10−9 substitutions per site per yr (Table 1).
Table 1.
Substitution rates for the different positions of the mtDNA molecule, calculated based on different groups of mouse strains
We produced a second tree (not shown) with complete mtDNA data from all common inbred strains, cell lines, and wild-derived mice, but excluding the rat. This analysis confirmed that all common inbred mice group with the M. m. domesticus representative. Based on the coalescence time for the main subspecies of Mus calculated in the above analysis, we obtained similar substitution rates to those derived from the previous group of strains (Table 1). Moreover, we could estimate a substitution rate for the control region of 5.6 × 10−8 per site per yr, which is about three to four times higher than the substitution rate of the first codon positions and eight to 11 times higher than that of the second codon positions. Third codon positions, however, remain the ones with a highest substitution rate, which is twice the value obtained for the control region.
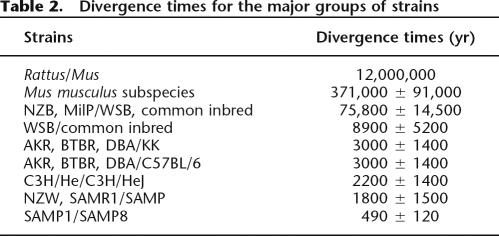
We estimated the divergence time for these common inbred mice and obtained older times than expected. The divergence of the common inbred strains (excluding the most divergent strains NZB and MilP) was dated at 3000 ± 1400 yr ago (Table 2), although it is documented that the first inbred strain was established only ∼100 yr ago (Rader 2004). Another example is the case of the SAMP strains that were not separated before 1968 (Takeda et al. 1997), though our estimate for their coalescence time was 490 ± 120 yr ago.
Table 2.
Divergence times for the major groups of strains
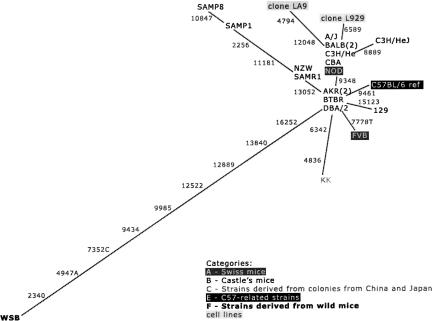
The mtDNA sequences of the common inbred mice show a star-like network (Fig. 2). The domesticus-derived WSB is the furthest away from the center. Except for the SAM strains, all the strains that belong to groups A (Swiss mice), B (Castle’s mice), and E (C57-related strains) are in the central groups or less than one polymorphism apart. Four inbred strains are two or more polymorphisms apart from the central groups: KK belongs to group C (strains derived from colonies from China and Japan), cell-line LA9, and the SAM strains. Although these belong to group B, they were crossed with mice from an unknown strain in 1968 (Beck et al. 2000).
Figure 2.
The single most parsimonious tree for complete mice mtDNA polymorphisms found in 16 different common inbred strains (19 sequences), two cell lines, and one wild-derived inbred strain (WSB).
Discussion
The maximum likelihood analysis of the wild-derived strains did not permit the establishment of the first split among the three major subspecies, and so we present a tree with a tricotomy at the base (Fig. 1): (1) M. m. castaneus, (2) M. m. musculus, and (3) M. m. domesticus. Not surprisingly, this ambiguity had already been unresolved in previous analyses with mitochondrial genes Cyt b and 12S (Lundrigan et al. 2002), while analyses with combined nuclear and mitochondrial genes disagree on which subspecies was the first to diverge (Lundrigan et al. 2002; Suzuki et al. 2004).
Our results from complete mtDNA sequences agree with others based on particular genes (Lundrigan et al. 2002) in placing M. m. molossinus closer to M. m. musculus than to other subspecies. This confirms not only that M. m. molossinus is a hybrid but also that its mtDNA shares a recent common ancestor with M. m. musculus.
A single female origin for lab mice strains
The extreme similarity among mtDNAs of the different common inbred strains is well illustrated by the small number of polymorphisms that were found (Fig. 2; Supplemental Table 1). The accumulation of polymorphisms agrees with published genealogies (Beck et al. 2000) and the history (Eppig et al. 2005) of the strains, particularly when individual groups (Swiss mice, Castle’s mice, strains derived from China and Japan, and C57-related strains) of strains are considered. Small deviations from the published genealogical trees can generally be justified by outbreeding somewhere back in the history of the strain (each strain is discussed in detail in the Supplemental data).
Genealogies do not suggest any relationship between the different groups (except for C57-related mice and Castle’s mice, which both descend from Abbie Lathrop’s stocks). Our results show, however, that all the common inbred strains appear clustered in the network, irrespectively of the group to which they belong. This finding suggests a common origin for all analyzed groups of common inbred strains. Furthermore, the fact that, in the phylogenetic tree, common inbred strains appear clustered with the M. m. domesticus-derived strain, and far from representatives of other subspecies, shows that the common mitochondrial ancestor of the inbred strains used today belonged to the M. m. domesticus wild subspecies.
Taking together our results and the history of inbred mice, it is likely that the same stocks of M. m. domesticus fancy mice were at the origin of all three groups; the maintenance of separate colonies in different regions would have resulted in the few differences that are now observed. The most divergent inbred strains were NZB and MilP as already reported by Bayona-Bafaluy et al. (2003). Different reasons may explain the divergence of these two strains: the MilP strain is not a true common inbred strain, as it descends from a wild female mouse caught in Italy (Loveland et al. 1990 in Bayona-Bafaluy et al. 2003), and NZB, although belonging to group B (Castle’s mice), did not originate from Abbie Lathrop’s stocks, but from unidentified “European & U.S. stocks,” and it was outbred during its development (Beck et al. 2000).
Although few polymorphisms were observed between the different common inbred strains, a couple of comments are worth making: (1) A polymorphism has occurred in the split between two substrains of C3H. C3H/He and C3H/HeJ can therefore be distinguished by the nucleotide at position 8889. (2) SAMP strains, although established only 30 yr ago, appear to have accumulated more substitutions than any other strain. This could be related to their senescence-accelerated phenotype, since mtDNA mutations have been proposed to be implicated in the aging process (Chinnery et al. 2002; Chomyn and Attardi 2003). Moreover, our results suggest a hypothesis about the unknown female ancestor (Mizutani et al. 2001) of SAM strains, by showing that NZW shares the same haplotype as SAMR1.
Different mtDNA evolution in wild and inbred strains
The substitution rates calculated based on wild and inbred mice data vary depending on the position. Third codon positions, unlike second positions, show a higher substitution rate probably because degeneracy of the genetic code makes these positions less prone to selective pressure: A change in a third position base will probably not replace the coded amino acid. Intriguingly, when analyzing the substitutions that occur in the inbred strains (Supplemental Table 1), we observe a low frequency (4/14) of substitutions involving third codon positions, and an even lower number (3/14) of synonymous variants. The same is observed for the WSB strain. However, this could be a consequence of performing the comparisons relative to an inbred strain (C57BL/6). Most (13) of the 15 substitutions found among inbred strains are transitions, and the overall transition/transversion rates were higher for all positions in the analysis with common inbred strains than in the one including the rat (data not shown). This higher accumulation of transversions in the wild was expected given the very old Mus/Rattus divergence when compared with the inbred strains.
Although data on RFLPs have placed the divergence inside the Mus musculus species 1–2 million yr ago (Yonekawa et al. 1981), long before the ∼370,000 yr ago that we obtained, our estimated coalescence times for the wild-derived strains fall within values obtained more recently with other markers (Suzuki et al. 2004). The estimates of coalescence times of the inbred strains, however, were much higher than the known divergence of the strains. It is known that the common inbred strains were first established at the beginning of the 20th century, but our estimate for coalescence of the inbred strains is ∼3000 yr ago. Even if, as is suggested by our network, all the common inbred strains (Castle’s mice, Swiss mice, strains derived from colonies from China and Japan, and C57-related strains) had the same ancestor earlier than the 20th century, in Japanese fancy mice (17th century, Rader 2004), values obtained in our estimates would still be very high. Furthermore, the divergence times for the splits between the different inbred strains are also much older in our estimates than what is documented.
Taken together, these estimates show a faster mtDNA evolution in inbred than in wild mice with a higher accumulation of replacement substitutions. Three hypotheses may account for these discrepancies: (1) the calibration point that we used (12 million yr for the Mus/Rattus divergence) is too distant relative to the young splits that we are trying to date; (2) mtDNA evolution in inbred strains, with such controlled conditions and restricted crosses, occurs faster than in wild environments; or (3) there are differences in generation times of inbred and wild strains.
The issue of the calibration points has been under debate in recent publications, and it has been suggested that substitution rates extrapolated across different time scales result in invalid date estimates (Ho and Larson 2006). In our case, using a 12-million-yr calibration point for a 100-yr-old estimate might be inappropriate. Since it was not possible to find a more suitable calibration point (paleontological data after the Mus/Rattus split are absent), we tried to overcome this limitation by assuming that our estimate for the Mus musculus divergence (∼370,000 yr ago) was correct, and from there estimating the substitution rate for the common inbred strains. Also, the fact that the coding region substitution rates that we obtained from the wild strain data are of the same order of magnitude as the values obtained for human mtDNA may suggest that our estimates for the substitution rates are not too far from the truth.
Therefore, a higher substitution rate in inbred mice may still stand, despite this rough time estimate. Curiously, mice cell lines included in our analyses have accumulated a similar number of substitutions to that observed for mouse inbred strains, although it had been reported that relaxation of negative selection resulted in a high substitution rate in HeLa cell lines, when compared with phylogenetically derived divergence of mtDNAs (Herrnstadt et al. 2002). Evolutionary rates of inbred mouse mtDNA appear to be closer to cell lines than to wild mice, suggesting that laboratory controlled conditions with strict bottlenecks may also account for the higher substitution rate.
A higher substitution rate could also be apparent if generation times of wild mice are longer than in inbred strains. It is possible that mice breeders cross mice before they would mate in the wild. For C57BL/6 mice it is documented (Genetic Background Resource Manual 2006, The Jackson Laboratory, http://jaxmice.jax.org/literature/manuals/mouse_genetics_resource_manual.pdf) that ∼120 generations have passed in the period from 1920 to 1970, resulting in 2.4 generations per yr, but other inbred strains may have shorter generation times. Wild mice are commonly assumed to have about two generations per yr. This difference could explain a slightly higher substitution rate, but it does not explain the high frequency of replacement substitutions.
Final remarks
Although the history of inbred mice reports at least three geographically separate origins for inbred mice, our data permit us to conclude that all common inbred strains that are now being resequenced originate from the same female ancestor, a M. m. domesticus mouse. Phylogenetic analyses suggest that two different processes of mtDNA evolution may occur depending on the environment: one in wild mice that results in a substitution rate similar to the one obtained for human mtDNA, and the other in the controlled environment of the laboratory strains, which gives rise to a faster evolution and a higher accumulation of replacement substitutions.
Methods
Samples and sequencing
Livers from 32 mice belonging to 16 inbred strains (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+tf/J, C3H/HeJ, C57BL/6J, DBA/2J, FVB/NJ, KK/HlJ, NOD/LtJ, NZW/LacJ, CAST/EiJ, MOLF/EiJ, PWD/PhJ, and WSB/EiJ) were provided as part of a collaboration with the Mouse Phenome Project (The Jackson Laboratory, Bar Harbor, ME). All mice were male and of about the same age. We sequenced DNA from two animals of each strain, belonging to proximate generations. Potential differences that could have been detected between the two mice would result in one of the following conclusions: (1) there was a sequencing error and/or contamination, (2) the strain was misidentified, or (3) the strain presented mtDNA diversity.
We extracted DNA from livers following a standard phenol-chloroform protocol (digested with proteinase K in buffer TE containing 0.5% SDS, purified with phenol-chlorophorm-isoamyl alcohol, and precipitated with ethanol). We amplified 34 overlapping fragments of ∼500 bp covering the entire mtDNA molecule, with the appropriate oligodeoxynucleotides and annealing temperatures (Supplemental Table 2). We purified the PCR products using Microspin S-300 HR columns (Amersham Biosciences), according to the manufacturer’s specifications. Sequence reactions were carried out using the Big-Dye Terminator Cycle Sequencing Ready Reaction kit (AB Applied Biosystems) with one primer (Supplemental Table 2), in forward and/or reverse directions. We purified the samples with Sephadex G-50 DNA Grade f (Amersham Biosciences) and ran them in an automatic sequencer ABI 3100 (AB Applied Biosystems). Sequences were compared using C57BL/6J as the reference sequence.
Construction of phylogenetic trees
For phylogenetic analyses, we combined our data with 13 complete sequences available online, totaling 20 different haplotypes. We aligned the mtDNA complete sequences with ClustalW (Chenna et al. 2003) and constructed a maximum parsimony phylogenetic network (NETWORK 4.1.1.2, Bandelt et al. 1995) of all strains, in order to suggest a number of possible branching orders.
We then performed maximum likelihood (PAML 3.15; Yang 1997) analyses using the rat (Rattus norvegicus) mitochondrial sequence as an outgroup, and assuming the HKY85 mutation model. Site heterogeneity was allowed via a gamma distribution of rates. Likelihood ratio tests were performed to test different models, namely: (1) to decide on the most likely branching order, (2) to decide on whether or not a uniform molecular clock was appropriate, and (3) to test whether different mutation parameters should be allowed for different genes. When the differences were not significant, the simpler model was chosen. The standard errors on time estimates and mutation rates come directly out of the ML analysis. The covariance matrix of model parameter estimates was derived from the second derivative of the log likelihood evaluated at the likelihood maximum. The standard errors then follow by the delta method.
Due to the difficulty in aligning the D-loop of rat and mouse sequences, the first analysis did not include the control region. After the tree was established, and because of the difference in evolutionary scale between the divergence of wild and inbred strains, we redid the analysis including only (1) the rat; (2) the wild-derived inbred strains; (3) two common inbred strains reported to be more divergent than the others, NZB and MilP; and (4) the C57BL/6 strain, as a representative of all other common inbred strains. Assuming from paleontological data (Jacobs et al. 1989; Jaeger et al. 1986 in Michaux et al. 2002; Suzuki et al. 2004) that the Mus/Rattus divergence occurred 12 million yr ago, we determined substitution rates and coalescence times for all the branches in the tree. We performed a separate maximum likelihood analysis excluding the rat sequence and including all common inbred strains, but considering the whole molecule (including the control region). Assuming that the divergence time of the Mus musculus subspecies calculated in the previous tree was correct, we recalculated substitution rates and coalescence times separately for this tree and compared the results. The two separate phylogenetic analyses allowed us to compare results obtained with and without the common inbred strains and to reduce the amount of difference in evolutionary scale of the strains used in each case.
We also constructed a maximum parsimony phylogenetic network including data from the complete mtDNAs from all common inbred strains and the M. m. domesticus representative (WSB). This summarizes all sequence data and illustrates the extreme lack of variation among the different common inbred strains.
Accession numbers
The published complete mtDNA sequences used in the analyses are available in GenBank, accession nos. AB042432, AB042523, AB042524, AB042809, AB049357, AJ489607, AJ512208, AY172335, AY339599, L07095, and L07096 (Bayona-Bafaluy et al. 2003), AY466499 (Moreno-Loshuertos et al. 2006), AY675564 (Akimoto et al. 2005), and DQ106412 (Kiebish and Seyfried 2005). The results have been submitted to the Mouse Phenome Database (http://www.jax.org/phenome) as part of the project MPD: 202 (Goios et al. 2006).
Acknowledgments
This work was partially supported by Fundação para a Ciência e a Tecnologia through a research grant to A.G. (SFRH/BD/16518/2004) and by “Programa Operacional Ciência, e Inovação 2010” (POCI 2010), VI Programa-Quadro (2002–2006).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. EF108330–EF108345.]
Article published online before print. Article and publication date are online at http://www.genome.org/cgi/doi/10.1101/gr.5941007
References
- Akimoto M., Niikura M., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Niikura M., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Ichikawa M., Yonekawa H., Nakada K., Honma Y., Hayashi J., Yonekawa H., Nakada K., Honma Y., Hayashi J., Nakada K., Honma Y., Hayashi J., Honma Y., Hayashi J., Hayashi J. Nuclear DNA but not mtDNA controls tumor phenotypes in mouse cells. Biochem. Biophys. Res. Commun. 2005;327:1028–1035. doi: 10.1016/j.bbrc.2004.12.105. [DOI] [PubMed] [Google Scholar]
- Bandelt H.-J., Forster P., Sykes B., Richards M., Forster P., Sykes B., Richards M., Sykes B., Richards M., Richards M. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby B.J., Shoubridge E.A., Shoubridge E.A. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 2001;10:2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy M.P., Acín-Pérez R., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Acín-Pérez R., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A., Fernández-Silva P., Bai Y., Enríquez J.A., Bai Y., Enríquez J.A., Enríquez J.A. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher E.M.C., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher E.M.C., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher E.M.C., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher E.M.C., Eppig J.T., Festing M.F.W., Fisher E.M.C., Festing M.F.W., Fisher E.M.C., Fisher E.M.C. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bishop C.E., Boursot P., Baron B., Bonhomme F., Hatat D., Boursot P., Baron B., Bonhomme F., Hatat D., Baron B., Bonhomme F., Hatat D., Bonhomme F., Hatat D., Hatat D. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature. 1985;315:70–72. doi: 10.1038/315070a0. [DOI] [PubMed] [Google Scholar]
- Bogue M. Mouse Phenome Project: Understanding human biology through mouse genetics and genomics. J. Appl. Physiol. 2003;95:1335–1337. doi: 10.1152/japplphysiol.00562.2003. [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Gibson T.J., Higgins D.G., Thompson J.D., Higgins D.G., Thompson J.D., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P.F., Samuels D.C., Elson J., Turnbull D.M., Samuels D.C., Elson J., Turnbull D.M., Elson J., Turnbull D.M., Turnbull D.M. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: Is there a common mechanism? Lancet. 2002;360:1323–1325. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- Chomyn A., Attardi G., Attardi G. MtDNA mutations in aging and apoptosis. Biochem. Biophys. Res. Commun. 2003;304:519–529. doi: 10.1016/s0006-291x(03)00625-9. [DOI] [PubMed] [Google Scholar]
- Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Kadin J.A., Richardson J.E., Blake J.A., Richardson J.E., Blake J.A., Blake J.A., the members of the Mouse Genome Database Group The Mouse Genome Database (MGD): From genes to mice—A community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goios A., Pereira L., Bogue M., Macaulay V., Amorim A., Pereira L., Bogue M., Macaulay V., Amorim A., Bogue M., Macaulay V., Amorim A., Macaulay V., Amorim A., Amorim A. Mitochondrial DNA sequence of 16 inbred strains. 2006. Mouse Phenome Database, Accession MPD: 202. The Jackson Laboratory, Bar Harbor, ME, USA. [Google Scholar]
- Herrnstadt C., Preston G., Andrews R., Chinnery P., Lightowlers R.N., Turnbull D.M., Kubacka I., Howell N., Preston G., Andrews R., Chinnery P., Lightowlers R.N., Turnbull D.M., Kubacka I., Howell N., Andrews R., Chinnery P., Lightowlers R.N., Turnbull D.M., Kubacka I., Howell N., Chinnery P., Lightowlers R.N., Turnbull D.M., Kubacka I., Howell N., Lightowlers R.N., Turnbull D.M., Kubacka I., Howell N., Turnbull D.M., Kubacka I., Howell N., Kubacka I., Howell N., Howell N. A high frequency of mtDNA polymorphisms in HeLa cell sublines. Mutat. Res. 2002;501:19–28. doi: 10.1016/s0027-5107(01)00304-9. [DOI] [PubMed] [Google Scholar]
- Ho S.Y.W., Larson G., Larson G. Molecular clocks: When times are a-changin’. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ho S.Y.W., Phillips M.J., Cooper A., Drummond A.J., Phillips M.J., Cooper A., Drummond A.J., Cooper A., Drummond A.J., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Howell N., Smejkal C.B., Mackey D.A., Chinnery P.F., Turnbull D.M., Herrnstadt C., Smejkal C.B., Mackey D.A., Chinnery P.F., Turnbull D.M., Herrnstadt C., Mackey D.A., Chinnery P.F., Turnbull D.M., Herrnstadt C., Chinnery P.F., Turnbull D.M., Herrnstadt C., Turnbull D.M., Herrnstadt C., Herrnstadt C. The pedigree rate of sequence divergence in the human mitochondrial genome: There is a difference between phylogenetic and pedigree rates. Am. J. Hum. Genet. 2003;72:659–670. doi: 10.1086/368264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs L.L., Winkler D.A., Murry P.A., Winkler D.A., Murry P.A., Murry P.A. Modern mammal origins: Evolutionary grades in the Early Cretaceous of North America. Proc. Natl. Acad. Sci. 1989;86:4992–4995. doi: 10.1073/pnas.86.13.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J.-J., Tong H., Buffetaut E., Tong H., Buffetaut E., Buffetaut E. The age of Mus–Rattus divergence: Paleontological data compared with the molecular clock. Compt. Rend. Acad. Sci. Paris Ser. 1986;2302:917–922. [Google Scholar]
- Kiebish M.A., Seyfried T.N., Seyfried T.N. Absence of pathogenic mitochondrial DNA mutations in mouse brain tumors. BMC Cancer. 2005;5:102–109. doi: 10.1186/1471-2407-5-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland B., Wang C.R., Yonekawa H., Hermel E., Lindahl K.F., Wang C.R., Yonekawa H., Hermel E., Lindahl K.F., Yonekawa H., Hermel E., Lindahl K.F., Hermel E., Lindahl K.F., Lindahl K.F. Maternally transmitted histocompatibility antigen of mice: A hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- Lundrigan B.L., Jansa S.A., Tucker P.K., Jansa S.A., Tucker P.K., Tucker P.K. Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst. Biol. 2002;51:410–431. doi: 10.1080/10635150290069878. [DOI] [PubMed] [Google Scholar]
- McKenzie M., Trounce I.A., Cassar C.A., Pinkert C.A., Trounce I.A., Cassar C.A., Pinkert C.A., Cassar C.A., Pinkert C.A., Pinkert C.A. Production of homoplasmic xenomitochondrial mice. Proc. Natl. Acad. Sci. 2004;101:1685–1690. doi: 10.1073/pnas.0303184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux J.R., Chevret P., Filippucci M.-G., Macholand M., Chevret P., Filippucci M.-G., Macholand M., Filippucci M.-G., Macholand M., Macholand M. Phylogeny of the genus Apodemus with a special emphasis on the subgenus Sylvaemus using the nuclear IRBP gene and two mitochondrial markers: Cytochrome b and 12S rRNA. Mol. Phylogenet. Evol. 2002;23:123–136. doi: 10.1016/S1055-7903(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., Brandon M., Easley K., Chen E., Brown M.D., Easley K., Chen E., Brown M.D., Chen E., Brown M.D., Brown M.D., et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani J., Chiba T., Tanaka M., Higuchi K., Mori M., Chiba T., Tanaka M., Higuchi K., Mori M., Tanaka M., Higuchi K., Mori M., Higuchi K., Mori M., Mori M. Unique mutations in mitochondrial DNA of senescence-accelerated mouse (SAM) strains. J. Hered. 2001;92:352–355. doi: 10.1093/jhered/92.4.352. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R., Acín-Pérez R., Fernández-Silva P., Movilla N., Pérez-Martos A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., Acín-Pérez R., Fernández-Silva P., Movilla N., Pérez-Martos A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., Fernández-Silva P., Movilla N., Pérez-Martos A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., Movilla N., Pérez-Martos A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., Pérez-Martos A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., de Cordoba S.R., Gallardo M.E., Enríquez J.A., Gallardo M.E., Enríquez J.A., Enríquez J.A. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Pearson H. Mouse sequencing plan aims to boost models. Nature. 2004;432:5. doi: 10.1038/432005a. [DOI] [PubMed] [Google Scholar]
- Rader K. Making mice: Standardizing animals for American biomedical research, 1900–1955. Princeton University Press; Princeton, NJ: 2004. [Google Scholar]
- Sakai T., Kikkawa Y., Miura I., Inoue T., Moriwaki K., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Kikkawa Y., Miura I., Inoue T., Moriwaki K., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Miura I., Inoue T., Moriwaki K., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Inoue T., Moriwaki K., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Moriwaki K., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Shiroishi T., Satta Y., Takahata N., Yonekawa H., Satta Y., Takahata N., Yonekawa H., Takahata N., Yonekawa H., Yonekawa H. Origins of mouse inbred strains deduced from whole-genome scanning by polymorphic microsatellite loci. Mamm. Genome. 2005;16:11–19. doi: 10.1007/s00344-004-3013-9. [DOI] [PubMed] [Google Scholar]
- Sligh J.E., Levy S.E., Waymire K.G., Allard P., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Levy S.E., Waymire K.G., Allard P., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Waymire K.G., Allard P., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Allard P., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C., Heckenlively J.R., MacGregor G.R., Wallace D.C., MacGregor G.R., Wallace D.C., Wallace D.C. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc. Natl. Acad. Sci. 2000;97:14461–14466. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Shimadaa T., Terashimaa M., Tsuchiyab K., Aplin K., Shimadaa T., Terashimaa M., Tsuchiyab K., Aplin K., Terashimaa M., Tsuchiyab K., Aplin K., Tsuchiyab K., Aplin K., Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Takeda T., Hosokawa M., Higuchi K., Hosokawa M., Higuchi K., Higuchi K. Senescence-accelerated mouse (SAM): A novel murine model of senescence. Exp. Gerontol. 1997;32:105–109. doi: 10.1016/s0531-5565(96)00036-8. [DOI] [PubMed] [Google Scholar]
- Wade C.M., Kulbokas E.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Kulbokas E.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J., Lander E.S., Lindblad-Toh K., Daly M.J., Lindblad-Toh K., Daly M.J., Daly M.J. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yonekawa H., Moriwaki K., Gotoh O., Hayashi J.-I., Watanabe J., Miyashita N., Petras M.L., Tagashira Y., Moriwaki K., Gotoh O., Hayashi J.-I., Watanabe J., Miyashita N., Petras M.L., Tagashira Y., Gotoh O., Hayashi J.-I., Watanabe J., Miyashita N., Petras M.L., Tagashira Y., Hayashi J.-I., Watanabe J., Miyashita N., Petras M.L., Tagashira Y., Watanabe J., Miyashita N., Petras M.L., Tagashira Y., Miyashita N., Petras M.L., Tagashira Y., Petras M.L., Tagashira Y., Tagashira Y. Evolutionary relationships among five subspecies of Mus musculus based on restriction enzyme cleavage patterns of mitochondrial DNA. Genetics. 1981;98:801–816. doi: 10.1093/genetics/98.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa H., Moriwaki K., Gotoh O., Miyashita N., Migita S., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Moriwaki K., Gotoh O., Miyashita N., Migita S., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Gotoh O., Miyashita N., Migita S., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Miyashita N., Migita S., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Migita S., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Bonhomme F., Hjorth J.P., Petras M.L., Tagashira Y., Hjorth J.P., Petras M.L., Tagashira Y., Petras M.L., Tagashira Y., Tagashira Y. Origins of laboratory mice deduced from restriction patterns of mitochondrial DNA. Differentiation. 1982;22:222–226. doi: 10.1111/j.1432-0436.1982.tb01255.x. [DOI] [PubMed] [Google Scholar]
- Yoshiki A., Moriwaki K., Moriwaki K. Mouse phenome research: Implications of genetic background. ILAR J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]