Abstract
Peptide hormones are small, processed, and secreted peptides that signal via membrane receptors and play critical roles in normal and pathological physiology. The search for novel peptide hormones has been hampered by their small size, low or restricted expression, and lack of sequence similarity. To overcome these difficulties, we developed a bioinformatics search tool based on the hidden Markov model formalism that uses several peptide hormone sequence features to estimate the likelihood that a protein contains a processed and secreted peptide of this class. Application of this tool to an alignment of mammalian proteomes ranked 90% of known peptide hormones among the top 300 proteins. An analysis of the top scoring hypothetical and poorly annotated human proteins identified two novel candidate peptide hormones. Biochemical analysis of the two candidates, which we called spexin and augurin, showed that both were localized to secretory granules in a transfected pancreatic cell line and were recovered from the cell supernatant. Spexin was expressed in the submucosal layer of the mouse esophagus and stomach, and a predicted peptide from the spexin precursor induced muscle contraction in a rat stomach explant assay. Augurin was specifically expressed in mouse endocrine tissues, including pituitary and adrenal gland, choroid plexus, and the atrio-ventricular node of the heart. Our findings demonstrate the utility of a bioinformatics approach to identify novel biologically active peptides. Peptide hormones and their receptors are important diagnostic and therapeutic targets, and our results suggest that spexin and augurin are novel peptide hormones likely to be involved in physiological homeostasis.
The study of peptide hormones has received considerable attention because of their role in modulating a wide range of physiological functions (Kastin 2006). A large group of peptide hormones serve as both hormones and neurotransmitters, being secreted into the bloodstream by endocrine cells and released into the synapse by neurons (Hökfelt 1991). Because of this dual function, peptide hormones often play important roles in the coordination of behavioral and somatic responses to environmental stimuli, and understanding their biology has helped advance our understanding of interactions between brain and body.
Peptide hormones are short peptides (<100 amino acids) produced by the proteolytic cleavage of pre-pro-hormone precursors. Following signal peptide removal by the signal peptidase complex, the pro-hormone undergoes cleavage at specific sites by pro-hormone convertases (Steiner 1998) or furin (Thomas 2002). In many cases, processed peptides undergo post-translational modification, with >50% of peptide hormones becoming amidated at their C terminus (Eipper et al. 1992). Mature peptides pass through the secretory pathway and are released into the extracellular space, where they can bind to specific cell surface receptors and modulate cellular functions.
Most peptide hormones are ligands for G-protein-coupled receptors (GPCR), via which they modulate intracellular signaling pathways and regulate cellular homeostasis. GPCRs belong to the seven transmembrane receptor family and share a high degree of sequence homology. As a result, in many organisms the complete set of GPCRs has been identified and classified (Vassilatis et al. 2003). The fraction of human GPCRs with known peptide ligands has been used to estimate that 27 orphan GPCRs are expected to have endogenous peptide ligands (Vassilatis et al. 2003). Although some of these missing ligands may turn out to be either previously characterized peptide hormones or novel peptides produced by known genes (Shichiri et al. 2003), including known peptide hormone genes (Zhang et al. 2005), some of these are likely to be produced by as yet uncharacterized genes.
Several methods have been used to identify new peptide hormones. Biochemical purification coupled with functional assays has been the predominant discovery method (for example, see Braun-Menendez et al. 1939; Burgus et al. 1969; Schmidt et al. 1991; Katafuchi et al. 2003). More recently, with the advent of genomic sequence, bioinformatics search strategies have been developed. Bioinformatics search strategies have the advantage over biochemical approaches that they are not biased against proteins with low or highly restricted expression and can be equally well applied to organisms in which biochemical purification of sufficient peptides is prohibitive. Most of these bioinformatics strategies relied on searches for single sequence features common to peptide hormones, such as C-terminal RF-amide (Hinuma et al. 2000; Chartrel et al. 2003; Jiang et al. 2003), dibasic cleavage sites (Duckert et al. 2004; Zhang et al. 2005), homology with known peptide hormones (Hsu 1999; Park et al. 2002), and other shared motifs (Baggerman et al. 2005). In at least one case, a combination of features was used to identify candidate peptide hormones (Shichiri et al. 2003). In this study, several independent sequence requirements including presence of signal peptide and pro-hormone cleavage sites, subcellular location, and precursor length were applied to retrieve a novel peptide hormone precursor from human cDNA databases. These studies demonstrated the success of sequence-based approaches to peptide hormone discovery and inspired us to develop a more systematic bioinformatics approach to address this problem.
We present here the development of a hidden Markov model (HMM) based search algorithm that integrates several peptide hormone sequence features for the discovery of novel peptide hormones. HMM techniques are well adapted to address sequence analysis problems because of their ability to handle variable sequence length signals and to implicitly integrate information from multiple dispersed signals in a sequence. As a result, HMMs have been applied successfully to both gene prediction (Burge and Karlin 1997; Birney et al. 2004) and protein domain finding, leading to domain databases such as Pfam (Krogh et al. 1994; Finn et al. 2006). In this study, we provide an HMM for all peptide hormones, more akin to gene prediction models that integrate biological processing signals than protein domain models that integrate homology signals. Application of our peptide hormone HMM to the human proteome allowed us to identify two novel candidate peptide hormones. Biochemical characterization of the candidates demonstrates that they are processed and secreted as predicted, and one of them has biological activity in a stomach contractility assay. These results demonstrate the power of a bioinformatics approach to find novel biologically active peptides.
Results
Development of peptide hormone search algorithm
Peptide hormones contain several common sequence features that distinguish them from other proteins. Peptide hormones all carry a signal peptide sequence and cleavage site at their N terminus, at least one pro-hormone cleavage site (generally occurring at a pair of basic residues), and amino acid residues that are typical for extracellular proteins and frequently include aromatic amino acids. Finally, peptide hormones do not contain transmembrane domains, and their processed products are short (<100 amino acids). We reasoned that these features could be used to identify novel peptide hormone genes.
Our strategy was to build a hidden Markov model (HMM) that would score proteins according to the likelihood that they encode a peptide hormone. An HMM assigns states to each amino acid in a protein sequence. Each state is associated with a probability distribution over amino acids and a set of transition probabilities to other states. Generally, these states correspond to protein sequence motifs, and as a result HMMs can be used to determine whether a protein contains a specific motif or series of motifs. The two main advantages of HMMs are the ability to handle variable length regions and the ability to integrate multiple signals in a biologically constrained manner.
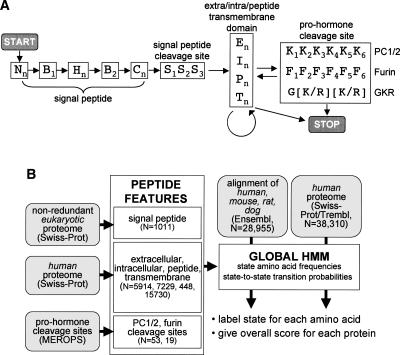
In our case, two steps were involved in using HMM for protein analysis. First, the HMM was trained on a set of proteins with well-characterized motifs in order to determine the amino acid frequencies and transition probabilities for each state. Second, the HMM was used to assign states to uncharacterized proteins and calculate a score based on how well the protein fits the HMM. The state architecture of our peptide hormone HMM is shown in Figure 1A. The HMM was assembled from three components each of which contains one or more states: (1) signal peptide, (2) extracellular/intracellular/peptide/transmembrane region, and (3) pro-hormone cleavage site. Several constraints were imposed on transitions between states so that, for example, the states for the signal peptide cleavage site (S1S2S3) had to follow the C-terminal signal peptide state (Cn). The scheme for building and running the peptide hormone HMM is shown in Figure 1B.
Figure 1.
Hidden Markov model (HMM) for the identification of peptide hormones. (A) State structure of the peptide hormone HMM with states indicated by letters and transitions between states indicated by arrows. States with numerical subscripts are single amino acid states, while states with the “n” subscript are multiple amino acid states whose length is determined by the transition probability between that state and other permitted states. Nn, B1, Hn, B2, Cn, and S1–3, are N terminus, border, hydrophobic, C terminus, and cleavage site states, respectively, of the signal peptide feature. En, In, Pn, and Tn are extracellular, intracellular, peptide, and transmembrane states, respectively, while K1–6 and F1–6 are pro-hormone cleavage site states and G[K/R][K/R] is a simple sequence motif. START and STOP mark entry and exit points of the HMM. (B) Protocol for building and running the peptide hormone HMM. HMM states for individual sequence features were built by learning amino acid frequencies and transition probabilities from sets of proteins or motifs with known features (N = size of training set). Signal peptide states were built using a previously curated set of eukaryotic SWISS-PROT proteins; extra/intra/peptide/transmembrane states were built using selected sets of human SWISS-PROT proteins; and pro-hormone cleavage sites were built using a set of PC1/2 and furin sites from the MEROPS database. The peptide hormone HMM was assembled with the state-to-state transition constraints outlined in A. Finally, the HMM was used to assign states and scores to either a set of alignments of human, mouse, rat, and dog proteins from Ensembl or a set of human proteins from SWISS-PROT/TrEMBL.
For the signal peptide feature, our state architecture was based on previous work (Nielsen and Krogh 1998; Zhang and Wood 2002) and comprised N-terminal, hydrophobic, and C-terminal states followed by a three-state cleavage site. To improve predictive accuracy, we added two intermediate boundary states (B1 and B2) (Fig. 1A). Frequencies and transition probabilities for each state were derived by training the HMM on a previously curated set of 1011 signal peptide containing proteins from SWISS-PROT downloaded from the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/ftp/signalp/euksig.red).
For the extracellular/intracellular/peptide/transmembrane features, frequencies and transition probabilities were built from sets of 5914, 7229, 448, and 15,730 sequences derived from human SWISS-PROT entries annotated as “extracellular,” “cytoplasmic,” “peptide,” and “transmembrane,” respectively. These features were modeled by a single state of variable length where the length distribution was encoded by the transition probability out of the state. Because the first-order HMM formalism produces length distributions that are geometric and that may not be best suited to model actual protein feature lengths, we used a modified HMM formalism that retains the efficiency of first-order HMM, while being able to model lengths more accurately (Ramesh and Wilpon 1992).
Finally, states for the pro-hormone cleavage site feature used three different cleavage site models. The first two included 6 states each (from −6 to −1 relative to the cleavage site) and were built from a training set of 53 pro-hormone convertase 1/2 (PC1/2) cleavage sites and 19 furin cleavage sites, respectively, derived from known and predicted eukaryotic cleavage sites collected in the MEROPS database (Rawlings et al. 2006). The third cleavage site model simply required the presence of the amino acid sequence G[K/R][K/R] and was added to ensure that this common cleavage site motif was not overlooked by the other two models.
Labeling by the peptide hormone HMM was achieved using the Viterbi algorithm, and scoring was performed by the forward–backward algorithm (Rabiner 1989). To ensure the exclusion of transmembrane domain–containing proteins, a maximal score was assigned to any protein containing a transmembrane state. As a result, our algorithm was not expected to detect peptides that are proteolytically released from transmembrane-containing pro-peptides, such as occurs for tumor necrosis factor (TNF) and CX3CL1.
Screening the human proteome for novel peptide hormones
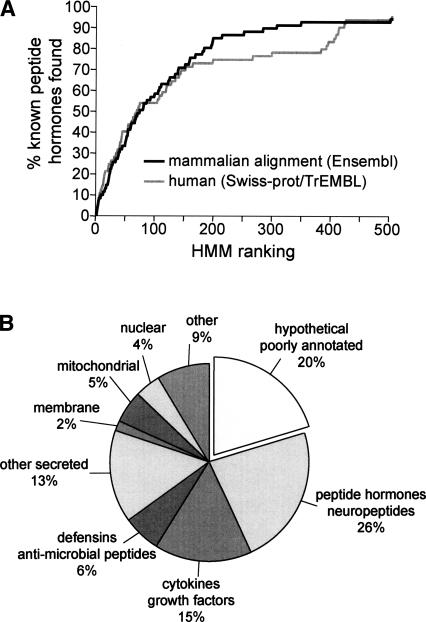
The peptide hormone HMM was applied to an alignment of nonredundant known and hypothetical proteins derived from the Ensembl database (human, mouse, rat, dog; Ntotal = 28,955), and proteins were ranked according to HMM scores. For each member of the set, multiple alignments of the human, rat, mouse, and dog orthologs were built using the Clustal W program with default settings (Thompson et al. 1994). An examination of the highest ranked sequences revealed that 90% of known peptide hormone precursors (66/75 proteins) (see Supplemental Material) were found in the top 300 proteins (Fig. 2A). An alternative HMM in which the PC1/2 and furin cleavage site models included only four states (from −4 to −1 relative to the cleavage site) performed slightly less well, returning 85% of known peptide hormones among the top 300 proteins (data not shown). Application of the peptide hormone HMM to human proteins from the SWISS-PROT/TrEMBL database (Ntotal = 38,310) was somewhat less efficient at recovering known peptide hormones, suggesting that screening the pre-aligned human proteome resulted in the recovery of fewer false positives (Fig. 2A). These findings demonstrate that our HMM successfully identified most known peptide hormones and suggests that as yet uncharacterized peptide hormones are likely to be found among the top scoring proteins in our list. The HMM Java application and supporting material are available at http://bioinfo.embl.it/.
Figure 2.
HMM successfully identified known peptide hormones. (A) Plot showing cumulative fraction of known peptide hormones (Ntotal = 77) (see Supplemental Table 1) identified among the top scoring 500 proteins; (solid line) aligned mammalian proteome as substrate; (gray line) human proteome as substrate. For the aligned proteome, 90% of known peptide hormones were found among the top 300 proteins. (B) Pie chart indicating percent composition of the top 300 proteins. Known peptide hormones, cytokines, growth factors, defensins, and other secreted proteins make up 61% of the proteins. Hypothetical and poorly annotated proteins make up 20% of the proteins and were submitted to further analysis to identify candidate novel peptide hormones.
At least 61% of the top 300 proteins belonged to several families of well-characterized secreted proteins, including peptide hormones, growth factors, cytokines, defensins, and antimicrobial peptides (Fig. 2B). A further 19% of the proteins were well-characterized membrane, mitochondrial, cytoplasmic, nuclear, or other nonsecreted proteins. The remaining 20% were hypothetical or poorly annotated proteins. In addition, we found four proteins (KISS1, TIP39, QRFP, OSTN) that had been recently reported to encode peptide hormones (Usdin et al. 1999; Ohtaki et al. 2001; Chartrel et al. 2003; Thomas et al. 2003) despite lacking annotations as processed and secreted proteins in SWISS-PROT at the time of our search (March 2004).
Next, we applied three additional criteria to the 61 hypothetical and poorly annotated proteins to determine whether novel peptide hormones might be included among this group. First, proteins in which at least one of the amino acids at each putative pro-hormone cleavage site was not conserved among orthologs were removed from the list. Second, proteins in which labeled cleavage sites formed part of a longer stretch of basic residues were removed. These regions were likely to be nuclear localization signals or other basic amino acid domains rather than pro-hormone cleavage sites. Finally, we required a significant change in amino acid homology surrounding at least one putative cleavage site. In known peptide hormones, pro-hormone cleavage sites typically separate highly from poorly conserved regions. For this calculation, a significant change was defined as >30% change in average homology index (Livingstone and Barton 1993) for the five amino acids preceding and following the putative cleavage site. Two out of 61 proteins satisfied all three criteria. These candidate peptide hormones ranked 41 and 276 in our list and were called spexin (Ensembl: ENSP00000256969) and augurin (Ensembl: ENSP00000238044), respectively. Spexin carries a SWISS-PROT annotation as containing a putative amidated peptide (http://www.expasy.org/uniprot/Q9BT56), and augurin was previously identified as a gene expressed in esophageal cancer cell lines (Su et al. 1998). However, to the best of our knowledge, our study is the first to argue that augurin encodes a peptide hormone.
Characterization of candidate peptide hormones
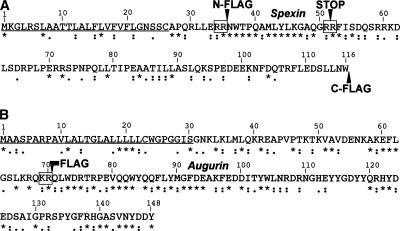
The primary structure of human spexin and augurin precursors is shown in Figure 3. Both proteins contain obvious signal peptide sequences and cleavage sites as well as at least one putative dibasic residue pro-hormone cleavage site. Spexin contains a small, 15 amino acid region flanked by putative dibasic pro-hormone cleavage sites that is highly conserved in mammals, birds, and fish (for a full alignment of spexin and augurin orthologs, see Supplemental Fig. S1). The presence of a glycine residue at the end of this putative peptide suggests that it is processed and amidated, a common feature of peptide hormones (Eipper et al. 1992). Augurin, on the other hand, contains a single putative pro-hormone cleavage site followed by a single, long putative peptide that is highly conserved in mammals and fish. Both spexin and augurin peptides contain many aromatic amino acids, a feature typical of peptide hormones. Finally, there is a significant increase in sequence conservation that coincides with the N-terminal putative pro-hormone cleavage site in both spexin and augurin, further supporting a biological role for these features.
Figure 3.
Primary structure of spexin and augurin. Sequence of spexin and augurin with the signal peptide underlined, pro-hormone cleavage sites boxed, and predicted processed peptide indicated in gray. Arrows indicate where the Flag antigen sequence (DYKDDDDK) was inserted to facilitate immunochemical detection of peptide products. Conservation among orthologs is shown below: (*) identity, (:) high homology; (·) low homology. The C-terminal glycine residue of the predicted spexin peptide is likely to be removed and the peptide amidated, a feature common to known peptide hormones. Both spexin and augurin peptides are enriched in aromatic amino acids.
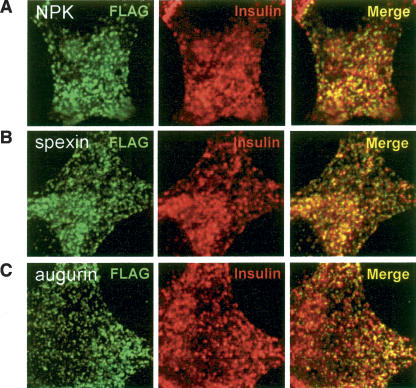
To study intracellular trafficking of spexin and augurin, the eight amino acid Flag antigen sequence (DYKDDDDK) was inserted just upstream and downstream of the putative spexin and augurin peptides (Fig. 3). As a control, Flag antigen was also inserted just upstream of neuropeptide K (NPK) in the human beta-preprotachykinin (TAC1) gene. Previous studies have shown that Flag sequences are compatible with proteolytic cleavage just N-terminal to the Flag sequence (Duguay et al. 1995). Immunocytochemistry with Flag antibodies following transfection of Flag-NPK, Flag-spexin, and Flag-augurin into a rat pancreatic cell line demonstrated colocalization of Flag antigen with endogenous insulin in punctate intracellular bodies (Fig. 4). This colocalization suggested that spexin and augurin, like neuropeptide K, underwent trafficking into dense core granules of the secretory pathway, a hallmark of peptide hormones.
Figure 4.
Colocalization of spexin and augurin with insulin in endocrine cells. Flag-tagged NPK, spexin, and augurin were transfected into rat pancreatic cells and fixed cells subjected to double immunofluorescence with Flag and insulin antibodies. (A) Neuropeptide K, (B) spexin, and (C) augurin show colocalization with insulin in small, cytoplasmic punctate structures. In all cases the majority of Flag immunoreactive puncta are also positive for insulin.
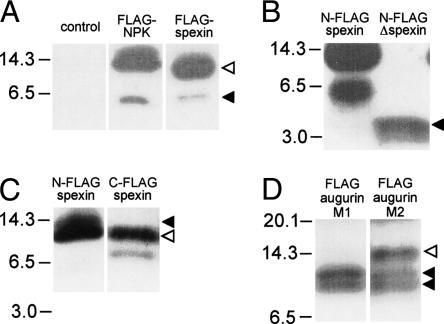
To determine whether spexin and augurin were processed and secreted, cell supernatants were collected from rat pancreatic cells transfected with Flag-NPK, N-Flag-spexin, C-Flag-spexin, and Flag-augurin. Western blotting of supernatant from N-Flag-spexin transfected cells, revealed three Flag-immunoreactive bands (13, 12, and 6 kDa), consistent with secretion of processed spexin products (Fig. 5A,B). Western blotting of supernatant from Flag-NPK transfected cells revealed processing and secretion of neuropeptide K, consistent with previous studies (Fig. 5A; Conlon et al. 1988). To determine whether the 6-kDa band could represent completely processed spexin peptide, we transfected cells with a truncated spexin protein, called N-Flag-Δspexin, in which a stop codon had been engineered to replace the C-terminal putative pro-hormone cleavage site (Fig. 3A). Western blotting of supernatants from N-Flag-Δspexin transfected cells revealed a 4-kDa band (Fig. 5B), suggesting that the 6-kDa band seen in N-Flag-spexin reflected cleavage at a site significantly C-terminal to the predicted GRR site (Fig. 3A). Processing of spexin was further assessed by Western blotting of supernatant from C-Flag-spexin transfected cells that revealed bands at 12 kDa and 8 kDa (Fig. 5C). The 12-kDa band corresponds to the 12-kDa band seen for N-Flag-spexin (Fig. 5C), while the 8-kDa band represents C-terminally cleaved spexin. The absence of the 13-kDa band for C-Flag-spexin supports the argument that processing N-terminal of spexin peptide occurred in both N-Flag- and C-Flag-spexin and that this appears to proceed more efficiently for C-Flag-spexin.
Figure 5.
Identification of spexin and augurin in cell supernatants. Vector control and Flag-tagged NPK, spexin, and augurin were transfected into rat pancreatic cells in culture, and cell supernatants were harvested and submitted to immunoblotting with a Flag antibody. (A) Supernatants from Flag-NPK and N-Flag-spexin transfected cells contained high (solid arrow) and low (open arrow) mobility bands that reflected processing of Flag-tagged products from these constructs. (B) Supernatant from N-Flag-Δspexin transfected cells contained a 4-kDa band, suggesting that the 6-kDa product seen for N-Flag-spexin was the result of cleavage significantly C-terminal to the spexin peptide. (C) Supernatant from C-Flag-spexin contained two bands (12 and 8 kDa), confirming C-terminal cleavage of spexin pro-peptide. A 12-kDa product is seen for both N-Flag and C-Flag spexin (open arrow), while a 13-kDa product is seen only in N-Flag-spexin and corresponds to incompletely N-terminally processed spexin (closed arrow). (D) The presence of two bands (10 and 8 kDa, solid arrows) in supernatant from Flag-augurin transfected cells probed with M1 Flag antibody demonstrated cleavage of augurin at the putative pro-hormone cleavage site as well as close to the C terminus of the pro-peptide. The same immunoblot probed with M2 Flag antibody revealed an additional low-mobility product, confirming cleavage at the predicted dibasic cleavage site immediately adjacent to the Flag tag (open arrow).
Western blotting of supernatant from Flag-augurin transfected cells revealed a pair of Flag-immunoreactive bands consistent with secretion of the pro-peptide and a processed variant (10 and 8 kDa) (Fig. 5D). Recognition of the Flag-augurin products by a Flag antibody that binds only N-terminal Flag antigen (M1) suggests that cleavage occurred at the predicted dibasic cleavage site just upstream of the Flag tag and supports the argument that the 8-kDa band reflects cleavage at a site near the C terminus of augurin. We speculate that this cleavage may occur at the noncanonical cleavage motif surrounding Arg132 (Fig. 3A). As expected, immunoblotting of the same supernatant with a Flag antibody that recognizes both N-terminal and embedded Flag epitopes (M2) revealed a high-molecular-weight product not recognized by the M1 antibody and corresponding to the full-length pro-peptide (Fig. 5D). Secretion and processing of Flag-augurin was confirmed in a second rat pancreatic cell line, RINm5f, that expresses high levels of insulin and forms distinct β-islet-like cell clusters (data not shown). These findings demonstrate that both spexin and augurin are processed and secreted when expressed in endocrine cells.
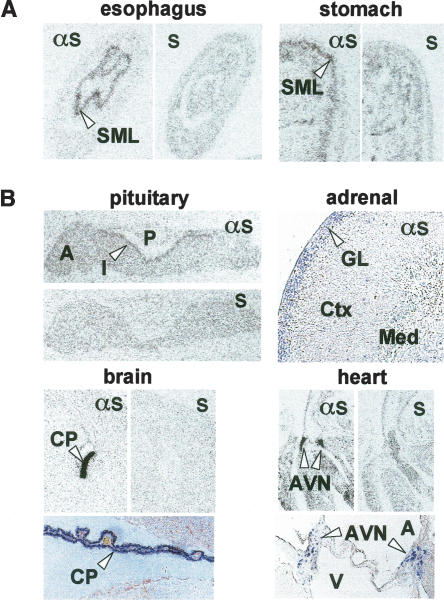
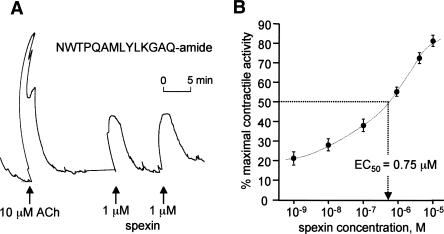
In situ hybridization localized spexin mRNA to the submucosal layer of esophagus and stomach fundus (Fig. 6A), a tissue containing the submucous plexus of the enteric nervous system and known to express several peptide hormones (e.g., gastrin-releasing peptide, vasoactive intestinal peptide) involved in the control of smooth muscle contractility (Costa et al. 2000). To examine whether the predicted peptide product of spexin could moderate smooth muscle contractility, a synthetic amidated spexin peptide, NWTPQAMLYLKGAQ-amide (Fig. 3A), was tested in a stomach explant contractility assay (Severini et al. 2000). The spexin peptide dose-dependently induced contraction of stomach muscle with an EC50 of 0.75 μM (Fig. 7). These findings demonstrate a biological activity for spexin and strongly support our hypothesis that spexin is a novel peptide hormone.
Figure 6.
Expression of spexin and augurin mRNA in mouse tissues. (A) In situ hybridization with antisense (αs) and sense (s) probes detected spexin mRNA in the submucosal layer (SML) of the esophagus and stomach fundus. (B) In situ hybridization with antisense (αs) and sense (s) probes detected augurin mRNA in the intermediate lobe (I) of the pituitary (A, anterior; P, posterior), glomerular layer (GL) of the adrenal cortex (Ctx, cortex; Med, medulla), choroid plexus (CP), and atrio-ventricular node of the heart (AVN) (A, aorta; V, ventricle).
Figure 7.
Spexin is a biologically active peptide hormone. (A) Representative muscle contractile response to 10 μM acetylcholine (ACh) and 1 μM spexin peptide (NWTPQAMLYLKGAQ-amide) in a rat stomach explant assay. Repeated administration of spexin peptide produced similar contactile responses. (B) Cumulative dose-response curve for contractile activity of spexin peptide on rat stomach explants (EC50 = 0.75 μM, N = 6). Error bars indicate standard error.
In situ hybridization revealed prominent augurin expression in mouse endocrine tissues, including the intermediate lobe of the pituitary, glomerular layer of the adrenal cortex, choroid plexus, and atrio-ventricular node of the heart (Fig. 6B). The intermediate lobe of the pituitary contains melanotrophs that produce alpha-melanocyte-stimulating hormone and beta-endorphin and whose role in mammalian physiology remains poorly understood (Mains and Eipper 1979; Saland 2001), while the glomerular layer of the adrenal cortex produces aldosterone and is involved in the regulation of salt homoeostasis (Connell and Davies 2005). In the heart, augurin mRNA localizes to a distinct set of cells that lie on either side of the ventricular valve and are likely to contribute to the cardiac conduction system. In situ hybridization on embryonic day 18.5 (E18.5) mouse revealed augurin mRNA in adrenal cortex, choroid plexus, and bone (data not shown). The expression pattern of augurin suggests that it is likely to express a secreted protein with a role in the modulation of salt and energy homeostasis, cardiovascular function, and cerebral spinal fluid composition.
Discussion
We have used a sequence-based approach to identify two candidate novel peptide hormones, which we called spexin and augurin. Both spexin and augurin were colocalized with insulin in the secretory pathway and were processed and secreted following transfection in endocrine cells. Furthermore, both spexin and augurin mRNA were expressed in endocrine tissues, and a predicted spexin peptide induced smooth muscle contractility in a stomach explant assay. Our findings confirm that most previously identified peptide hormones in the human proteome can be identified using a sequence-based screening approach. Our discovery of two novel peptide hormones suggests that our method is useful for the systematic screening of proteomes for biologically active peptides.
Several factors are likely to have prevented us from identifying additional candidate peptide hormones. First, we based our search on annotated protein databases that depend heavily on ESTs and full-length cDNAs for gene prediction. Given the poor expression level and restricted expression pattern of many known peptide hormones, it is possible that some peptide hormone genes are not present in these databases. Second, we decided to focus on hypothetical or poorly annotated proteins and did not apply our HMM to search for novel peptides produced by previously characterized, well-known genes. Recently, the peptide hormones obestatin and salusin were discovered to be produced by the ghrelin (Zhang et al. 2005) and Torsin 2A (Shichiri et al. 2003) genes, respectively, and further examples of such peptide hormone symbiosis are likely to exist. Finally, we used stringent criteria based on sequence conservation to identify spexin and augurin from among 61 high scoring candidates. Many proteins in this group failed the criterion requiring a shift in conservation across at least one cleavage site simply because they lacked distant orthologs and thus sufficient information to draw conclusions based on sequence conservation. Thus, it is possible that additional peptide hormones were overlooked among the top 61 high scoring proteins.
We believe that the HMM approach presented here could be extended to provide better sensitivity and specificity. First, the peptide hormone HMM could be combined with a DNA sequence HMM to create a peptide hormone-specific gene prediction method. Second, our use of orthology information was somewhat ad hoc, and integrating protein homology data internally into each state in the manner of phylogenetic HMMs in DNA sequence (Pedersen and Hein 2003; Siepel et al. 2005) could be envisioned. Third, it is clear that our background model (in this case, a simple one-state distribution of average amino acid content) is not rich enough to capture other features in real protein sequences, which may mislead the HMM. Nevertheless, this initial HMM, coupled with some downstream computational screens, has already provided several candidates for further biochemical screens and compares favorably to other experimental screening approaches.
Although there is considerable scope for improvement of the HMM, our initial results suggest that there is a low number (<15) of undiscovered peptide hormone precursors in the existing set of cDNA- and EST-supported genes (26% of 61 hypothetical or poorly annotated top scoring proteins) (see Fig. 2B). A more sophisticated HMM with less reliance on cDNA/EST based predictions will allow us to more confidently establish whether we have captured most peptide hormones with this biological model. The combination of computational screens and targeted biochemical verification will be a main route for further discoveries of peptide hormones.
Methods
Bioinformatics
Protein sequence data sets for the training of HMM states were retrieved from public databases using SRS (Sequence Retrieval System, http://www.expasy.org/srs5/) and Perl scripts—signal peptide: a previously curated set of 1011 nonredundant eukaryotic signal peptide-containing proteins (http://www.cbs.dtu.dk/ftp/signalp/euksig.red); extracellular: 5914 human SWISS-PROT entries with FtDescription = “extracellular”; intracellular: 7229 human SWISS-PROT entries with FtDescription = “cytoplasmic”; peptide: 448 human SWISS-PROT entries with FtKey = “peptide”; transmembrane: 15,730 human SWISS-PROT entries with FtKey = “transmem.” Signal peptides were aligned using their hydrophobic and predicted cleavage sites features using a custom Perl script. The hydrophobic region was defined as the stretch of amino acids where the number of hydrophobic residues (AILFVMWY)/length was maximal. Amino acid frequencies and lengths for the signal peptide states were derived from this alignment. For pro-hormone convertase 1/2 and furin cleavage sites, data sets were retrieved from the MEROPS database and aligned at the cleavage site using a custom Perl script. Amino acid frequencies and lengths for the other feature states were directly derived from the relevant protein sets. This information was used to build the observation and transition matrices. Labeling and scoring were performed using Viterbi and forward–backward algorithms (Rabiner 1989), respectively, in Java. For the In, En, Pn, and Tn states, we modified the Viterbi algorithm to allow transition probabilities to depend on current state duration. This modification enabled us to model nongeometric transition probabilities (Ramesh and Wilpon 1992). Selection of candidate peptide hormones from the top 300 proteins was carried out by hand with the aid of a custom Java tool that displayed the score and assigned states of each protein. All custom scripts are available at http://bioinfo.embl.it/.
Cell culture and secretion assays
Unless otherwise noted, all cell culture was carried out in rat pancreatic β-TC3 cells (Efrat et al. 1988) in growing media (DMEM, 15% horse serum, 2.5% FBS). Forty-eight hours after transfection, cells were switched to serum-free RPMI media, and supernatant was collected for 24 h. In the case of Figure 5A, growing media was replaced immediately following transfection, and supernatant was collected for 48 h and immunoprecipitated with Flag antibodies. RINm5f cells (Gazdar et al. 1980) were grown in RPMI, 10% FBS. Supernatants were precipitated with acetone prior to immunoblotting. Transfection was carried out using Lipofectamine 2000 (Invitrogen) with transfection efficiency controlled by spiking 1:20 with a GFP expression plasmid (pLP-EGFP-C1 plasmid; Clontech). Human spexin (IMAGp958N21321), mouse augurin (IRAVp968F095D6), and human TAC1 (IRATp970E0722D6) cDNA were obtained from RZPD. Flag-tagged expression constructs were designed with the Flag sequence (DYKDDDDK) inserted precisely at the beginning or end of the putative processed peptide. For neuropeptide K, Flag was inserted at residue 72 just before the first amino acid of neuropeptide K. For Flag-Δspexin, a stop codon was engineered just following the glycine residue of the putative spexin peptide. M2, and where indicated M1, Flag antibody was used (Sigma).
Immunocytochemistry
Rat pancreatic RINm5f cells were cultured in serum-containing RPMI medium, transfected with Flag-tagged NPK, spexin, and augurin, and grown for 48 h before fixation. Double fluorescent immunolabelling was performed with M2 Flag (Sigma) and insulin antibodies (Dako) following established protocols and visualized by confocal microscopy. Goat anti-mouse Alexa-488 and Goat anti-guinea pig Alexa-568 (Invitrogen) secondary antibodies were used.
In situ hybridization
Tissues and E18.5 embryos were dissected, fixed overnight in 4% paraformaldehyde, and embedded in paraffin. In situ hybridization using digoxigenin-labeled or 35S-CTP-labeled probes on 8-μm paraffin sections was performed according to procedures previously described (Neubuser et al. 1995; Niederreither and Dolle 1998). Briefly, sections were dewaxed, rehydrated, digested with proteinase K, and hybridized with probe at 65°C. Post-hybridization washes in 20% formamide, 0.5× SSC were done at 60°C. The spexin probe was a 0.3-kb cDNA fragment cloned from mouse brain RNA (primers: 5′-ACAGGGTCGGAACATGAAGGG, 3′-AAGAGTCTGTCTTCCAAGAGTTCGC). The augurin probe was a 0.4-kb fragment amplified from mouse adrenal RNA (primers: 5′-CACCATGAGCACCTCGTCTGCG, 3′-TCTGTGGGCACC TCAGGG).
Explant assay
Albino Wistar female rats (250–350 g; Charles River) were sacrificed by inspiration of 75% CO2, and stomach fundus muscles strips were isolated, washed in fresh Tyrode’s solution (137 mM NaCl, 5.4 mM KCl, 0.5 mM MgCl2, 1.8 mM CaCl2, 10 mM glucose, 11.9 mM NaHCO3, 0.4 mM NaH2PO4 at pH 7.4), mounted vertically in a 5-mL organ bath in oxygenated (95% O2, 5% CO2) Tyrode’s solution, and maintained at 37°C. The segments were stretched to a tension of 2.0 g and allowed to equilibrate for 30–60 min, with the superfusion buffer changed every 15–20 min. At the beginning of each experiment, acetylcholine chloride (ACh 10−5 M) was applied to achieve a maximal control contraction. The potency of contractions was recorded isometrically by a strain gauge transducer (DY 1; Ugo Basile) and displayed on a recording microdynamometer (Unirecord; Ugo Basile). When reproducible responses to ACh were obtained, increasing concentrations (from 10−9 to 10−5 M) of synthetic amidated spexin peptide (NWTPQAMLYLKGAQ-amide; Primm) were applied every 2 min to establish a cumulative dose-response curve followed by washing and recovery for minimum 20 min. The EC50 was calculated by interpolation from the cumulative dose-response curve. Consecutively, single doses of spexin 10−6 M were applied until reproducible responses were obtained.
Acknowledgments
We thank W. Witke and F. Jönsson for antibodies and immunocytochemistry expertise, E. Lara-Pezzi for help with cell culture, D. Tosh for the gift of RINm5f cells, and S. Kang and P. Pilo Boyl for helpful suggestions and discussions. This work was supported by funds from the European Commission (N.R.). Manuscript charges were covered by EMBL.
Footnotes
[Supplemental material is available online at www.genome.org and at http://bioinfo.embl.it/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5755407
References
- Baggerman G., Liu F., Wets G., Schoofs L., Liu F., Wets G., Schoofs L., Wets G., Schoofs L., Schoofs L. Bioinformatic analysis of peptide precursor proteins. Ann. N. Y. Acad. Sci. 2005;1040:59–65. doi: 10.1196/annals.1327.006. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. Genewise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Menendez E., Fasciolo J.C., Leloir L.F., Muñoz J.M., Fasciolo J.C., Leloir L.F., Muñoz J.M., Leloir L.F., Muñoz J.M., Muñoz J.M. Hypertensin: The substance causing renal hypertension. Nature. 1939;144:980–981. [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burgus R., Dunn T.F., Desiderio D., Guillemin R., Dunn T.F., Desiderio D., Guillemin R., Desiderio D., Guillemin R., Guillemin R. Molecular structure of the hypothalamic hypophysiotropic TRF factor of ovine origin: Mass spectrometry demonstration of the PCA-His-Pro-NH2 sequence. C. R. Hebd. Seances Acad. Sci. 1969;269:1870–1873. [PubMed] [Google Scholar]
- Chartrel N., Dujardin C., Youssef Anouar J.L., Decker A., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Dujardin C., Youssef Anouar J.L., Decker A., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Youssef Anouar J.L., Decker A., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Decker A., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Vandesande F., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Llorens-Cortes C., Costentin J., Beauvillain J.-C., Costentin J., Beauvillain J.-C., Beauvillain J.-C., et al. Identification of 26RFa, a hypothalamic neuropeptide of the RFamide peptide family with orexigenic activity. Proc. Natl. Acad. Sci. 2003;100:15247–15252. doi: 10.1073/pnas.2434676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon J.M., Deacon C.F., Grimelius L., Cedermark B., Murphy R.F., Thim L., Creutzfeldt W., Deacon C.F., Grimelius L., Cedermark B., Murphy R.F., Thim L., Creutzfeldt W., Grimelius L., Cedermark B., Murphy R.F., Thim L., Creutzfeldt W., Cedermark B., Murphy R.F., Thim L., Creutzfeldt W., Murphy R.F., Thim L., Creutzfeldt W., Thim L., Creutzfeldt W., Creutzfeldt W. Neuropeptide K-(1-24)-peptide: Storage and release by carcinoid tumors. Peptides. 1988;9:859–866. doi: 10.1016/0196-9781(88)90134-9. [DOI] [PubMed] [Google Scholar]
- Connell J.M.C., Davies E., Davies E. The new biology of aldosterone. J. Endocrinol. 2005;186:1–20. doi: 10.1677/joe.1.06017. [DOI] [PubMed] [Google Scholar]
- Costa M., Brookes S.J.H., Hennig G.W., Brookes S.J.H., Hennig G.W., Hennig G.W. Anatomy and physiology of the enteric nervous system. Gut. 2000;47:iv15–iv19. doi: 10.1136/gut.47.suppl_4.iv15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P., Brunak S., Blom N., Brunak S., Blom N., Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Duguay S.J., Lai-Zhang J., Steiner D.F., Lai-Zhang J., Steiner D.F., Steiner D.F. Mutational analysis of the insulin-like growth factor 1 prohomone processing site. J. Biol. Chem. 1995;270:17566–17574. doi: 10.1074/jbc.270.29.17566. [DOI] [PubMed] [Google Scholar]
- Efrat S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Grant S., Hanahan D., Baekkeskov S., Hanahan D., Baekkeskov S., Baekkeskov S. β-Cell lines derived from transgenic mice expressing a hybrid insulin gene oncogene. Proc. Natl. Acad. Sci. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper B.A., Stoffer D.A., Mains R.E., Stoffer D.A., Mains R.E., Mains R.E. The biosynthesis of neuropeptides: Peptide α-amidation. Annu. Rev. Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- EUKSIG Center for Biological Sequence Analysis . [Google Scholar]
- Finn R.D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Moxon S., Marshall M., Khanna A., Durbin R., Marshall M., Khanna A., Durbin R., Khanna A., Durbin R., Durbin R., et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar A.F., Chick W.L., Oie H.K., Sims H.L., King D.L., Weir G.C., Lauris V., Chick W.L., Oie H.K., Sims H.L., King D.L., Weir G.C., Lauris V., Oie H.K., Sims H.L., King D.L., Weir G.C., Lauris V., Sims H.L., King D.L., Weir G.C., Lauris V., King D.L., Weir G.C., Lauris V., Weir G.C., Lauris V., Lauris V. β-Cell lines derived from transgenic mice expressing a hybrid insulin gene oncogene. Proc. Natl. Acad. Sci. 1980;6:3519–3523. [Google Scholar]
- Hinuma S., Shintani Y., Fukusumi S., Iijima N., Matsumoto Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Shintani Y., Fukusumi S., Iijima N., Matsumoto Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Fukusumi S., Iijima N., Matsumoto Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Iijima N., Matsumoto Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Matsumoto Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Hosoya M., Fujii R., Watanabe T., Kikuchi K., Terao Y., Fujii R., Watanabe T., Kikuchi K., Terao Y., Watanabe T., Kikuchi K., Terao Y., Kikuchi K., Terao Y., Terao Y., et al. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat. Cell Biol. 2000;400:703–708. doi: 10.1038/35036326. [DOI] [PubMed] [Google Scholar]
- Hökfelt T. Neuropeptides in perspective: The last ten years. Neuron. 1991;7:867–879. doi: 10.1016/0896-6273(91)90333-u. [DOI] [PubMed] [Google Scholar]
- Hsu S.Y. Cloning of two novel mammalian paralogs of relaxin/insulin family proteins and their expression in testis and kidney. Mol. Endocrinol. 1999;13:2163–2174. doi: 10.1210/mend.13.12.0388. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Luo L., Gustafson E.L., Yadav D., Laverty M., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Luo L., Gustafson E.L., Yadav D., Laverty M., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Gustafson E.L., Yadav D., Laverty M., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Yadav D., Laverty M., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Laverty M., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Murgolo N., Vassileva G., Zeng M., Laz T.M., Behan J., Vassileva G., Zeng M., Laz T.M., Behan J., Zeng M., Laz T.M., Behan J., Laz T.M., Behan J., Behan J., et al. Identification and characterization of a novel RF-amide peptide ligand for orphan G-protein-coupled receptor SP9155. J. Biol. Chem. 2003;278:27652–27657. doi: 10.1074/jbc.M302945200. [DOI] [PubMed] [Google Scholar]
- Kastin A., editor. Handbook of biologically active peptides. Academic Press; New York: 2006. [Google Scholar]
- Katafuchi T., Kikumoto K., Hamano K., Kangawa K., Matsuo H., Minamino N., Kikumoto K., Hamano K., Kangawa K., Matsuo H., Minamino N., Hamano K., Kangawa K., Matsuo H., Minamino N., Kangawa K., Matsuo H., Minamino N., Matsuo H., Minamino N., Minamino N. Calcitonin receptor-stimulating peptide, a new member of the calcitonin gene-related peptide family. J. Biol. Chem. 2003;278:12046–12054. doi: 10.1074/jbc.M207970200. [DOI] [PubMed] [Google Scholar]
- Krogh A., Brown M., Mian I.S., Sjölander K., Haussler D., Brown M., Mian I.S., Sjölander K., Haussler D., Mian I.S., Sjölander K., Haussler D., Sjölander K., Haussler D., Haussler D. Hidden Markov models in computational biology. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- Livingstone C., Barton G., Barton G. Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Mains R.E., Eipper B.A., Eipper B.A. Synthesis and secretion of corticotropins, melanotropins, and endorphins by rat intermediate pituitary cells. J. Biol. Chem. 1979;16:7885–7894. [PubMed] [Google Scholar]
- Neubuser A., Koseki H., Balling R., Koseki H., Balling R., Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Proc. Natl. Acad. Sci. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Niederreither K., Dolle P., Dolle P. In situ hybridization with 35S-labeled probes for retinoid receptors. Methods Mol. Biol. 1998;89:247–267. doi: 10.1385/0-89603-438-0:247. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Krogh A., Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G., Engelbrecht J., Brunak S., von Heijne G., Brunak S., von Heijne G., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Matsumoto H., Hori A., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Hori A., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Kanehashi K., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Yasuko Terao S.K., Takatsu Y., Masuda Y., Ishibashi Y., Takatsu Y., Masuda Y., Ishibashi Y., Masuda Y., Ishibashi Y., Ishibashi Y., et al. Metastasis suppressor gene KISS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Park Y., Kim Y.-J., Adams M.E., Kim Y.-J., Adams M.E., Adams M.E. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand–receptor coevolution. Proc. Natl. Acad. Sci. 2002;99:11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Hein J., Hein J. Gene finding with a hidden Markov model of genome structure and evolution. Bioinformatics. 2003;19:219–227. doi: 10.1093/bioinformatics/19.2.219. [DOI] [PubMed] [Google Scholar]
- Rabiner L.R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Ramesh P., Wilpon J.G., Wilpon J.G. IEEE International Conference on Acoustics, Speech, and Signal Processing ICASSP-92. Vol. 1. San Francisco, CA: 1992. Modeling state durations in hidden Markov models for automatic speech recognition; pp. 381–384. [Google Scholar]
- Rawlings N.D., Morton F.R., Barrett A.J., Morton F.R., Barrett A.J., Barrett A.J. The peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saland L.C. The mammalian pituitary intermediate lobe: An update on innervation and regulation. Brain Res. Bull. 2001;54:587–593. doi: 10.1016/s0361-9230(01)00471-3. [DOI] [PubMed] [Google Scholar]
- Schmidt W.E., Kratzin H., Eckart K., Drevs D., Mundkowski G., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Kratzin H., Eckart K., Drevs D., Mundkowski G., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Eckart K., Drevs D., Mundkowski G., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Drevs D., Mundkowski G., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Mundkowski G., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Clemens A., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Katsoulis S., Schäfer H., Gallwist B., Kreutzfeldt W., Schäfer H., Gallwist B., Kreutzfeldt W., Gallwist B., Kreutzfeldt W., Kreutzfeldt W., et al. Isolation and primary structure of pituitary human galanin, a 30-residue non-amidated neuropeptide. Proc. Natl. Acad. Sci. 1991;88:11435–11439. doi: 10.1073/pnas.88.24.11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severini C., Salvadori S., Guerrini R., Falconieri-Erspamer G., Mignogna G., Erspamer V., Salvadori S., Guerrini R., Falconieri-Erspamer G., Mignogna G., Erspamer V., Guerrini R., Falconieri-Erspamer G., Mignogna G., Erspamer V., Falconieri-Erspamer G., Mignogna G., Erspamer V., Mignogna G., Erspamer V., Erspamer V. Parallel bioassay of 39 tachykinins on 11 smooth muscle preparations. Structure and receptor selectivity/affinity relationship. Peptides. 2000;21:1587–1595. doi: 10.1016/s0196-9781(00)00290-4. [DOI] [PubMed] [Google Scholar]
- Shichiri M., Ishimaru S., Ota T., Nishikawa T., Isogai T., Hirata Y., Ishimaru S., Ota T., Nishikawa T., Isogai T., Hirata Y., Ota T., Nishikawa T., Isogai T., Hirata Y., Nishikawa T., Isogai T., Hirata Y., Isogai T., Hirata Y., Hirata Y. Salusins: Newly identified bioactive peptides with hemodynamic and mitogenic activities. Nat. Med. 2003;9:1166–1172. doi: 10.1038/nm913. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner D.F. The preprotein convertases. Curr. Opin. Chem. Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- Su T., Liu H., Lu S., Liu H., Lu S., Lu S. Cloning and identification of cDNA fragments related to human esophageal cancer. Zhonghua Zhong Liu Za Zhi. 1998;20:254–257. [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Moffatt P., Salois P., Gaumond M.-H., Gingras R., Godin E., Miao D., Goltzman D., Lanctot C., Moffatt P., Salois P., Gaumond M.-H., Gingras R., Godin E., Miao D., Goltzman D., Lanctot C., Salois P., Gaumond M.-H., Gingras R., Godin E., Miao D., Goltzman D., Lanctot C., Gaumond M.-H., Gingras R., Godin E., Miao D., Goltzman D., Lanctot C., Gingras R., Godin E., Miao D., Goltzman D., Lanctot C., Godin E., Miao D., Goltzman D., Lanctot C., Miao D., Goltzman D., Lanctot C., Goltzman D., Lanctot C., Lanctot C. Osteocrin, a novel bone-specific secreted protein that modulates the osteoblast phenotype. J. Biol. Chem. 2003;278:50563–50571. doi: 10.1074/jbc.M307310200. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin T.B., Hoare S.R.J., Wang T., Mezey E., Kowalak J.A., Hoare S.R.J., Wang T., Mezey E., Kowalak J.A., Wang T., Mezey E., Kowalak J.A., Mezey E., Kowalak J.A., Kowalak J.A. TIP39: A new neuropeptide and PTH2-receptor agonist from hypothalamus. Nat. Neurosci. 1999;2:941–943. doi: 10.1038/14724. [DOI] [PubMed] [Google Scholar]
- Vassilatis D.K., Hohmann J.G., Zeng H., Li F., Ranchalis J.E., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Hohmann J.G., Zeng H., Li F., Ranchalis J.E., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Zeng H., Li F., Ranchalis J.E., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Li F., Ranchalis J.E., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Ranchalis J.E., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Marty T., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Mortrud A.B., Rodriguez S.S., Weller J.R., Wright A.C., Rodriguez S.S., Weller J.R., Wright A.C., Weller J.R., Wright A.C., Wright A.C., et al. The G-protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wood W.I., Wood W.I. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics. 2002;19:307–308. doi: 10.1093/bioinformatics/19.2.307. [DOI] [PubMed] [Google Scholar]
- Zhang J.V., Ren P.-G., Avsian-Kretchmer O., Luo C.-W., Rauch R., Klein C., Hsueh A.J.W., Ren P.-G., Avsian-Kretchmer O., Luo C.-W., Rauch R., Klein C., Hsueh A.J.W., Avsian-Kretchmer O., Luo C.-W., Rauch R., Klein C., Hsueh A.J.W., Luo C.-W., Rauch R., Klein C., Hsueh A.J.W., Rauch R., Klein C., Hsueh A.J.W., Klein C., Hsueh A.J.W., Hsueh A.J.W. Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science. 2005;310:996–999. doi: 10.1126/science.1117255. [DOI] [PubMed] [Google Scholar]