Abstract
Eimeria tenella is an intracellular protozoan parasite that infects the intestinal tracts of domestic fowl and causes coccidiosis, a serious and sometimes lethal enteritis. Eimeria falls in the same phylum (Apicomplexa) as several human and animal parasites such as Cryptosporidium, Toxoplasma, and the malaria parasite, Plasmodium. Here we report the sequencing and analysis of the first chromosome of E. tenella, a chromosome believed to carry loci associated with drug resistance and known to differ between virulent and attenuated strains of the parasite. The chromosome—which appears to be representative of the genome—is gene-dense and rich in simple-sequence repeats, many of which appear to give rise to repetitive amino acid tracts in the predicted proteins. Most striking is the segmentation of the chromosome into repeat-rich regions peppered with transposon-like elements and telomere-like repeats, alternating with repeat-free regions. Predicted genes differ in character between the two types of segment, and the repeat-rich regions appear to be associated with strain-to-strain variation.
Eimeria tenella is an obligate, intracellular protozoan parasite that infects epithelial cells of the intestinal tract (caeca) of the domestic fowl (Gallus gallus). Severe infections are common and give rise to coccidiosis, an enteritis that impedes growth, presents a high morbidity, and may increase the mortality rate of the affected flocks. Eimeria species are ubiquitous, and the poultry industry relies extensively on prophylactic medication with anticoccidial drugs, or on vaccination, to control infection in the 30 billion chickens reared annually worldwide. The combined cost of control and of losses caused by coccidiosis is estimated at £2 billion annually (Shirley et al. 2004). New control measures are urgently required because of the frequent emergence of drug-resistant strains, changing attitudes to in-feed medication, and the difficulties of producing safe, cost-effective, live-attenuated vaccines. The development of new control agents is likely to be based on a detailed understanding of the biology and genomics of Eimeria.
Eimeria spp. are also of wider scientific interest, falling as they do in the same phylum (Apicomplexa) as the malarial parasites (Plasmodium spp.), the zoonotic parasites Toxoplasma gondii and Cryptosporidium spp., and the cattle parasites Theileria spp. Comparative analyses of these latter organisms are already illuminating many aspects of apicomplexan biology, and the genome sequence of E. tenella, a non-cyst-forming coccidian, will add a new dimension to these analyses.
The pioneering work of Tyzzer (1929) established E. tenella as a favored model species for study, and it offers many advantages including ubiquity in the field, high virulence and replication within the host, good accessibility of both its transmission (oocysts) and invasive (zoites) stages, and a tractable genetic system. It is the only Eimeria species from the chicken that can be propagated in vitro. The Houghton (H) strain of E. tenella, isolated in the United Kingdom in 1949 and first cloned in 1986, is one of a small number of defined reference strains used for laboratory studies (Chapman and Shirley 2003). Its molecular karyotype comprises 14 chromosomes of between 1 and >7 Mbp, and an 8.4-fold shotgun sequence for the ∼55-Mbp genome is now available (http://www.sanger.ac.uk/Projects/E_tenella/). The haploid:diploid life cycle has enabled the construction of a genetic linkage map defined by 443 polymorphic DNA markers (Shirley and Harvey 2000).
Complementing the genome-wide shotgun sequencing, parallel projects have been undertaken to generate the complete sequences of chromosomes 1 (∼1 Mb) and 2 (∼1.2 Mb), which carry genetic markers associated with resistance to the anticoccidial drug arprinocid, and with precocious development, respectively (Shirley and Harvey 2000). Here we report the sequencing of chromosome 1 and the analysis of its content and organization.
Results
Mapping, sequencing, and assembly
The sequence was assembled primarily from reads taken from chromosome-specific small-insert plasmid libraries, prepared from pulsed-field gel electrophoresis- (PFGE-) purified chromosome 1 (Chr1) of E. tenella (Houghton strain) (Chapman and Shirley 2003). Further information was drawn from the ongoing chromosome 2 (Chr2) and whole-genome shotgun (WGS) sequencing projects of the same strain (http://www.sanger.ac.uk/Projects/E_tenella/), both of which contain some Chr1 reads, and from end sequences of bacterial artificial chromosome (BAC) and fosmid clones (see Methods).
To check the assembly, assist the resolution of repetitive regions, and orientate sequence scaffolds and isolated contigs, a high-resolution HAPPY map was constructed (see Methods). A total of 173 markers (taken from the early chromosome-specific sequence reads or, later, from contigs of the assembly) were mapped to a single linkage group. Markers that failed to link to the remainder were inferred to be predominantly contaminating sequences from other chromosomes; the majority appear to lie on Chr2 (data not shown), and the inferred level of contamination was typical for that seen when purifying chromosomes by PFGE.
The remaining gaps in the assembly are classified as sequence gaps (spanned by at least two small-insert plasmid clones) and physical gaps (spanned by fewer than two clones). Where possible, sequence gaps were closed by resequencing and primer walking, or subcloning and sequencing at least one bridging clone. Several sequencing and physical gaps were also closed by skimming of BAC and fosmid clones.
Details of the final assembly and the comparison with the HAPPY map are given in the Supplemental Material. The strong concordance between the order of markers on the HAPPY map and that found in the sequence implies a substantially correct assembly. The absence of any linked HAPPY markers beyond the ends of the assembly suggests that the assembly (including gaps) spans almost the full length of the chromosome, as does the presence of telomeric motifs at each end of the sequence (see below). The assembly consists of 889,314 bp of sequence, but spans 1,347,714 bp including the physical and sequence gaps. In each case, the maximum possible gap size (based on the sizes of bridging plasmid or BAC clones) is indicated in the EMBL entry—an overestimate in most cases. When reasonable estimates are made (based on the actual lengths of bridging clones where known, or on the distribution of insert sizes in the clone libraries, and taking into account the overlaps between the clones and the linked sequences) of the actual lengths of the gaps, the total span of the assembly is 1015 kb, in close agreement with the chromosomal size of 1050 kb measured by PFGE.
In calculating the content of the chromosome (e.g., base composition or density of genes and other features), we consider only the sequenced bases of the assembly.
Segmental structure of the chromosome
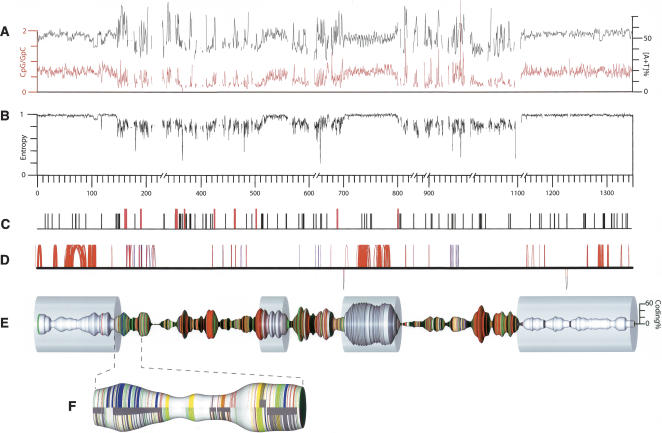
Even a cursory examination reveals that the chromosome is organized into two types of sequence, which differ markedly in their characteristics (Fig. 1). We define these two types of sequence as feature-rich (R) and feature-poor (P) segments. We identify three major R-segments of between approximately 140 and 370 kb in length and totaling 854 kb (63% of the assembly), separated and flanked by four P-segments of between approximately 40 and 200 kb, those nearest the chromosomal termini being the largest.
Figure 1.
E. tenella chromosome 1. (A) The [A+T] content of the sequence (black; 1-kb sliding window) and the ratio of CpG to GpC dinucleotides (red; 1-kb sliding window). (B) The information content (second-order Markov entropy) of the sequence, in a 1-kb sliding window. Values are normalized such that fully repetitive sequence has an information content of 0, and random sequence has an information content of 1. (C) The locations of TGCATGCA motifs (black vertical ticks) or longer [TGCA] tandem repeats (red). (D) Perfect segmental duplications of >50 bp in length along the chromosome (represented by the solid horizontal line); hoops above the line link sequences are duplicated in the same orientation; those below the link are inverted duplications. The color of the hoops indicates the proportion of simple tandem repeats in the duplicated element—(blue) more repetitive, (red) less repetitive; duplicates involving only simple tandem repetitive sequence are not shown. Note that many duplications have only a very short intervening sequence and hence appear as vertical lines. (E) Chromosome 1 with the thickness of the spindle corresponding to the coding density (proportion of nucleotides encoding amino acids; 20-kb center-weighted sliding window; scale at right). Colored bands indicate CAG repeats (red), telomere-like AGGGTTT repeats (green), other simple-sequence repeats (yellow), LINE-related sequences (blue), and gaps (black). The transparent cylinders indicate the feature-poor (P) segments. Physical gaps that are represented as >10 kb in the assembly (and in the corresponding GenBank record) have been condensed to 10 kb in this representation; the distance scale is broken to reflect this. (F) A representative section of a feature-rich (R) segment expanded to show the arrangement of repeat elements (color coding as for panel B) and genes (dark gray; solid segments near the center line indicate complete genes, and narrower wrap-around bands indicate the individual exons). Features are depicted separately for the forward strand (upper) and reverse strand (lower).
Sequence composition and information content
Overall, the chromosome is 49.7% A+T. Although the nucleotide compositions of the P- and R-segments are similar (52.4% and 48.7% A+T, respectively), the P-segments have a fairly uniform composition, whereas the A+T content of the R-segments fluctuates widely along their length and mirrors the distribution of coding sequences (Fig. 1).
The dinucleotide CpG is considerably under-represented in the R-segments: it occurs only 31% as often as its isomer GpC. Such under-representation is usually indicative of cytosine methylation at CpG sequences (since deamination of 5′-methyl cytosine leads to its replacement with thymidine unless actively preserved by selection), although there is no other evidence for or against methylation (which can regulate gene expression) in Eimeria. In contrast, CpG is only slightly under-represented in the P-segments, being present 64% as frequently as GpC.
Looking at longer sequences (trinucleotides and above), the R-segments show a strongly skewed composition, with heavy over-representation of a small subset of sequences (Fig. 2). In contrast, the sequence of the P-segments is much more similar to random sequence, with no grossly over- or under-represented short sequences. These skews are also reflected in the information content (entropy) of the sequence (Fig. 1). The entropy of the P-segments is as high as that of random sequence, while that in the R-segments is lower and variable, reflecting a mixture of highly repetitive and motif-rich sequence.
Figure 2.
Hexanucleotide content of R- and P-regions. For the R-segment sequence (A) and the P-segment sequence (B), the plots show the relative frequencies (vertical axes) of each possible hexanucleotide. Hexanucleotide sequences are arranged in the horizontal plane, with the homopolymers (T)6, (A)6, (C)6, and (G)6 at the corners of the plane as indicated. The sequences of the most abundant hexamers in the R-segment sequence (all circular permutations of CAGCAG or its complement) are indicated in A; the peak labeled “Tel” and others of similar height are hexamers from the telomere-like motif AGGGTTT, its complement, and circular permutations thereof.
Simple-sequence repeats and LINE-like elements
A striking feature of the chromosome is the abundance of long tandem repeats of the trinucleotide CAG and of the heptamer AGGGTTT. Both are present almost exclusively in the R-segments, where together they make up ∼14% of the sequence (Fig. 1). CAG repeats are found preferentially in the predicted exons of the R-segments, where they occur about once every 200 bp on average. The heptamer motif has been identified as a telomeric repeat unit in Plasmodium (for review, see Figueiredo and Scherf 2005); tracts of this sequence are found at each end of the E. tenella Chr1 assembly, although that at the left end (as oriented in Fig. 1) could not be unambiguously assembled and is therefore not included in the sequence. More surprisingly, clusters of telomere-like repeats are also common in the intronic and intergenic regions of the R-segments.
Other simple-sequence repeats (SSRs) are also confined almost exclusively to the R-segments; indeed, the P-segments appear to have been “swept clean” of SSRs, containing only about one per 12 kb, as compared to one per 170 bp in the R-segments, one per 200 bp in Plasmodium falciparum (Gardner et al. 2002), or one per 2 kb in human (International Human Genome Sequencing Consortium 2001). Further details are given in the Supplemental Material.
The chromosome also has 57 regions with significant similarity to known LINE transposons, exclusively in the R-segments. There is some indication of clustering, especially of closely related elements, and with adjacent elements tending to lie in the same orientation. Most of these regions are small, and do not appear to encode functional genes.
Apicomplexan-specific palindromic octamer motif
The palindromic octamer TGCATGCA has been found as an abundant motif not only in the genome of E. tenella but also in those of several other apicomplexans: P. falciparum, Toxoplasma gondii, and Cryptosporidium parvum (Bankier et al. 2003). On E. tenella chromosome 1, the octamer occurs 157 times, as compared to ∼14 instances expected in random sequence of similar length and base composition (Fig. 1). Most of these octamers are found in the R-segments, and are more abundant in intergenic sequence than within introns or exons. This motif appears so far to be unique to the Apicomplexa.
Segmental duplications
The chromosome contains many segmental duplications (Fig. 1), mostly within the P-segments and often containing predicted genes. Duplications within the R-segments are almost always short and often involve telomere-like repeats. Notably, inverted segmental duplications are rare (only two instances), as are duplications bridging more than a few tens of kilobases, suggesting that unequal crossover or unequal sister-chromatid exchange in meiosis accounts for the majority of duplication events.
Predicted transcripts
Two approaches were used to predict encoded proteins. In the first, automated gene prediction software tools trained on E. tenella genes were used to predict transcripts, which were then subjected to thorough manual refinement with reference to Eimeria ESTs and to known genes in other apicomplexans. In the second, alignment of genomic sequence with Eimeria ESTs and BLAST searches of the Eimeria chromosomal sequence against proteins from other apicomplexans were used to find further genes not identified by the prediction tools; these also were subjected to manual refinement (for details, see Methods and Supplemental Material).
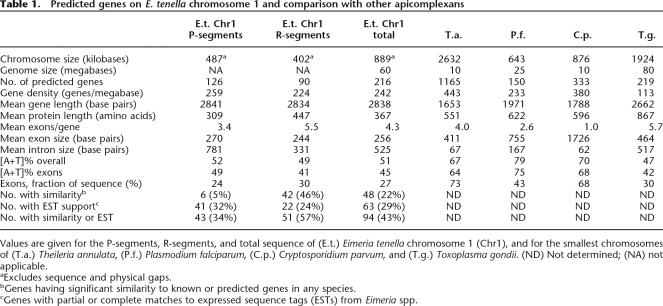
The chromosome encodes 216 predicted proteins, distributed approximately equally between the repeat-rich R-segments and the P-segments. However, the characteristics of the genes in these two regions differ (Table 1).
Table 1.
Predicted genes on E. tenella chromosome 1 and comparison with other apicomplexans
Values are given for the P-segments, R-segments, and total sequence of (E.t.) Eimeria tenella chromosome 1 (Chr1), and for the smallest chromosomes of (T.a.) Theileria annulata, (P.f.) Plasmodium falciparum, (C.p.) Cryptosporidium parvum, and (T.g.) Toxoplasma gondii. (ND) Not determined; (NA) not applicable.
aExcludes sequence and physical gaps.
bGenes having significant similarity to known or predicted genes in any species.
cGenes with partial or complete matches to expressed sequence tags (ESTs) from Eimeria spp.
The predicted genes in the R-segments (“R-genes”) typically encode long proteins (447 amino acids on average) and have several fairly small introns, and the [A+T] content of their exons is lower than the average for the chromosome. About half of these proteins have predicted transmembrane domains, and, of these, about half also have predicted signal peptides. Many candidates for surface proteins are therefore included in this group. Although only 46% of these predicted proteins have similarity to proteins in other organisms, this is not unusual in newly sequenced genomes. Matches to ESTs from Eimeria species support 24% of these predictions, and, overall, 57% of the predictions are supported by either similarity or by EST data (or by both). Most (60%) of these predictions were made by two or more of the automated prediction tools. Therefore, we are confident that the majority of R-gene predictions are essentially correct.
The genes in the P-segments (“P-genes”) encode shorter proteins (309 amino acids on average) and have long introns, and their exons have a high [A+T] content. (The third P-segment is atypical, but it contains several large imperfect tandem duplications, which may skew the coding content of this segment.) Fewer of the P-genes (about one-quarter) have predicted transmembrane domains than is the case for R-genes, and far fewer (only six) have predicted signal peptides. The gene predictions in the P-segments are less well supported than those in the R-segments: only 43 (34%) are supported by similarities to other species or by EST data (mostly the latter), and the majority were predicted by only one of the automated prediction programs (predominantly GlimmerHMM). However, a detailed analysis (see Supplemental Material) suggests that GlimmerHMM is the most efficient prediction tool in this genome. It is possible that the P-genes in general are more likely to be Eimeria-specific, accounting for their lower support from similarity. Therefore, we cannot dismiss the majority of P-gene predictions as erroneous, although we suspect that mispredictions are more common among these than among the R-genes.
E. tenella genes are typically much larger than those of Theileria annulata, P. falciparum or C. parvum (Table 1), owing mainly to numerous long introns, leading to a lower overall coding density. T. gondii has an equally low protein-coding fraction (and, like Eimeria, a relatively large genome), although this is due more to large intergenic regions than to large introns. We note that there is little apparent synteny between E. tenella Chromosome 1 and the genomes of other sequenced Apicomplexans (see Supplemental Material).
No tRNAs or other non-protein transcripts were identified, but, as this chromosome represents only 1/60th of the genome, we assume that they lie on other chromosomes.
Simple-sequence repeats in predicted proteins and in expressed sequences
The (CAG)n repeats that abound in the R-segments are found preferentially in many of the predicted exons, giving rise to homopolymer tracts of glutamine (CAG), serine (AGC), alanine (GCA or GCT), leucine (CTG), or cysteine (TGC) depending on the orientation and reading frame of the DNA repeat. Homopolymers of all of these amino acids occur with roughly comparable frequencies (between 21 and 51 instances of each type, alanine repeats being the most abundant), with the exception of cysteine (only six instances). Curiously, not all of the homopolymer amino acid tracts are encoded by perfect trinucleotide repeats. For example, many CAG (glutamine) repeats are peppered with CAA (glutamine) codons. This prevalence of silent mutations suggests that selective pressure is preserving homopolymer amino acid tracts even as the DNA repeat degenerates.
Low-complexity amino acid tracts (e.g., LLLLQLLLLQLL LQQLLLLLLLQLLLLLQ) are also common, and are encoded by (CAG)n nucleotide repeats that have undergone frameshifting mutations or inversions. Other nucleotide simple-sequence repeats are rarely found in the predicted exons: neither the telomere-like repeat (AGGGTTT)n nor its complement occurs.
These simple-sequence amino acid repeats and low-complexity tracts are confined almost exclusively to the proteins in the repeat-rich R-segments: only one P-segment protein contains such a region. Of the 90 predicted R-proteins, two-thirds (58) contain one or more repeats or low-complexity tracts, and most of these proteins contain several. We see no obvious general differences in the functions of repeat-containing and repeat-free proteins; however, the number of proteins with well-established functions is probably too small to make such a distinction.
It is important to determine whether this abundance of apparent coding repeats is reflected in transcripts. Among the R-genes, support for predictions from EST or BLAST data is about equal for the repeat-containing transcripts (16/58, or 28% supported) and the non-repeat-containing transcripts (11/32, or 34% supported). Repeats are also abundant in ESTs from all developmental stages of E. tenella, and in other species of Eimeria (see Supplemental Material). We are therefore confident that abundant amino acid repeats are a real feature of Eimeria proteins, and not an artifact of the gene prediction process.
Interstrain variation associated with R-segments
Chromosome 1 is known (Shirley 1994) to manifest size polymorphisms between the virulent Houghton wild-type strain and two lines derived from it (attenuated by adaptation to egg passage and by selection for precocious development). Polymorphism in several chromosomes has also been detected in a panel of wild-type parasites including the Weybridge and Wisconsin strains (Shirley 1994; Chapman and Shirley 2003), suggesting that the genome is markedly plastic.
We tried to determine whether the strain-to-strain variation in Chr1 was associated with the repeat-rich R-segments or with the repeat-poor P-segments. Present genetic maps of E. tenella (Shirley and Harvey 2000) are not detailed enough to resolve this question. Genomic DNA from three strains of E. tenella (Houghton, Weybridge, and Wisconsin) was Southern blotted after digestion with each of five different restriction enzymes, and hybridized with probes against each of four different nonrepetitive sequences lying in two of the P-segments and four lying in two of the R-segments of Chr1.
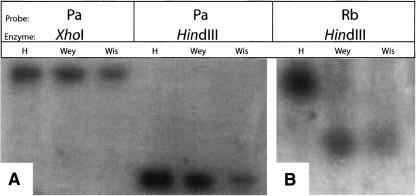
The results are shown in Figure 3 and summarized in Table 2. None of the four P-segment probes revealed any difference between the three strains when digested with any of the five restriction enzymes tested. In marked contrast, all four R-segment probes revealed differences between the strains in one of the five digests.
Figure 3.
Restriction fragment length polymorphisms in P- and R-segments. The figure shows two representative blots. Genomic DNA of E. tenella strains Houghton (H), Weybridge (Wey), or Wisconsin (Wis) was digested with the indicated restriction enzymes, electrophoresed, blotted, and hybridized with radiolabeled probes for P- or R-segment sequences (probe names at top). (A) Shows no size polymorphism between the three strains in the XhoI or HindIII fragments detected by probe Pa; (B) shows a size polymorphism in the HindIII fragment detected by probe Rb.
Table 2.
Analysis of restriction-fragment length polymorphisms in P- and R-segments
Sizes (in kilobases) are given for the restriction fragments (enzymes listed across top) generated from genomic DNA of E. tenella strains Houghton (H), Weybridge (Wey), or Wisconsin (Wis), as detected by hybridization with probes to P- and R-segment sequences (named at left; Segment refers to the segment of chromosome 1 within which the probe is expected to hybridize). Polymorphisms (fragments that differ in size between the strains) are highlighted in bold. (NA) Not applicable; (ND) not determined. “Total P” and “Total R” refer to the total sizes of restriction fragments (as measured on the Houghton strain) hybridized by the probes used. Further details of the probes are given in the Supplemental Material.
Discussion
Eimeria species are the most important global parasites of intensively reared livestock, causing coccidiosis in poultry, cattle, and sheep. Coccidiosis has most impact in the intensive poultry industry, where all flocks become infected with some or all of the seven species of avian Eimeria. As well as having a large economic impact, coccidiosis is a severe welfare problem causing weight loss, diarrhea, hemorrhage, anemia, and death. Resistance to all classes of anticoccidial drugs is universal, no new drugs are in the pipeline, and live-attenuated vaccines are relatively expensive. It is hoped, therefore, that genomic and post-genomic analysis will offer new insights into the biology of this parasite and reveal new targets for the development of drugs and vaccines.
The most striking feature of the chromosome is its segmented organization, which is reflected in all aspects of its content. About half the chromosome is in R-segments, which are rich in repeats of several types; the remainder is in P-segments, which are relatively featureless. An important question is whether this segmental organization is typical of the remainder of the genome or peculiar to this, the smallest chromosome. We find (data not shown) a closely similar segmentation on Chr2: a preliminary assembly reveals two R-segments, each of ∼280 kb, divided and flanked by P-segments of between about 120 and 260 kb. The largest contigs of the ongoing whole-genome shotgun assembly also display this bipartite character, although, of course, they do not reveal the long-range order of P- and R-segments. We are therefore confident that the segmentation found on Chr1 is typical of the remainder of the genome.
We suspect that the unusual genome organization may serve to facilitate rapid evolution and diversification, a strategy of benefit to a parasite. Rearrangements within R-segments may be facilitated by the abundant CAG repeats between and within genes. E. tenella Chr1 has none of the characteristic subtelomeric regions that, in parasites such as P. falciparum and Trypanosoma brucei, harbor dynamic populations of genes involved in evading the host immune system (Gardner et al. 2002; Berriman et al. 2005). However, the telomere-like repeats that pepper the R-segments in Eimeria may reflect an analogous process for genome shuffling.
In support of this model, strong evidence for genome plasticity comes from known size polymorphisms between different strains of E. tenella (Shirley 1994). We have shown that, on Chr1, restriction fragment length polymorphisms between strains are associated exclusively with the R-segments, at least in all of the instances we tested.
Turning to the predicted proteins, a question remains over the accuracy of predictions in the P-segments, which have less support from interspecies similarity than do those in the R-segments. However, we cannot dismiss these as mis-predictions: perhaps the P-genes are peculiar to Eimeria accounting for the paucity of BLAST and EST support. We hope that this question will be resolved by analysis of the whole-genome shotgun data and by further studies of Eimeria transcripts.
The R-segment genes are more robustly predicted, and their proteins are striking in containing numerous repetitive amino acid tracts arising from CAG repeats within exons. There is little doubt that these tracts are real (and not artifacts of misprediction), as they are abundant in the ESTs of several Eimeria species (A. Gruber and A.M.B.N. Madeira, unpubl.). Although amino acid repeats are rare in vertebrate proteins (and are often associated with disease) (Gatchel and Zoghbi 2005), they are common in several lower eukaryotes, including Dictyostelium discoideum (Eichinger et al. 2005) and P. falciparum (Gardner et al. 2002; Hall et al. 2002). In all instances, the amino acid repeats arise from nucleotide repeats that are characteristic of the genome in question, and tend to be conserved at the amino acid level even when the original nucleotide repeat degenerates. The function (if any) of protein repeats in these genomes remains a mystery. They may contribute to the genome plasticity discussed above by encouraging recombination (see Verstrepen et al. 2005; Verstrepen and Klis 2006). It has also been suggested (Anders 1986; Schofield 1991) that such repeats in the proteins of Plasmodium and other protozoans may provide an “immunological smokescreen” to assist host immune system evasion. While this “smokescreen” theory is attractive for bloodstream parasites, it is not so appealing for intestinal parasites like Eimeria and even less so for the free-living Dictyostelium. Clearly, the nature and role of amino acid repeats in lower eukaryotes warrant further investigation.
In conclusion, the segmental organization and repeat-richness of this chromosome (and, apparently, of the rest of the genome) raises many questions, only some of which can be tentatively answered. Comparison with other isolates of E. tenella or with other members of the genus may reveal whether this organization is associated with a dynamic, adaptable genome as we postulate. Forthcoming genome-wide analysis of E. tenella (albeit a shotgun sequence rather than assembled chromosomes) may more clearly reveal broad differences between the genes that populate the P- and R-segments. Finally, we note that the unusual long-range organization of this chromosome presents a strong argument for the completion and assembly of genome sequences beyond the shotgun level.
Methods
HAPPY mapping
HAPPY mapping was performed on a panel of subgenomic aliquots of DNA prepared from E. tenella Houghton, essentially as described previously (Konfortov et al. 2000; Glöckner et al. 2002; Bankier et al. 2003). Sequences for mapping (markers) were selected mainly from the data generated by the chromosome-specific sequencing project. Initially, arbitrarily chosen sequences from this data set were used; later, sequences were chosen preferentially from contigs of the assembly wherever needed to validate the contig structure or to link contigs. Some early markers were derived from Chr1-enriched libraries made from the Wis strain of E. tenella. Details of the method are given in the Supplemental Material.
Library construction
Chromosomal DNA from sporozoites of E. tenella (Houghton) was prepared in agarose blocks as previously described (Shirley et al. 1990), and chromosomes were resolved by pulsed-field gel electrophoresis (PFGE). After electrophoresis, Chr1 was excised and eluted from the agarose gel, sonicated, and cloned into the SmaI-digested pTrueBlue vector (Genomics One). Two small-insert libraries (with insert size ranges of 1.0–2.0 kb and 2.0–4.0 kb) were constructed for the whole-chromosome shotgun sequencing strategy.
Sequencing, assembly, and gap closure
Details of the sequence assembly and gap closure are provided as Supplemental Material. Briefly, 23,560 reads (from 11,780 randomly selected clones) were generated from plasmid ends using the ABI PRISM BigDye Terminator v3.1 (Applied Biosystems) chemistry on Applied Biosystems machines. Initial assembly was carried out using the Staden-based PHRAP (P. Green, unpubl.). Reads from the whole-genome shotgun (WGS) sequencing project (http://www.sanger.ac.uk/Projects/E_tenella/) were incorporated into the database by using a directed method that employed GAP4 (Bonfield et al. 1995); additional reads from the closely migrating Chr2 were also incorporated.
Contigs were ordered based on at least two consistent paired reads before being subjected to BLASTN against BAC-end (ftp.sanger.ac.uk/pub/pathogens/Eimeria/tenella/BAC/) and fosmid-end (ftp.sanger.ac.uk/pub/pathogens/Eimeria/tenella/fosmid/) sequences. Relevant BAC and fosmid clones were obtained from the WTSI, and the clones were sized using PFGE. BAC-end and fosmid-end sequences and HAPPY markers were used to order the contigs into scaffolds as well as superscaffolds. Sequence gaps were closed where possible using primer walking, and difficult regions were sequenced using alternative chemistries. A variety of approaches (including subcloning and transposon-mediated sequencing of bridging BACs or fosmids, or long-range PCR) were also used to close gaps.
Prediction of transcripts, generation of gene models, and annotation
Prediction of coding regions was carried out initially using GlimmerHMM (Majoros et al. 2004), SNAP (Korf 2004), and Genefinder (P. Green, unpubl.), each trained on a similar set of 424 E. tenella genes. The training genes were either derived from mRNA sequences or were predicted genes that had been manually annotated. Predictions were also generated with Twinscan using T. gondii genome sequence as a template. All automated predictions were screened by BLAST against the Uniprot database (http://www.uniprot.org) to identify the best gene prediction for each locus. Putative E. tenella gene sequence was then screened by BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) against the proteins of Plasmodium spp., Theileria spp., Cryptosporidium spp., and the predicted genes of Toxoplasma spp. to try to extend the E. tenella coding sequence.
Multiple alignment between E. tenella putative open reading frames and apicomplexan counterparts was performed to identify protein motifs conserved in apicomplexan homologs but missing from the initial E. tenella gene models. Each model was edited with reference to EST matches from Eimeria spp. using Exonerate (Slater and Birney 2005) after repetitive and low-complexity sequence had been masked using RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) and Dust (R.L. Tatusov and D.J. Lipman, in prep.). Overlapping EST matches were merged using a custom post-processing script. EST data for this analysis were taken from Wan et al. (1999); Ng et al. (2002); Li et al. (2003); the Eimeria ORESTES Project (A. Gruber and A.M.B.N. Madeira, unpubl.) at http://www.coccidia.icb.usp.br/eimeria; Merck Research Laboratories and Washington University, USA; Wellcome Trust Sanger Institute and Institute for Animal Health Compton Laboratory, UK; and Universiti Kebangsaan Malaysia, Malaysia.
Additional models (not originating from automated predictions) were generated from EST mapping using a “sim4” output parsed with a custom Perl script (X. Wu, unpubl.) applying the following criteria: (1) The aligned part of the EST must be a single continuous sequence. (2) This aligned sequence must be at least two-thirds the length of the EST. (3) If the aligned part of the genomic DNA is also continuous (a single-exon alignment), the identity of the alignment must be >90%, and it must be ≥50 bp long. (4) If the aligned part of the genomic DNA is discontinuous (a spliced alignment across ≥2 exons), the identity of the terminal exons must be >90%, while that for internal exons must be >95%, the exon length in either case being >10 bp.
Signal peptides and transmembrane helices were predicted using Phobius (Kall et al. 2004), and GPI cleavage sites using DGPI (Kronegg and Buloz 1999). InterProScan (Quevillon et al. 2005) was used to find matches to protein signatures in the predicted proteins, with the Interpro2go setting allowing automated assignment of Gene Ontology terms.
tRNAscan-SE (Lowe and Eddy 1997) was used to check for tRNA genes. Genes encoding structural RNAs were sought by searching against the Rfam database (Griffiths-Jones et al. 2003).
Analysis of simple tandem repeats
Chr1 sequence was processed by Tandem Repeats Finder 4.00 (Benson 1999) using the following set of parameters: (2, 1000, 1000, 80, 10, 25, 1000). Very high mismatch and indel penalties (second and third values) guaranteed that only perfect repeats would be found. TRF output was then processed and filtered by TRAP (Sobreira et al. 2006).
Information content analysis
Information content was calculated as the second-order Markov entropy of the sequence, as described in Pasechnik et al. (2005). The algorithm is broadly similar to that used in Artemis (Rutherford et al. 2000).
Other analyses
Other analyses (information content, coding content) were performed using custom software (P.H. Dear, unpubl.); the graphical representations in Figure 1 were produced primarily using this custom software and Cinema4D-9.5 (Maxon Computer GmbH). Software is available at http://www.mrc-lmb.cam.ac.uk/happy/HappyGroup/happyhomepage.html, and further information is contained in the Supplemental Material.
Restriction-fragment length polymorphism analysis
BlastN comparison of P- and R-segments with the E. tenella genome shotgun sequence (http://www.genedb.org/genedb/etenella/) identified eight sequences, four in each segment type, which were unique within the genome. RepeatMasker (http://www.repeatmasker.org) confirmed the absence of simple repeat sequences. Fragments of 227–499 bp were amplified from each sequence by PCR in reactions containing 5 ng of E. tenella genomic DNA purified from oocysts as described by Blake et al. (2003), 20 pmol of relevant forward and reverse primers (see primer sequences in Supplemental Material), 0.5 U of Taq polymerase (Invitrogen), 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl, and 0.2 mM dNTPs; cycling conditions were: 95°C for 1 min; followed by 30 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 1 min; then 72°C for 10 min. Amplimers were gel-purified (QIAGEN) and radiolabeled using standard protocols.
Five hundred nanograms of each genomic DNA (Houghton, Wisconsin, or Weybridge strains, purified as above) was digested using BamHI, BglII, EcoRI, HindIII, or XhoI (Invitrogen) and resolved by gel electrophoresis (1.0% [w/v] agarose in 1× TBE, 20 V, 16 h). DNA was Southern blotted and hybridized with the relevant labeled probes using standard protocols (Sambrook and Russell 2001).
Acknowledgments
The Malaysian investigators thank Nor Muhammad Mahadi for his contribution and the staff of the Malaysia Genome Institute for their support. The Sanger Institute investigators thank David Harper and Paul Mooney for assistance with sequence analysis and exchange, and Bob Plumb for assistance with sequencing. P.H.D. thanks Derek Gatherer for helpful discussions on information content analysis. This work was supported by a Top-Down Grant (IRPA 09-02-02-002-BTK/TD/003) from the Ministry of Science, Technology and Innovation (MOSTI) in Malaysia and by FAPESP and CNPq in Brazil. The investigators of this study at the Sanger Institute were supported by the Wellcome Trust through their funding of the Pathogen Sequencing Unit; the sequencing was partly funded by BBSRC grant S17754.
Footnotes
[Supplemental material is available online at www.genome.org. Software is available at http://www.mrc-lmb.cam.ac.uk/happy/HappyGroup/happyhomepage.html. The sequence data from this study have been submitted to EMBL under accession no. AM269894.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5823007
References
- Anders R.F. Multiple cross-reactivities amongst antigens of Plasmodium falciparum impair the development of protective immunity against malaria. Parasite Immunol. 1986;8:529–539. doi: 10.1111/j.1365-3024.1986.tb00867.x. [DOI] [PubMed] [Google Scholar]
- Bankier A.T., Spriggs H.F., Fartmann B., Konfortov B.A., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Spriggs H.F., Fartmann B., Konfortov B.A., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Fartmann B., Konfortov B.A., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Konfortov B.A., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Vogel C., Teichmann S.A., Ivens A., Dear P.H., Teichmann S.A., Ivens A., Dear P.H., Ivens A., Dear P.H., Dear P.H. Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genomics. 2003;1:1787–1799. doi: 10.1101/gr.1555203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Lennard N.J., Caler E., Hamlin N.E., Haas B., Caler E., Hamlin N.E., Haas B., Hamlin N.E., Haas B., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Blake D.P., Smith A.L., Shirley M.W., Smith A.L., Shirley M.W., Shirley M.W. Amplified fragment length polymorphism analyses of Eimeria spp.: An improved process for genetic studies on recombinant parasites. Parasitol. Res. 2003;90:473–475. doi: 10.1007/s00436-003-0890-x. [DOI] [PubMed] [Google Scholar]
- Bonfield J.K., Smith K.F., Staden R., Smith K.F., Staden R., Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.D., Shirley M.W., Shirley M.W. The Houghton strain of Eimeria tenella: A review of the type strain selected for genome sequencing. Avian Pathol. 2003;32:115–127. doi: 10.1080/0307945021000071588. [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J.A., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Pachebat J.A., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Song J., Olsen R., Szafranski K., Xu Q., Olsen R., Szafranski K., Xu Q., Szafranski K., Xu Q., Xu Q., et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo L., Scherf A., Scherf A. Plasmodium telomeres and telomerase: The usual actors in an unusual scenario. Chromosome Res. 2005;13:517–524. doi: 10.1007/s10577-005-0996-3. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Carlton J.M., Pain A., Nelson K.E., Bowman S., Pain A., Nelson K.E., Bowman S., Nelson K.E., Bowman S., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchel J.R., Zoghbi H.Y., Zoghbi H.Y. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Glöckner G., Eichinger E., Szafranski K., Pachebat J.A., Bankier A.T., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Eichinger E., Szafranski K., Pachebat J.A., Bankier A.T., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Szafranski K., Pachebat J.A., Bankier A.T., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Pachebat J.A., Bankier A.T., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Bankier A.T., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Dear P.H., Lehmann R., Baumgart C., Parra G., Abril J.F., Lehmann R., Baumgart C., Parra G., Abril J.F., Baumgart C., Parra G., Abril J.F., Parra G., Abril J.F., Abril J.F., et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Marshall M., Khanna A., Eddy S.R., Khanna A., Eddy S.R., Eddy S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall N., Pain A., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Pain A., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Mungall K., Bowman S., Atkin R., Baker S., Bowman S., Atkin R., Baker S., Atkin R., Baker S., Baker S., et al. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L., Krogh A., Sonnhammer E.L., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Konfortov B.A., Cohen H.M., Bankier A.T., Dear P.H., Cohen H.M., Bankier A.T., Dear P.H., Bankier A.T., Dear P.H., Dear P.H. A high-resolution HAPPY map of Dictyostelium discoideum chromosome 6. Genome Res. 2000;10:1737–1742. doi: 10.1101/gr.141700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronegg J., Buloz D., Buloz D. Detection/prediction of GPI cleavage site (GPI-anchor) in a protein (DGPI) 1999. [Google Scholar]
- Li L., Brunk B.P., Kissinger J.C., Pape D., Tang K., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Brunk B.P., Kissinger J.C., Pape D., Tang K., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Kissinger J.C., Pape D., Tang K., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Pape D., Tang K., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Tang K., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Cole R.H., Martin J., Wylie T., Dante M., Fogarty S.J., Martin J., Wylie T., Dante M., Fogarty S.J., Wylie T., Dante M., Fogarty S.J., Dante M., Fogarty S.J., Fogarty S.J., et al. Gene discovery in the Apicomplexa as revealed by EST sequencing and assembly of a comparative gene database. Genome Res. 2003;13:443–454. doi: 10.1101/gr.693203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros W.H., Pertea M., Salzberg S.K., Pertea M., Salzberg S.K., Salzberg S.K. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Ng S.-T., Jangi M.S., Shirley M.W., Tomley F.M., Wan K.-L., Jangi M.S., Shirley M.W., Tomley F.M., Wan K.-L., Shirley M.W., Tomley F.M., Wan K.-L., Tomley F.M., Wan K.-L., Wan K.-L. Comparative EST analyses provide insights into gene expression in two asexual developmental stages of Eimeria tenella. Exp. Parasitol. 2002;101:168–173. doi: 10.1016/s0014-4894(02)00109-1. [DOI] [PubMed] [Google Scholar]
- Pasechnik A., Mylläri A., Salakoski T., Mylläri A., Salakoski T., Salakoski T. Dynamical visualization of the DNA sequence and its nucleotide content. In: Bounsaythip C., et al., editors. Proceedings of KRBIO ’05, International Symposium on Knowledge Representation in Bioinformatics. Espoo, Finland: 2005. pp. 47–50. [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Harte N., Mulder N., Apweiler R., Lopez R., Mulder N., Apweiler R., Lopez R., Apweiler R., Lopez R., Lopez R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.-A., Barrell B., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.-A., Barrell B., Crook J., Horsnell T., Rice P., Rajandream M.-A., Barrell B., Horsnell T., Rice P., Rajandream M.-A., Barrell B., Rice P., Rajandream M.-A., Barrell B., Rajandream M.-A., Barrell B., Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D., Russell D. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schofield L. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol. Today. 1991;7:99–105. doi: 10.1016/0169-4758(91)90166-l. [DOI] [PubMed] [Google Scholar]
- Shirley M.W. The genome of Eimeria tenella: Further studies on its molecular organisation. Parasitol. Res. 1994;80:366–373. doi: 10.1007/BF00932373. [DOI] [PubMed] [Google Scholar]
- Shirley M.W., Harvey D.A., Harvey D.A. A genetic linkage map of the apicomplexan protozoan parasite Eimeria tenella. Genome Res. 2000;10:1587–1593. doi: 10.1101/gr.149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley M.W., Kemp D.J., Pallister J., Prowse S.J., Kemp D.J., Pallister J., Prowse S.J., Pallister J., Prowse S.J., Prowse S.J. A molecular karyotype of Eimeria tenella as revealed by contour-clamped homogenous electric field gel electrophoresis. Mol. Biochem. Parasitol. 1990;38:169–174. doi: 10.1016/0166-6851(90)90020-m. [DOI] [PubMed] [Google Scholar]
- Shirley M.W., Ivens A., Gruber A., Madeira A.M., Wan K.L., Dear P.H., Tomley F.M., Ivens A., Gruber A., Madeira A.M., Wan K.L., Dear P.H., Tomley F.M., Gruber A., Madeira A.M., Wan K.L., Dear P.H., Tomley F.M., Madeira A.M., Wan K.L., Dear P.H., Tomley F.M., Wan K.L., Dear P.H., Tomley F.M., Dear P.H., Tomley F.M., Tomley F.M. The Eimeria genome projects: A sequence of events. Trends Parasitol. 2004;20:199–201. doi: 10.1016/j.pt.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Slater G.S., Birney E., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;15:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira T.J., Durham A.M., Gruber A., Durham A.M., Gruber A., Gruber A. TRAP: Automated classification, quantification, and annotation of tandemly repeated sequences. Bioinformatics. 2006;22:361–362. doi: 10.1093/bioinformatics/bti809. [DOI] [PubMed] [Google Scholar]
- Tyzzer E.E. Coccidiosis in gallinaceous birds. Am. J. Hyg. 1929;10:269–283. [Google Scholar]
- Verstrepen K.J., Klis F.M., Klis F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- Verstrepen K.J., Jansen A., Lewitter F., Fink G.R., Jansen A., Lewitter F., Fink G.R., Lewitter F., Fink G.R., Fink G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K.-L., Chong S.-P., Ng S.-T., Shirley M.W., Tomley F.M., Jangi M.S., Chong S.-P., Ng S.-T., Shirley M.W., Tomley F.M., Jangi M.S., Ng S.-T., Shirley M.W., Tomley F.M., Jangi M.S., Shirley M.W., Tomley F.M., Jangi M.S., Tomley F.M., Jangi M.S., Jangi M.S. A survey of genes in Eimeria tenella merozoites by EST sequencing. Int. J. Parasitol. 1999;29:1885–1892. doi: 10.1016/s0020-7519(99)00160-5. [DOI] [PubMed] [Google Scholar]