Abstract
Rice is an important model species for the Poaceae and other monocotyledonous plants. With the availability of a near-complete, finished, and annotated rice genome, we performed genome level comparisons between rice and all plant species in which large genomic or transcriptomic data sets are available to determine the utility of cross-species sequence for structural and functional annotation of the rice genome. Through comparative analyses with four plant genome sequence data sets and transcript assemblies from 185 plant species, we were able to confirm and improve the structural annotation of the rice genome. Support for 38,109 (89.3%) of the total 42,653 nontransposable element-related genes in the rice genome in the form of a rice expressed sequence tag, full-length cDNA, or plant homolog from our comparative analyses could be found. Although the majority of the putative homologs were obtained from Poaceae species, putative homologs were identified in dicotyledonous angiosperms, gymnosperms, and other plants such as algae, moss, and fern. A set of rice genes (7669) lacking a putative homolog was identified which may be lineage-specific genes that evolved after speciation and have a role in species diversity. Improvements to the current rice gene structural annotation could be identified from our comparative alignments and we were able to identify 487 genes which were mostly likely missed in the current rice genome annotation and another 500 genes for structural annotation review. We were able to demonstrate the utility of cross-species comparative alignments in the identification of noncoding sequences and in confirmation of gene nesting in rice.
The Poaceae (or grass family) is the most economically important family of plants as the majority of food for human diet or feed food are obtained from species within the family including rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), sorghum (Sorghum bicolor), oats (Avena sativa), millet (Eleusine coracana), and rye (Secale cereale) (http://faostat.fao.org). At the genome level, gene content and gene order are well conserved among the Poaceae (Gale and Devos 1998; Goff et al. 2002; Sorrells et al. 2003; The Rice Chromosome 3 Sequencing Consortium 2005), and colinearity at the sequence level (i.e., micro-colinearity) is conserved in spite of gene loss, inversion, duplication, and local genome rearrangements (Bennetzen 2000).
The map-based sequence of the rice genome (O. sativa ssp. japonica var. Nipponbare) was completed in 2005 (International Rice Genome Sequencing Project 2005) and a draft sequence of the indica subspecies (var. 93–11) has also been made available (Yu et al. 2002, 2005). Annotation of the rice genome has been performed by a number of laboratories and consortia (Sakata et al. 2002; Zhao et al. 2004; Ito et al. 2005; Yuan et al. 2005; Ohyanagi et al. 2006). Yuan et al. (2005) reported a structural and functional annotation pipeline in which 43,719 nontransposable element (TE)-related genes were described. This annotation has been updated in which 42,653 non-TE-related genes representing 49,472 gene models have been annotated (Release 4, January 2006, http://rice.tigr.org). Of these, 21,403 have a putative or known function, 6913 were annotated as encoding an expressed protein (transcript support only), and 14,337 annotated as encoding a hypothetical protein. In addition, 13,237 TE-related genes were identified.
As rice is the first finished cereal genome, the rice genome annotation will be used extensively in the annotation of genes in other cereals and grass species. As with other eukaryotic species, annotation of the rice genome was initiated using gene predictions from ab initio gene finders and further improved by using cDNA and expressed sequence tags (ESTs) (Yuan et al. 2005). Currently, more than 30,000 full-length cDNAs (Kikuchi et al. 2003; Xie et al. 2005) and ∼1.2 million ESTs are available for rice. Even with the availability of a large transcript sequence data set, a subset of predicted rice genes still lack transcript data support and are derived solely from ab initio gene predictions. Other evidence such as protein similarity and comparative alignments can be used to either support or amend gene models predicted by the ab initio gene finders and has been demonstrated to be useful in genome-wide analysis in metazoan eukaryotes (Ureta-Vidal et al. 2003).
In this study, we performed large-scale comparative genome analyses with rice using all available major plant sequence data sets. Genome sequence data sets include the finished sequence of the model dicotyledonous (dicot) plant, Arabidopsis thaliana (Arabidopsis Genome Initiative 2000), a draft sequence of poplar (Populus trichocarpa; Tuskan et al. 2006), a woody dicot perennial, as well as gene-rich genomic sequences of two monocotyledonous (monocot) Poaceae species, maize (Palmer et al. 2003; Whitelaw et al. 2003) and sorghum (Bedell et al. 2005), which were generated using the strategies of methylation filtration (Rabinowicz et al. 1999; Rabinowicz and Bennetzen 2006) and/or high C0t selection (Peterson et al. 2002; Yuan et al. 2003). In addition, transcript sequence data in the form of ESTs are available from over 400 plant species (9.8 million ESTs in total, dbEST Release dated on 6/26/2006) which provide an estimation of the transcriptome from a wide range of plant species. To improve the structural annotation of the rice genome and further extend our understanding of sequence conservation among plants and, specifically, the Poaceae, we compared the rice genome and its predicted proteome with the genomes or the genomic sequences of Arabidopsis, maize, sorghum, and poplar along with transcript data from 185 plant species. One goal in our study was to improve the current rice genome annotation while the other goal was to build linkages between rice genome annotations and other plant species, especially other cereal species, to facilitate the annotation of those species.
Results
Support for current rice genome annotation
Support in the form of rice transcripts or putative homologs of the 55,890 total rice genes were identified by searching against sequence data sets from 185 plant species which collectively represents 2670 Mb of sequence. The sequence data included (1) genomic sequences from A. thaliana, P. trichocarpa, Z. mays, and S. bicolor, (2) the Arabidopsis proteome, and (3) 185 plant transcript data sets which are clustered assemblies of ESTs, mRNAs, and full-length cDNAs (Table 1; http://plantta.tigr.org; Childs et al. 2007). The numbers of transcript sequences used in the build of the TIGR Plant Transcript Assemblies (TAs) were variable; of these 185 species, 17 species have more than 100,000 transcript sequences included in the build, which cumulatively represent 72.7% of the total plant transcript sequences (Supplemental Table 1). The Poaceae represent 44% of all of the transcripts in the 185 species TA collection, with rice having more transcript sequences than any other plant species, representing ∼16% of all the plant transcripts (Table 1; Supplemental Fig. 1). Clearly, the numbers of putative rice homologs within the Plant TAs will vary based on both the representation of the transcriptome and the evolutionary distance between rice and each species.
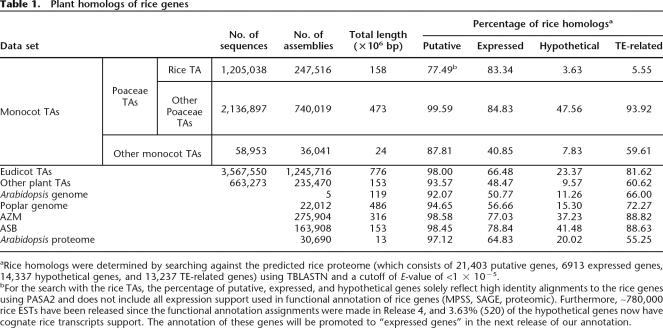
Table 1.
Plant homologs of rice genes
aRice homologs were determined by searching against the predicted rice proteome (which consists of 21,403 putative genes, 6913 expressed genes, 14,337 hypothetical genes, and 13,237 TE-related genes) using TBLASTN and a cutoff of E-value of <1 × 10−5.
bFor the search with the rice TAs, the percentage of putative, expressed, and hypothetical genes solely reflect high identity alignments to the rice genes using PASA2 and does not include all expression support used in functional annotation of rice genes (MPSS, SAGE, proteomic). Furthermore, ∼780,000 rice ESTs have been released since the functional annotation assignments were made in Release 4, and 3.63% (520) of the hypothetical genes now have cognate rice transcripts support. The annotation of these genes will be promoted to “expressed genes” in the next release of our annotation.
In this study, the TA and genomic sequences were placed into 10 groupings based on the type of data source and taxonomic distance relative to rice: (1) rice TA, (2) Other Poaceae TAs (excluding Oryza sativa; 23 species), (3) Other Monocot TAs (excluding Poaceae species; eight species), (4) Eudicotyledons (Eudicot) TAs (121 species), (5) Other Plant TAs (32 species, such as basal angiosperms, algae, mosses, and ferns), (6) Assembled Zea mays (AZMs) genomic sequences, which are assembled methylation filtration and high C0t reads from the pilot maize gene enrichment sequencing project (Whitelaw et al. 2003), (7) Assembled Sorghum bicolor (ASBs) genomic sequences, which are assembled methylation filtration reads from the sorghum gene enrichment sequencing project (Bedell et al. 2005), (8) near-complete, finished Arabidopsis genome sequence, (9) poplar genomic assemblies, and (10) Arabidopsis-predicted proteome. Rice genes supported by cognate rice transcripts were identified using the Program to Assemble Spliced Alignments (PASA2) program (Haas et al. 2003). As ∼780,000 ESTs have been released since functional annotation of Release 4 of our annotation and Massively Parallel Signature Sequencing (MPSS), Serial Analysis of Gene Expression (SAGE), and proteomic data were utilized in functional annotation of Release 4 models (http://rice.tigr.org), there are some inconsistencies between the function assignment of the gene models in Release 4 and the data presented in this study. For example, hypothetical genes should lack transcript support. However, in this study, we identified 520 (3.6%) hypothetical genes with cognate transcripts due to the recent rice EST release (Table 1), which should be promoted in their annotation to “expressed gene”. In this study, only 83.3% of the rice genes annotated with expression support in Release 4 have cognate EST and/or full-length cDNA transcript support, indicating that the remaining 16.7% genes annotated with expression support in Release 4 were obtained through MPSS, SAGE, and peptide evidence data types.
Among the 10 groupings, homologs for rice genes in all gene categories (i.e., known/putative, expressed, hypothetical, and TE-related; Table 1) were most frequently identified within the Other Poaceae TA data sets, which is consistent with previous reports of high sequence identity among the Poaceae (Ware and Stein 2003). By combining the Other Poaceae TAs with the cereal genomic sequence data sets (i.e., AZM and ASB), more putative homologs for the rice loci could be identified than with any other data set combination. Fewer putative homologs for the known/putative rice genes were identified with the Other Monocot TA data sets compared to the Eudicot TA data sets, which was mainly attributable to the relatively low abundance of the sequence data from non-Poaceae monocots (Table 1; Supplemental Fig. 1). Surprisingly, over 50% of hypothetical genes could be supported by sequence data from the other species, primarily Poaceae sequence data (Table 1). Although we did employ a flexible cutoff of the TBLASTN/BLASTP E-value in this study, these data suggest that many of the hypothetical genes encode “real” genes for which cognate transcript evidence in rice is currently lacking.
The prevalence of homologs from diverse clades of the plant kingdom suggests that most of these “core plant genes” may be important housekeeping genes that are not only constitutively expressed and detectable through EST sampling methods but also conserved in function. In contrast, the inability to detect a homolog for 7669 rice genes (including 30 known genes, 895 expressed genes, and 6744 hypothetical genes) in the 2512 Mb of non-rice genomic and transcriptomic sequence available to date suggests the presence of lineage specific genes in rice, which may have evolved after speciation and have a role in species diversity. Alternatively, these, or a subset of these genes, may be artifacts of our annotation methods or encode pseudogenes or transposable elements that we have failed to identify properly.
Distribution of support for rice gene models throughout the plant kingdom
The above analysis suggests that, with the exception of rice itself, Poaceae sequences provide the best support of the rice genes compared to sequence data from the other plant species. However, it is not clear how many of the rice loci are supported solely by Poaceae sequences, non-Poaceae monocots, or other plant species. To identify the breakdown of support, the plant sequence data sets (TAs, genomic assemblies, predicted proteome) were divided into three groups: Poaceae (including 24 Poaceae TAs, AZMs, and ASBs), non-Poaceae monocots (eight non-Poaceae monocot TAs), and all other plant species group (153 plant TAs, genomic sequences of Arabidopsis and poplar, and the Arabidopsis predicted proteome).
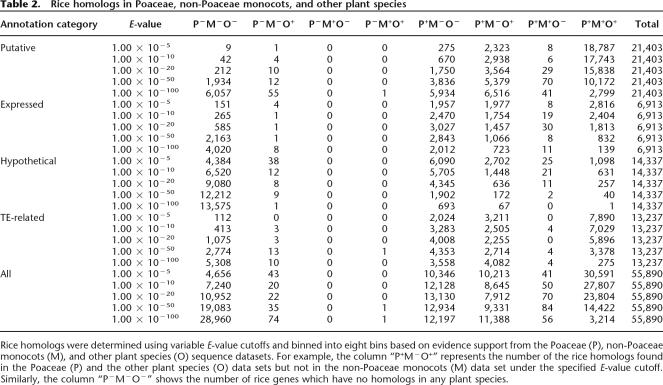
As shown in Table 2, the majority of the rice genes have Poaceae evidence support and only a very small number (i.e., P−M−O+ + P−M+O− + P−M+O+ = 43 + 0 + 0 = 43) of rice genes are supported solely by non-Poaceae sequence data. Noticeably, a mere 4544 (10.6%) of the total 42,653 non-TE-related loci have no evidence support under the significance level (E-value cutoff <1 × 10−5) used in this study, of which 4384 of these unsupported loci are hypothetical genes. Overall, evidence support could be identified for 69.4% (Total − P−M−O− = 14,337 –4384 = 9953; Table 2) of the 14,337 hypothetical genes using an E-value cutoff of <1 × 10−5 and 2116 loci (= 14,337 – 12,221) had distinct support under the more stringent E-value cutoff of <1 × 10−50. All of the hypothetical genes are the result of the prediction of the program FGENESH (Salamov and Solovyev 2000). Preliminary analysis of rice genes using full-length cDNA-supported gene models showed that the accuracy of FGENESH is <82% at the exon level and 45% at the whole-gene level (B. Haas and W. Zhu, unpubl.). While this level of specificity could be improved, the identification of putative homologs of a large percentage of the hypothetical genes in other Poaceae species suggests that the structure of those hypothetical genes could be improved by homologous evidence using similarity-based gene finders (Mathe et al. 2002).
Table 2.
Rice homologs in Poaceae, non-Poaceae monocots, and other plant species
Rice homologs were determined using variable E-value cutoffs and binned into eight bins based on evidence support from the Poaceae (P), non-Poaceae monocots (M), and other plant species (O) sequence datasets. For example, the column “P+M−O+” represents the number of the rice homologs found in the Poaceae (P) and the other plant species (O) data sets but not in the non-Poaceae monocots (M) data set under the specified E-value cutoff. Similarly, the column “P−M−O−” shows the number of rice genes which have no homologs in any plant species.
Frequency of homologs in gene-enriched genomic versus transcript Poaceae sequences
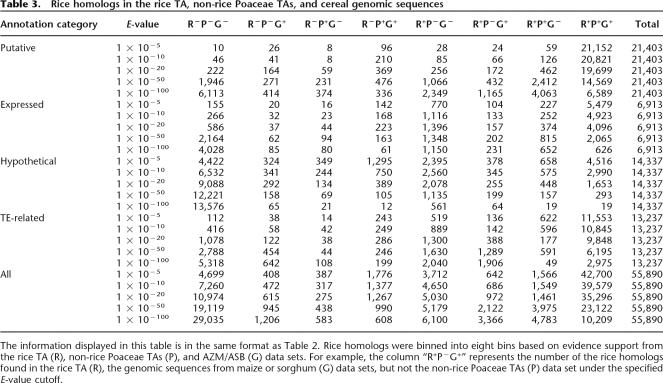
The above analyses indicated that Poaceae sequence data are a valuable resource for annotating the rice genome. The Poaceae data set contains 24 TAs, AZMs, and ASBs. Although it is well known that transcript sequence data are the most important resource in the gene identification, we were interested in ascertaining the contribution of the Poaceae (excluding rice) TAs and maize/sorghum genomic sequences relative to the rice TAs in providing support for genome annotation. Not surprisingly, the rice TA data set yielded the best contribution among the three major Poaceae data resources (Table 3). Interestingly, the non-rice Poaceae TAs had a comparable number of rice homologs as the maize and sorghum genome assemblies, suggesting broad representation of the Poaceae transcriptome in the collective Poaceae TA data set. Indeed, ∼93.8% [(19,699 + 369)/21,403] of the known/putative rice genes have a potential homolog in both the non-rice Poaceae TAs and AZM/ASB sequences at a high significance level (E-value cutoff of <1 × 10−20). AZMs and ASBs have homologs in 92.4% (19,780) and 89.1% (19,070) of the known/putative rice genes, respectively. Over 98% (21,071) coverage would be reached if the significance was lowered to 1 × 10−5, consistent with reports that the gene-rich sequencing strategy provides significant coverage of the maize and sorghum gene space (Palmer et al. 2003; Whitelaw et al. 2003; Bedell et al. 2005). Certainly, the homologs with lower E-value are more likely to offer better support in the gene structure identification. Using the significance E-value of <1 × 10−100, as shown in Table 3, a total of 1224 (i.e., R−P−G+ + R−P+G− + R−P+G+ = 414 + 374 + 336) known/putative rice genes are supported not by a rice TA sequence but by sequences from the other Poaceae species, suggesting that cross-species comparative analysis could provide additional support for this subset of known genes.
Table 3.
Rice homologs in the rice TA, non-rice Poaceae TAs, and cereal genomic sequences
The information displayed in this table is in the same format as Table 2. Rice homologs were binned into eight bins based on evidence support from the rice TA (R), non-rice Poaceae TAs (P), and AZM/ASB (G) data sets. For example, the column “R+P−G+” represents the number of the rice homologs found in the rice TA (R), the genomic sequences from maize or sorghum (G) data sets, but not the non-rice Poaceae TAs (P) data set under the specified E-value cutoff.
Overall, 90,039 (32.6%) of the total 275,904 AZMs had BLASTZ alignments with the rice genome. Of these, 51,403 AZMs have representative alignments that cover 54.5 Mb of the rice genome. Similarly, 65,233 (39.8%) of the total 163,908 ASBs had BLASTZ alignments to the rice genome with representative alignments from 39,885 ASBs spanning 69 Mb of the rice genome. In total, the genic regions of over two thirds of the total rice genes were covered at least partially by the AZM and ASB genomic alignments, including 31,690 (74%) non-TE-related genes.
Noncognate transcripts
Comparative analyses can be applied not only across species but also within the species. In Release 4 of the TIGR rice genome annotation, only the best hit of each rice transcript sequence in the rice genome with ≥95% sequence identity and ≥90% coverage was used in the annotation process to ensure that only the cognate transcript was associated with its respective gene. A large portion of rice genes originated from the large-scale segmental duplication (or polyploidization) that occurred in the rice genome about 70 MYA (Paterson et al. 2004; Wang et al. 2005a), and a recent study showed that 95% of the introns have been conserved among segmentally duplicated rice genes (Lin et al. 2006). These noncognate rice transcripts provide a valuable resource that can be exploited to improve the structural annotation of paralogous genes. Using an E-value cutoff of <1 × 10−5, approximately one-quarter [R+P−G−/(Total − R−P−G−) = 2395/(14,337 – 4422) = 24.2%] of the hypothetical rice genes supported by Poaceae data were supported by rice transcripts only (Table 3). Despite the 3.6% (520) hypothetical rice genes with cognate transcripts from the recent rice EST release, the remaining 20.6% (2395 – 520 = 1875 R+P−G−) rice hypothetical genes can be supported by the transcripts from putative rice paralogs.
Improvement of gene prediction using comparative alignments
The rice genes in Release 4 were identified on the basis of either coding potential using the program FGENESH and/or the spliced alignment of the cognate transcripts by PASA2 (Yuan et al. 2005), and thus those rice genes with limited transcript data or low coding potential were not likely to be identified in the process of the genome annotation. Use of genomic comparisons between rice and other plant genomic sequences can contribute to the identification of newly identified genes in conserved intergenic regions and could be utilized to improve the structure of those rice genes lacking cognate rice transcript data, especially hypothetical genes. The refinement could be achieved in two nonexclusive ways: by generating gene predictions using the similarity-based program like TWINSCAN (Korf et al. 2001), N-SCAN (Gross and Brent 2006), AUGUSTUS+ (Stanke et al. 2006), and EUGENE'HOM (Foissac et al. 2003), or by manual inspection using a genome annotation tool. We utilized cross-species spliced and genomic alignments to improve our structural annotation as well as to identify “unannotated” genes.
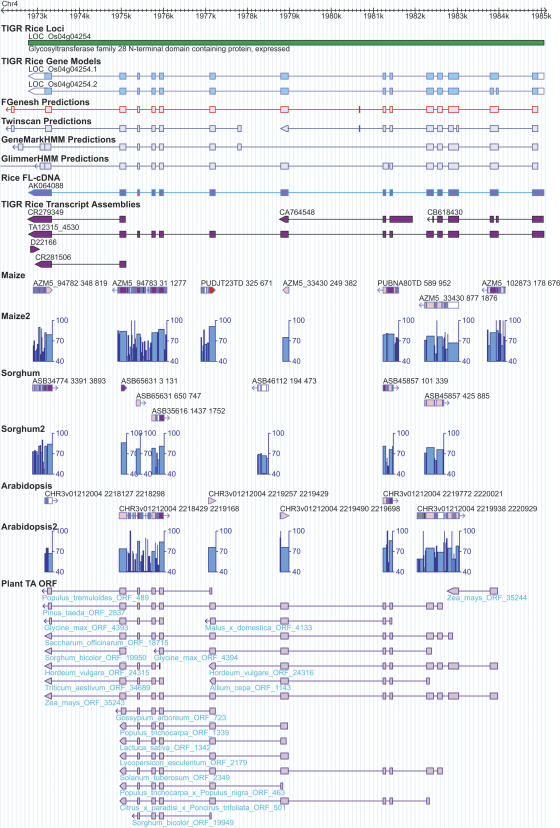
Cross-species spliced alignments can be used to corroborate predicted gene structures (Fig. 1) and amend gene predictions (Brendel et al. 2004). A total of 217,269 filtered cross-species alignments were assembled into 40,935 assemblies using PASA2 (Haas et al. 2003). PASA2 was utilized to compare the assemblies with the gene models, and 25,268 (61.7%) alignments could be successfully incorporated into 15,654 distinct rice genes. Among the 15,654 rice genes supported by these transcript assemblies, 12,548 are known/putative genes, 2460 are expressed genes, 314 are hypothetical genes, and 332 are TE-related genes. While these data strongly support our annotation, many of the assemblies were not completely consistent with the existing gene models.
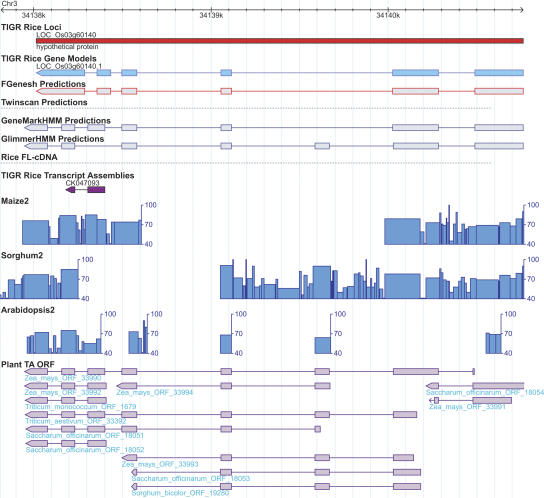
Figure 1.
Example of a rice gene (LOC_Os04g04254) supported by additional evidence derived from the comparative analyses. The green bar represents the TIGR rice locus with the locus name above and the putative function assignment below the track. There are two gene models (i.e., alternative splicing isoforms) predicted for this locus as shown in the track “TIGR Rice Gene Models,” in which exons are represented by rectangles and introns by the intervening horizontal thin lines. The coding regions of the gene model are in light blue, distinguished from the untranslated regions, which are in white. The genes prediction program output (FGENESH, TWINSCAN, GeneMark.hmm, and GlimmerHMM) and the spliced alignments of the rice cognate full-length cDNAs and TAs are displayed in a similar manner as the gene models. Above the transcript alignment tracks are the GenBank accession numbers or TA identifiers to indicate the sequence data source. Below the transcript alignment tracks are the BLASTZ alignments between rice and maize, sorghum, Arabidopsis. Each BLASTZ alignment is composed of one or more gap-free blocks interspersed with gaps (or insertion/deletions). Each block is represented by a rectangle with the color for the average sequence identity within the block: red color (>90% identity), purple (80%–90% identity), pink (70%–80% identity), light gray (60%–70% identity), gray (50%–60% identity), and black (<50% identity). The gaps (not introns) between the flanking blocks are represented by the horizontal thin line. Each BLASTZ alignment feature is also labeled with the sequence identifier, which is followed by two numbers marking the start and end points of the match in the assembly. For example, ASB assembly ASB45857 has two matches with the genic region of the rice gene LOC_Os04g04254 which are separated by a 1-kb intron in the rice genome. The labels “ASB45857 101 339” and “ASB45857 425 885” indicate that the two matched regions have a gap of 85 (= 425 – 339 – 1) bp in the sorghum genome. The tracks “Maize2,” “Sorghum2,” and “Arabidopsis2” displayed the exact same BLASTZ alignment data but in alternative manners: The sequence identity is represented by the height of the column and the block size is represented by the column width. The cross species spliced alignments are displayed in the last track “Plant TA ORF” (for details, see Methods). As shown in the figure, the exonic regions have higher sequence identity than the neighboring intronic regions in the BLASTZ alignments. The gene structure of the gene LOC_Os4g04254 is well supported by rice full-length cDNA and rice TAs as well as the TAs from other plant species such as apple (Malus x domestica), barley (Hordeum vulgare), loblolly pine (Pinus taeda), lettuce (Lactuca sativa), maize (Zea mays), onion (Allium cepa), soybean (Glycine max), and sugarcane (Saccharum officinarum).
As exon-intron boundaries defined by spliced alignments of heterologous transcripts are not as reliable as those by cognate transcripts, more stringent criteria were employed to refine our analysis. First, a putative exon had to be supported by at least three alignments. As a result, 75,111 putative exons were predicted. Second, we compared these cross-species putative exons with existing exons within our annotation. To avoid confounding our results due to alternative splicing and UTR exons, we focused on “novel” exons in genic regions, which did not overlap with existing exons in Release 4, i.e., “novel” exons in annotated intronic regions and in which the annotated intron is not supported by any transcript (rice or heterologous). A total of 500 genes (including 395 known genes, 66 expressed genes, and 39 hypothetical genes) with potential new exons were identified through cross-species alignments in which the exon had to be supported by at least three independent alignments. For 477 of the 500 genes with new exons, cross-species alignments were from more than one species. Manual inspection showed that most of these genes have incorrect gene structures (see Figs. 2, 3; Supplemental Figs. 2, 3), suggesting that known genes and expressed genes can be improved through comparative analyses. In addition to refinement of gene structures, comparison using PASA2 identified 1854 assemblies, which supported unannotated or “missed” genes. Using a stringent set of criteria (≥300 bp in length and ≥3 exons), we conservatively identified 388 assemblies located in 255 distinct intergenic regions as candidate unannotated genes.
Figure 2.
Gene structure of the expressed gene LOC_Os12g10200 corrected by the cross-species spliced alignments. Symbols are as in Fig. 1. Several rice transcripts cover a portion of the expressed gene LOC_Os12g10200 which were stitched into the FGENESEH prediction to create the existing gene model by the program PASA2. The cross-species spliced alignments suggest that two exons have been missed.
Figure 3.
Gene structure of the hypothetical gene LOC_Os03g60140 corrected by the cross-species spliced alignments. Symbols are as in Fig. 1. It appears that there is an exon missed in the second intron of LOC_Os03g60140 and the correct gene structure is shown via cross-species alignments but not correct by any gene finders.
Using BLASTZ alignments of the AZMs and ASBs to the rice genome, many alignments between rice genes and putative homologs were identified that spanned multiple exons. Not surprisingly, the identity of the alignment in intronic regions was significantly lower than the flanking exonic regions, appearing as a banded pattern in the genome browser display (Fig. 1). For example, maize, sorghum, and even Arabidopsis genomic comparisons indicated a potential gene upstream of LOC_Os04g45820 that was not predicted by FGENESH and in which only short cognate rice EST sequences and two cross-species spliced alignments are available (Supplemental Fig. 4). By combining the partial gene structure provided by rice EST spliced alignments, exon patterns in the genomic alignments with AZM5_17958, AZM_5_84956, ASB44489, ASB71162 and ASB45539, and cross-species spliced alignments from wheat and maize, we can construct a gene model consistent with the gene prediction from TWINSCAN (Korf et al. 2001). In addition, the last exon of the new gene model could be further extended in the 3′-UTR exon region as indicated by the rice transcript assembly TA26377_4530.
Genomic comparisons can also indicate the existence of novel genes. Using the AZMs and ASBs, numerous BLASTZ alignments were located in “intergenic” regions, which may lead to the identification of the unannotated genes. Each continuous intergenic region was regarded as one unit in the analysis to simplify the computation (which may contain more than one gene). In total, there were 1145 and 830 intergenic regions over 1000 bp length containing alignments with AZMs and ASBs, respectively. Overall, 1614 distinct intergenic regions were covered and 361 of them were covered by matches from both an AZM and an ASB sequence. The conserved regions were then searched against the TIGR Oryza Repeat and the UniProt databases, resulting in 493 and 339 non-TE-related conserved intergenic sequences identified from maize and sorghum, respectively. Even when the significance was increased to an E-value cutoff of <1 × 10−50, there were still 291 and 175 potentially new genes identified from maize and sorghum, respectively, which could be merged into 324 distinct intergenic regions. Further analyses showed that many of those regions encode genes not contained in the current rice genome annotation (data not shown). As our filtering criteria were stringent in that they required similarity to annotated proteins, other conserved regions may also encode genes which have not been previously identified. Indeed, by removing the filter of UniProt similarity yet retaining the repetitive sequence filter, we identified 800 additional candidate new genes. Some conserved regions may contain multiple genes (Supplemental Fig. 5), while others may contain coding regions of the neighboring genes missed in the annotation process and not new genes. Nevertheless, these conserved “intergenic regions” can be used to improve the current rice genome annotation.
Conserved noncoding regions
The analyses described above concentrated on the identification of protein coding regions; however, conservation is not restricted within protein coding regions but also exists in noncoding regions. In addition to the regular protein coding genes, there are numerous non-protein coding RNA (ncRNA) molecules encoded in the genome that have been found in eukaryotes, eubacteria, archaebacteria, and viruses (Eddy 2001; Dennis and Omer 2005; Liu et al. 2005). ncRNAs can be classified into different groups such as transfer RNA (tRNA), small nucleolar RNAs (snoRNA), ribosomal RNAs (rRNA), and microRNAs (miRNA). miRNAs are a recently identified type of ncRNA, a short (∼21 nucleotides) single-stranded RNA excised from a long self-complementary precursor (Bartel 2004). A majority of miRNAs are conserved between Arabidopsis and rice (Reinhart et al. 2002; Sunkar and Zhu 2004; Sunkar et al. 2005; Jones-Rhoades et al. 2006; Zhang et al. 2006) and, in this study, we examined whether conserved regions identified among the cereal genomes could be ascribed to ncRNA genes. We downloaded all 153 rice miRNAs from the miRBase Sequence Database (Release 8.1; ftp://ftp.sanger.ac.uk/pub/mirbase/sequences/CURRENT/genomes/osa.gff). Ninety-eight of the 153 miRNAs were found in the alignments between rice and the AZMs while 85 were detected in alignments between rice and the ASBs. Of these, 73 were commonly shared among rice, maize, and sorghum. Furthermore, 13 out of the 153 miRNAs could be found in the alignments between Arabidopsis and rice, and 12 were shared with either maize or sorghum. Intriguingly, nine miRNAs were conserved between the three cereal genomes and the Arabidopsis genome.
Although our study utilized simple genomic alignments and did not employ algorithms dedicated to finding miRNAs (Adai et al. 2005; Wang et al. 2005b; Berezikov et al. 2006), we were able to demonstrate that miRNAs loci are more conserved compared to their neighboring regions (Fig. 4A). Indeed, sequence conservation was observed in the miRNA target sites between rice, sorghum, and Arabidopsis. Six target sites of the miRNA family miR399 were found in the 5′-UTR region of LOC_Os05g48390 and four target sites were found in the 5′-UTR region of the Arabidopsis orthologous gene At2g33770 (Supplemental Fig. 6; see also Fig. 4 in Sunkar and Zhu 2004). In addition to the conservation examined in the miRNA genes and miRNA target sites, conservation was also observed for other ncRNAs such as tRNAs and snoRNAs (Fig. 4B,C).
Figure 4.
Conserved noncoding RNAs: (A) miRNA, (B) snoRNA, and (C) tRNA. (A) The precursor miRNA transcript osa-miR394 is represented by the red bar with the arrow for the orientation and the mature miRNA sequence is highlighted in yellow color which is significantly more conserved in maize, sorghum, and Arabidopsis than the neighboring intergenic or intronic regions. The snoRNA (B) and tRNA (C) features are represented by arrowed bars in orange and blue, respectively.
Discussion
Our analyses show that comparative analyses are extremely useful in the annotation of the rice genome even when more than one million rice transcript sequences are available. Furthermore, we show that the completed rice genome sequence and its annotation provide a valuable data resource for genomic research in other grass species and will certainly facilitate the ongoing maize genome annotation or play an even more important role for those cereal species with only limited sequence data such as oat and rye.
Through our comparative analyses, we were able to identify 255 and 324 unannotated candidate genes which were missed in Release 4, by cross-species spliced alignments and genomic comparison, respectively, of which, 92 were found by both methods. In total, 487 distinct candidates were identified. Further analysis showed that, although there are FGENESH predictions in 350 (72%) of these conserved “intergenic regions”, in most cases, the FGENESH algorithm predicted a single, long gene model that spanned two valid neighboring genes with an intron in a relatively short intergenic region, i.e., a merged model. As full-length cDNAs are available to support one gene in the merged FGENESH model, the long FGENESH prediction is truncated by the PASA2 program which heavily weights full-length cDNA evidence over ab initio gene finder output. Consequently, the other exons within the merged FGENESH model that lack cDNA support are deleted and not included in the final model or gene set. Of the remaining 137 unannotated gene candidates, 43 (31%) likely originate from organellar insertions (data not shown; Supplemental Fig. 5). This analysis suggests that a modified update strategy for the PASA2 program to capture the deprecated portion of merged FGENESH models, coupled with integration of an organellar gene finder into our annotation pipeline, should undercover a majority (80.7%) of these two classes of missed genes.
The BLASTZ alignments between rice and maize or sorghum were able to span short introns and clear, distinct alignments were apparent; however, these alignments might be split by long introns. Clearly, these genomic comparisons are able to reveal gene structures, although it may be still difficult for a curator to determine the exact exon-intron boundaries without additional information. To address this problem, spliced alignments from paralogous and heterologous transcripts could be employed to identify the exact exon-intron boundaries. It was shown that 25,258 (61.7%) of the cross-species spliced alignment assemblies can be incorporated into the genes annotated in Release 4. Many assemblies may reveal the right gene structure (Figs. 2, 3; Supplemental Figs. 2, 3); however, most of them are problematic due to low sequence similarity or gene structure alternation subsequent to speciation. Therefore, additional filters are needed to improve the quality of the spliced alignments. For example, establishing a requirement that each exon-intron boundary in cross-species spliced alignment assemblies be supported by at least three or more alignments may permit more automated incorporation of cross-species alignments data into an annotation pipeline.
We also show that genomic comparisons can shed light on the evolution of gene structure and organization. Some rice genes are intervened by a short intergenic region and synteny of not only gene order but also intergenic regions, which can be seen with rice, maize, and sorghum (data not shown). However, it is unclear whether the conservation of short intergenic regions has a biological function role. Alternative splicing is a common feature in plants (Wang and Brendel 2006), and conservation of alternative splicing isoforms across species may indicate that some of the alternative splicing events have biological significance and therefore are preserved after species divergence. For example, the rice gene LOC_Os07g43950 has a dominant exon-skipping isoform and the skipped exon was conserved in both maize and sorghum. Intriguingly, cross-species alignments indicate that the same alternative splicing event may occur in maize and sugarcane (Supplemental Fig. 7), consistent with the report that 25% of human alternatively skipped exons are alternatively spliced in mouse (Sorek et al. 2004).
Comparative analyses can also be applied to the study of the transposable elements. Some mutator-like transposable elements (MULEs), for example, can capture fragments from host gene and are referred to as Pack-MULEs (Jiang et al. 2004). Recent studies reported that there are several thousand Pack-MULEs in the rice genome (Jiang et al. 2004; Juretic et al. 2005). There is an interesting example of gene nesting which is likely caused by a MULE. Cross-species alignments confirmed that the gene structure supported by the full-length cDNA AK059758 is likely to be accurate, which contains an intron ∼5 kb in length (Supplemental Fig. 8). Rice EST evidence clearly indicates that there is a gene encoded within the long intron, suggestive of gene nesting. The alignments of the ASB assembly ASB37 indicate that the rice gene encoded by the AK059758 is conserved in sorghum while the length of the corresponding intron is only 100 bp in sorghum (Supplemental Fig. 8). Additional information from rice repetitive sequences and trans-duplicated MULEs (Juretic et al. 2005) suggest that this particular case of the nested gene was likely to be mediated by the mutator-like elements.
In this study, we have shown the value of comparative alignments in improving structural and functional annotation of the rice genome, which can be attributed in large part to the deep representation of genomic and transcriptomic sequence for the Poaceae. Clearly, the depth of sequence data is not evenly distributed among taxa in the plant kingdom, and increased efforts in sequencing non-Poaceae monocots may shed light not only on the evolution of the Poaceae genome but also on the divergence of monocots from eudicots.
Methods
TIGR rice genome annotation
Release 4.0 of the TIGR rice genome annotation (available at http://rice.tigr.org/; Yuan et al. 2005) was utilized in this study in which genes were identified using the ab initio gene finder FGENESH and amended based on rice transcript evidence (full-length cDNAs and ESTs) with PASA2 (Haas et al. 2003). Transposable element-related genes in Release 4 were identified as described in Yuan et al. (2005). Putative, known, and hypothetical genes were annotated as described previously (Yuan et al. 2005). Genes were annotated as encoding expressed proteins if the gene lacked protein support but had expression support from MPSS, SAGE, EST/full-length cDNA, or proteomic data sets.
Other plant genomes
Maize genomic assemblies (Release 5.0), derived from methylation filtration and high C0t reads and termed AZMs were downloaded from TIGR (ftp://ftp.tigr.org/pub/data/MAIZE/AZMs/release_5.0; Chan et al. 2006). There are 275,904 assemblies in the AZM data set (Table 1), with lengths ranging from 65 bp to 16,340 bp. Over 90% of AZM assemblies are shorter than 2200 bp. The sorghum genomic sequences derived from methylation filtration reads (Bedell et al. 2005) were downloaded from GenBank and assembled into ASBs in a similar way as the maize assemblies at TIGR (available at ftp://ftp.tigr.org/pub/data/MAIZE/Sorghum_assembly/ASB.gz). The length distribution of ASBs is similar to that of AZMs. The Populus trichocarpa assembled scaffolds (Release 1.0) were downloaded from the Joint Genome Institute (JGI, ftp://ftp.jgi-psf.org/pub/JGI_data/Poplar/assembly/v1.0/poplar.masked.fasta.gz). The Arabidopsis pseudomolecules and the genome annotation (Release 6) were downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair).
Plant transcript assemblies
Transcript assemblies (TAs; Release 1.0; 185 total) were downloaded from the Web site of the TIGR plant transcript assembly (ftp://ftp.tigr.org/pub/data/plantta/update_08152005), where transcript assemblies were constructed for all plant species with >1000 ESTs in NCBI dbEST (as of August 15, 2005). The rice proteome was searched against the non-rice plant genome sequence data sets using TBLASTN.
Cross-species spliced alignments
To annotate rice gene structures using the Plant TA sequences, all Plant TAs (except rice) were aligned to the TIGR Release 4 pseudomolecules using the program GeneSeqer (Usuka et al. 2000; Brendel et al. 2004). In order to improve the efficiency, the source code of the program GeneSeqer was modified and a new command-line option for the minimum coverage was inserted so that the optimal spliced alignment would be generated only when the high-scoring segment pairs span a specified length coverage of the transcript sequence (Brendel et al. 2004). In addition to the default GeneSeqer setting, we empirically set the minimum coverage (i.e., the new command option) at 40%. Due to the low sequence similarity between divergent species, the exon-intron boundaries defined by those cross-species spliced alignments are likely to be partially, if not completely, erroneous. Further complicating the interpretation is the possibility that there may have been an alteration of the gene structure after speciation and/or gene duplication. Therefore, three stringent criteria were applied to remove regions of poor quality from spliced alignments: (1) The matched rice genome segments had to contain a long, open reading frame (≥150 bp), (2) the open reading frame had to contain at least one intron, and (3) all intron boundaries had to contain the canonical splices sites GT/AG. The qualified regions were further assembled using the PASA2 program (Haas et al. 2003) and compared to the current rice genome annotation.
Genomic comparisons
The TIGR Release 4 pseudomolecules were aligned with the genomic assemblies of maize, sorghum, Arabidopsis, and poplar using the program BLASTZ (Schwartz et al. 2003). The BLASTZ options “H = 2200 C = 2” were employed for maize and sorghum, whereas slightly different options “H = 2200 C = 0” were used for Arabidopsis and poplar. A simple algorithm was applied to identify “representative alignments” for each species. For each species, by sorting the BLASTZ alignment scores in descending order, if the rice genomic region lacked any BLASTZ alignments, the alignment was selected as the “representative alignment” and no further alignments were allowed in this region. If an alignment was already “assigned” to this genomic region, the alignment was skipped. Therefore, each selected alignment represents the best alignment for a specified rice genomic region with no overlap permitted among the representative alignments from the same species. Those representative alignments were displayed as feature tracks in the TIGR Rice Genome Browser (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/; also see Fig. 1).
Unannotated genes
To identify “intergenic” regions that might represent candidate unannotated genes, a lower cutoff for the match length between cross-species alignments was empirically set to 1 kb and the putative conserved intergenic regions were further searched using the BLAST package against the TIGR Oryza Repeat Database (Ouyang and Buell 2004) and the UniProt database (http://www.ebi.uniprot.org/index.shtml). The conserved regions lacking a significant match in the TIGR Oryza Repeat Database but having a significant match with an entry in the UniProt database were selected as candidates for newly identified genes. We also searched for unannotated genes using cross-species spliced alignments. We conservatively selected each spliced alignment in the intergenic regions with at least 300 bp length and three exons as an unannotated gene candidate.
Data availability
All alignments are viewable on the TIGR Rice Genome Browser (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/) through selection of the appropriate tracks. Additional data sets are made available through supplemental files associated with the on-line version of this manuscript. Supplemental File 1 contains a list of potential new genes identified in intergenic regions using comparative alignments. Supplemental File 2 contains a list of potential new exons within gene models identified through cross-species spliced alignments. Supplemental File 3 lists the loci that are supported or unsupported by our comparative analysis. These are binned into putative/known, expressed, hypothetical, and transposable-element related. They are further separated into two sets: those with rice TA support and those without rice TA support. We have provided a set of files of interest for download at ftp://ftp.tigr.org/pub/data/rice/GENOME_2006_058818/, including a FASTA formatted file of the CDS and proteins of the new exons added to genes.
Acknowledgments
We thank members of the rice annotation team at TIGR for critical comments on the manuscript, and B. Haas for technical assistance on the configuration of the PASA2 pipeline. This work was supported by a National Science Foundation Plant Genome Research Program grant to C.R.B. (DBI-0321538).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5881807
References
- Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Manocha V., Vance V., Sundaresan V., Vance V., Sundaresan V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bedell J.A., Budiman M.A., Nunberg A., Citek R.W., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Budiman M.A., Nunberg A., Citek R.W., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Nunberg A., Citek R.W., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Citek R.W., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Jones J., Flick E., Rholfing T., Fries J., Bradford K., Flick E., Rholfing T., Fries J., Bradford K., Rholfing T., Fries J., Bradford K., Fries J., Bradford K., Bradford K., et al. Sorghum genome sequencing by methylation filtration. PLoS Biol. 2005;3:e13. doi: 10.1371/journal.pbio.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. Approaches to microRNA discovery. Nat. Genet. 2006;38 (Suppl. 1):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- Brendel V., Xing L., Zhu W., Xing L., Zhu W., Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- Chan A.P., Pertea G., Cheung F., Lee D., Zheng L., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Pertea G., Cheung F., Lee D., Zheng L., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Cheung F., Lee D., Zheng L., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Lee D., Zheng L., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Zheng L., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Whitelaw C., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., Pontaroli A.C., SanMiguel P., Yuan Y., Bennetzen J., SanMiguel P., Yuan Y., Bennetzen J., Yuan Y., Bennetzen J., Bennetzen J., et al. The TIGR Maize Database. Nucleic Acids Res. 2006;34:D771–D776. doi: 10.1093/nar/gkj072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.L., Hamilton J., Zhu W., Ly E., Cheung F., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Hamilton J., Zhu W., Ly E., Cheung F., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Zhu W., Ly E., Cheung F., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Ly E., Cheung F., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Cheung F., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Hank W., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P., Town C.D., Buell C.R., Chan A.P., Buell C.R., Chan A.P., Chan A.P. The TIGR Plant Transcript Assemblies Database. Nucleic Acids Res. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.P., Omer A., Omer A. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 2005;8:685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Foissac S., Bardou P., Moisan A., Cros M.J., Schiex T., Bardou P., Moisan A., Cros M.J., Schiex T., Moisan A., Cros M.J., Schiex T., Cros M.J., Schiex T., Schiex T. EUGENE'HOM: A generic similarity-based gene finder using multiple homologous sequences. Nucleic Acids Res. 2003;31:3742–3745. doi: 10.1093/nar/gkg586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M.D., Devos K.M., Devos K.M. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Glazebrook J., Sessions A., Oeller P., Varma H., Sessions A., Oeller P., Varma H., Oeller P., Varma H., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gross S.S., Brent M.R., Brent M.R. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Mount S.M., Wortman J.R., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Wortman J.R., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Smith R.K., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Maiti R., Ronning C.M., Rusch D.B., Town C.D., Ronning C.M., Rusch D.B., Town C.D., Rusch D.B., Town C.D., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ito Y., Arikawa K., Antonio B.A., Ohta I., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Arikawa K., Antonio B.A., Ohta I., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Antonio B.A., Ohta I., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Ohta I., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., Shimano A., Masukawa M., Shibata M., Yamamoto M., Masukawa M., Shibata M., Yamamoto M., Shibata M., Yamamoto M., Yamamoto M., et al. Rice Annotation Database (RAD): A contig-oriented database for map-based rice genomics. Nucleic Acids Res. 2005;33:D651–D655. doi: 10.1093/nar/gki083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R., Bao Z., Zhang X., Eddy S.R., Wessler S.R., Zhang X., Eddy S.R., Wessler S.R., Eddy S.R., Wessler S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B., Bartel D.P., Bartel B., Bartel B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Juretic N., Hoen D.R., Huynh M.L., Harrison P.M., Bureau T.E., Hoen D.R., Huynh M.L., Harrison P.M., Bureau T.E., Huynh M.L., Harrison P.M., Bureau T.E., Harrison P.M., Bureau T.E., Bureau T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Yazaki J., Ishikawa M., Yamada H., Ooka H., Ishikawa M., Yamada H., Ooka H., Yamada H., Ooka H., Ooka H., et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M.R., Flicek P., Duan D., Brent M.R., Duan D., Brent M.R., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17 (Suppl. 1):S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Lin H., Zhu W., Silva J.C., Gu X., Buell C.R., Zhu W., Silva J.C., Gu X., Buell C.R., Silva J.C., Gu X., Buell C.R., Gu X., Buell C.R., Buell C.R. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Zhang Y., Bu D., Zhao Y., Chen R., Bu D., Zhao Y., Chen R., Zhao Y., Chen R., Chen R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe C., Sagot M.F., Schiex T., Rouze P., Sagot M.F., Schiex T., Rouze P., Schiex T., Rouze P., Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyanagi H., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Antonio B.A., Nagamura Y., Imanishi T., Nagamura Y., Imanishi T., Imanishi T., et al. The Rice Annotation Project Database (RAP-DB): Hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., Buell C.R., Buell C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer L.E., Rabinowicz P.D., O'Shaughnessy A.L., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., Rabinowicz P.D., O'Shaughnessy A.L., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., O'Shaughnessy A.L., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R., de la Bastide M., Martienssen R.A., McCombie W.R., Martienssen R.A., McCombie W.R., McCombie W.R. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A., Bowers J.E., Chapman B.A., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.G., Schulze S.R., Sciara E.B., Lee S.A., Bowers J.E., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Schulze S.R., Sciara E.B., Lee S.A., Bowers J.E., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Sciara E.B., Lee S.A., Bowers J.E., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Lee S.A., Bowers J.E., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Bowers J.E., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Nagel A., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Jiang N., Tibbitts D.C., Wessler S.R., Paterson A.H., Tibbitts D.C., Wessler S.R., Paterson A.H., Wessler S.R., Paterson A.H., Paterson A.H. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 2002;12:795–807. doi: 10.1101/gr.226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz P.D., Bennetzen J.L., Bennetzen J.L. The maize genome as a model for efficient sequence analysis of large plant genomes. Curr. Opin. Plant Biol. 2006;9:149–156. doi: 10.1016/j.pbi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Rabinowicz P.D., Schutz K., Dedhia N., Yordan C., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A., Schutz K., Dedhia N., Yordan C., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A., Dedhia N., Yordan C., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A., Yordan C., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A., Stein L., McCombie W.R., Martienssen R.A., McCombie W.R., Martienssen R.A., Martienssen R.A. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Rhoades M.W., Bartel B., Bartel D.P., Bartel B., Bartel D.P., Bartel D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Rice Chromosome 3 Sequencing Consortium Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 2005;15:1284–1291. doi: 10.1101/gr.3869505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K., Nagamura Y., Numa H., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Nagamura Y., Numa H., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Numa H., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Shimizu Y., Horiuchi I., Matsumoto T., Horiuchi I., Matsumoto T., Matsumoto T., et al. RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 2002;30:98–102. doi: 10.1093/nar/30.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A.A., Solovyev V.V., Solovyev V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Shamir R., Ast G., Shamir R., Ast G., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Sorrells M.E., La Rota M., Bermudez-Kandianis C.E., Greene R.A., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., La Rota M., Bermudez-Kandianis C.E., Greene R.A., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Bermudez-Kandianis C.E., Greene R.A., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Greene R.A., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Miftahudin Mahmoud A., Ma X., Gustafson P.J., Ma X., Gustafson P.J., Gustafson P.J., et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Schoffmann O., Morgenstern B., Waack S., Schoffmann O., Morgenstern B., Waack S., Morgenstern B., Waack S., Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.K., Girke T., Jain P.K., Zhu J.K., Jain P.K., Zhu J.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Putnam N., Ralph S., Rombauts S., Salamov A., Ralph S., Rombauts S., Salamov A., Rombauts S., Salamov A., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A., Ettwiller L., Birney E., Ettwiller L., Birney E., Birney E. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Usuka J., Zhu W., Brendel V., Zhu W., Brendel V., Brendel V. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics. 2000;16:203–211. doi: 10.1093/bioinformatics/16.3.203. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V., Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi X., Hao B., Ge S., Luo J., Shi X., Hao B., Ge S., Luo J., Hao B., Ge S., Luo J., Ge S., Luo J., Luo J. Duplication and DNA segmental loss in the rice genome: Implications for diploidization. New Phytol. 2005a;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang J., Li F., Gu J., He T., Zhang X., Li Y., Zhang J., Li F., Gu J., He T., Zhang X., Li Y., Li F., Gu J., He T., Zhang X., Li Y., Gu J., He T., Zhang X., Li Y., He T., Zhang X., Li Y., Zhang X., Li Y., Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005b;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Ware D., Stein L., Stein L. Comparison of genes among cereals. Curr. Opin. Plant Biol. 2003;6:121–127. doi: 10.1016/s1369-5266(03)00012-8. [DOI] [PubMed] [Google Scholar]
- Whitelaw C.A., Barbazuk W.B., Pertea G., Chan A.P., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Barbazuk W.B., Pertea G., Chan A.P., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Pertea G., Chan A.P., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Chan A.P., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., van Heeringen S., Karamycheva S., Bennetzen J.L., Karamycheva S., Bennetzen J.L., Bennetzen J.L., et al. Enrichment of gene-coding sequences in maize by genome filtration. Science. 2003;302:2118–2120. doi: 10.1126/science.1090047. [DOI] [PubMed] [Google Scholar]
- Xie K., Zhang J., Xiang Y., Feng Q., Han B., Chu Z., Wang S., Zhang Q., Xiong L., Zhang J., Xiang Y., Feng Q., Han B., Chu Z., Wang S., Zhang Q., Xiong L., Xiang Y., Feng Q., Han B., Chu Z., Wang S., Zhang Q., Xiong L., Feng Q., Han B., Chu Z., Wang S., Zhang Q., Xiong L., Han B., Chu Z., Wang S., Zhang Q., Xiong L., Chu Z., Wang S., Zhang Q., Xiong L., Wang S., Zhang Q., Xiong L., Zhang Q., Xiong L., Xiong L. Isolation and annotation of 10828 putative full length cDNAs from indica rice. Sci. China C Life Sci. 2005;48:445–451. doi: 10.1360/062004-90. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Deng Y., Dai L., Zhou Y., Zhang X., Dai L., Zhou Y., Zhang X., Zhou Y., Zhang X., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Yu J., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Zhou J., Ni P., Dong W., Hu S., Zeng C., Ni P., Dong W., Hu S., Zeng C., Dong W., Hu S., Zeng C., Hu S., Zeng C., Zeng C., et al. The Genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., SanMiguel P.J., Bennetzen J.L., SanMiguel P.J., Bennetzen J.L., Bennetzen J.L. High-Cot sequence analysis of the maize genome. Plant J. 2003;34:249–255. doi: 10.1046/j.1365-313x.2003.01716.x. [DOI] [PubMed] [Google Scholar]
- Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Hamilton J., Haas B., Sultana R., Cheung F., Haas B., Sultana R., Cheung F., Sultana R., Cheung F., Cheung F., et al. The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cannon C.H., Cobb G.P., Anderson T.A., Pan X., Cannon C.H., Cobb G.P., Anderson T.A., Cannon C.H., Cobb G.P., Anderson T.A., Cobb G.P., Anderson T.A., Anderson T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang J., He X., Huang X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., Wang J., He X., Huang X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., He X., Huang X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., Huang X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., Dai M., Wei S., Fu J., Chen Y., Ren X., Wei S., Fu J., Chen Y., Ren X., Fu J., Chen Y., Ren X., Chen Y., Ren X., Ren X., et al. BGI-RIS: An integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All alignments are viewable on the TIGR Rice Genome Browser (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/) through selection of the appropriate tracks. Additional data sets are made available through supplemental files associated with the on-line version of this manuscript. Supplemental File 1 contains a list of potential new genes identified in intergenic regions using comparative alignments. Supplemental File 2 contains a list of potential new exons within gene models identified through cross-species spliced alignments. Supplemental File 3 lists the loci that are supported or unsupported by our comparative analysis. These are binned into putative/known, expressed, hypothetical, and transposable-element related. They are further separated into two sets: those with rice TA support and those without rice TA support. We have provided a set of files of interest for download at ftp://ftp.tigr.org/pub/data/rice/GENOME_2006_058818/, including a FASTA formatted file of the CDS and proteins of the new exons added to genes.