Abstract
High-density SNP microarrays provide insight into the genomic events that occur in diseases like cancer through their capability to measure both LOH and genomic copy numbers. Where currently available methods are restricted to the use of fresh frozen tissue, we now describe the design and validation of copy number measurements using the Illumina BeadArray platform and the application of this technique to formalin-fixed, paraffin-embedded (FFPE) tissue. In fresh frozen tissue from a set of colorectal tumors with numerous chromosomal aberrations, our method measures copy number patterns that are comparable to values from established platforms, like Affymetrix GeneChip and BAC array-CGH. Moreover, paired comparisons of fresh frozen and FFPE tissues showed nearly identical patterns of genomic change. We conclude that this method enables the use of paraffin-embedded material for research into both LOH and numerical chromosomal abnormalities. These findings make the large pathological archives available for genomic analysis, which could be especially relevant for hereditary disease where fresh material from affected relatives is rarely available.
Genomic copy number variations (CNVs) and allelic imbalances are common characteristics of cancer and other diseases (Rajagopalan and Lengauer 2004; Pinkel and Albertson 2005). For the detection of these features, different microarray technologies have been used, such as classical CGH, BAC array-based comparative genomic hybridization (array-CGH), cDNA array-CGH, and high-density single-nucleotide polymorphism (SNP) arrays (Kallioniemi et al. 1993; Pinkel et al. 1998; Pollack et al. 1999; Lindblad-Toh et al. 2000; Primdahl et al. 2002; Bignell et al. 2004; Janne et al. 2004). These techniques allow high-resolution mapping of deletions and amplifications, and eventually identification of the underlying disease-causing genes, as was recently demonstrated for the MITF gene in malignant melanoma (Garraway et al. 2005). In addition to CNV analysis, only SNP arrays offer the benefit of detecting Loss of Heterozygosity (LOH) (Zhou et al. 2004b) and, consequently, copy neutral mitotic recombination (Bignell et al. 2004). Moreover, the combination of CNV and LOH status with the parental origin of the aberrant allele could lead to the identification of the genes involved in hereditary cancer (Mao et al. 1999; Tomlinson et al. 1999).
Genome-wide SNP array CNV and LOH profiles have been reported for two different SNP typing platforms: Affymetrix GeneChip arrays and Illumina BeadArrays (Oliphant et al. 2002; Matsuzaki et al. 2004; Lips et al. 2005; Shen et al. 2005). The profiles were generated for several cancers, including breast, colorectal, and lung cancers, and for several cancer cell lines (Lindblad-Toh et al. 2000; Primdahl et al. 2002; Dumur et al. 2003; Bignell et al. 2004; Janne et al. 2004; Zhao et al. 2004; Zhou et al. 2004a; Lips et al. 2005; Irving et al. 2005). Both platforms were originally designed for high-throughput genotyping. After array hybridization, thousands of SNP genotypes are extracted from allele-specific signal intensities. The underlying methodologies of the platforms, however, are fundamentally different. The GeneChip whole-genome sampling assay (WSGA) (Kennedy et al. 2003) is based on restriction enzyme digestion of high-quality genomic DNA, followed by linker adapter ligation and PCR. The GoldenGate assay for BeadArrays, on the other hand, is based on allele-specific primer extension directly on genomic DNA with primers directly surrounding the SNP. Subsequent ligation generates allele-specific artificial PCR templates (Fan et al. 2003). This requires only short intact genomic segments of ∼40 bp flanking each SNP of interest. Consequently, the GoldenGate assay can be used with partially degraded DNA, and we have shown that it is suitable for reliable genotyping and LOH detection on DNA from archival formalin-fixed, paraffin-embedded (FFPE) tissue when compared to fresh frozen tumors and leukocyte DNA (Lips et al. 2005). Although the generation of copy number and LOH profiles from FFPE DNA has been reported for GeneChips, concordance was low and the signal showed high variability (Thompson et al. 2005).
In this study, we have developed a method to measure DNA copy numbers from FFPE tumors on Illumina BeadArrays and compared the outcome to copy number profiles from fresh frozen tumors. Tumors from different hospitals were included, from which both normal and tumor FFPE tissue, fresh frozen tumor, and normal leukocyte DNA were available. We determined reliability and reproducibility for all types of tissue and compared copy number patterns from fresh frozen tumor with FFPE tumor.
For the reliable detection of regions with CNVs, accurate normalization algorithms are essential to identify only real aberrations. For GeneChips, several algorithms have been reported (Lieberfarb et al. 2003; Lin et al. 2004; Herr et al. 2005; Ishikawa et al. 2005; Nannya et al. 2005). In order to analyze the BeadArray data, we developed an algorithm for normalization and representation of the copy number and LOH profiles. These were validated by comparison with 10K SNP GeneChip arrays and a 3700 probe BAC array.
We show here that the signal intensity values for BeadArrays can be used to create reliable copy number profiles from FFPE colorectal tumors with very high reproducibility between experiments, high concordance with frozen tissue from the same tumor, and a high degree of agreement to other methods.
Results
Copy number calculations
Genotypes from BeadArrays were computated from allele-specific signal intensities (Fan et al. 2003). For the construction of copy number profiles, we explored the data properties of the Illumina BeadArrays. We studied the issues of background correction, normalization, and combination of allele-specific signals into a single, locus-specific copy number. Finally, our findings were validated by comparing results from frozen and FFPE tumors and performing cross-platform comparisons with GeneChip and BAC arrays.
Channel properties
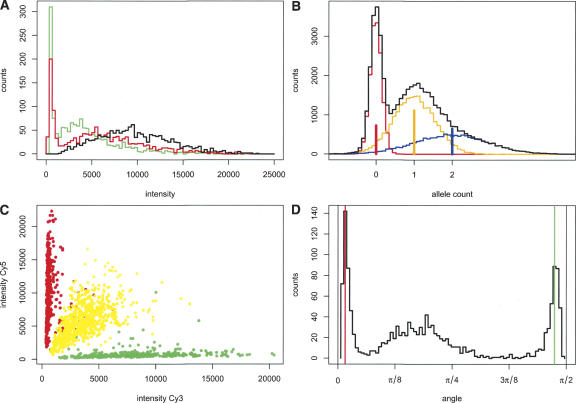
Figure 1A shows the differences in the distributions of the signals in the red (Cy5) and green (Cy3) channels. The median of the red signal is almost twice that of the green signal. This is a dye effect since the proportions of alleles per dye are nearly equal. Therefore, we treated the channels separately and tested the effect of normalization between the channels within the samples.
Figure 1.
Exploration of BeadArray signal properties. (A) Example of the distributions of the green (Cy3) and red (Cy5) fluorescent signals from a normal sample. The combined intensity (sum of both alleles of a probe) is shown in black. Note that the combined signal is not simply the addition of both signals because of the reciprocal relationship between the alleles. (B) Simulation of the distribution of single channel signal intensities derived from SNPs with zero (red), one (orange), and two (blue) copies of an allele. The frequency for each allele type was taken from the red signal of panel A. The separate distributions were modeled as Gaussian distributions with mean = copy number and SD = copy number × 0.4 + 0.15. The black line indicates the combined signal distribution plot. (C) Scatter plot of raw intensities for homozygous (red and green) and heterozygous (yellow) SNPs. (D) Histogram of raw intensities in polar coordinates. The red line indicates the most prevalent angle for homozygous Cy5-labeled SNPs; the green line, homozygous Cy3-labeled SNPs.
Background correction
Background correction is often an essential step in data processing. Since the scanning software does not provide direct estimates of background intensity for each measurement, we tested three types of background estimation based on observations of the signal properties.
First, the background intensity can be estimated as the minimal signal intensity in a channel. Second, the first mode of the intensity histogram can be used. In samples without CNVs, SNPs have three possible states per allele: zero, one, or two copies. For probes that have one or two copies in the sample under investigation, the variability of the measured intensity is determined by the PCR, hybridization, the measurement properties of that probe, and noise. For probes that have zero copies, the variability is only determined by noise. A simulation of this model, with Gaussian distributions for probe properties and noise, is shown in Figure 1B. The distribution of the zero alleles shows a narrow, distinguishable peak, implying that the first mode of the signal can be used as an estimation of the background signal.
The third approach is based on the observation that the population of homozygous SNPs is slightly slanted inward on a scatter plot (Fig. 1C); the signal intensity of absent alleles is higher at higher intensity of the present alleles. This effect could be due to crosstalk or spectral overlap between the fluorescent dyes. In order to correct for this, we chose to convert the green and red intensities for each SNP into polar coordinates and to use the angle value of the two peaks adjacent to the quadrant boundaries at 0 and π/2 (Fig. 1D) to estimate the background intensity of the contralateral allele. For background correction, the estimated allele-specific background was subtracted from the measured signal.
Within-array sample normalization
Sample normalization must generally be performed to even out differences in the DNA input of the samples. Allele-specific measurements present a number of challenges with regard to normalization. A high proportion of zero signals will put the mean and median intensity at a lower, probably unstable, value. Also, the presence of CNVs will affect the DNA content of the cell. LOH is often seen in regions with copy number changes. We explored whether the heterozygous SNPs in a sample could be used as the fraction that represents the unaffected regions of the genome best. Since CNVs will also affect the ability of the genotyping algorithm to reliably determine a genotype, we also tested whether including only high-quality heterozygous SNPs in the invariant set would improve normalization.
Between-array locus normalization
Finally, data were normalized by locus. A typical experiment contains 24 samples because of the 96-well microtiterplate format of the Sentrix arrays. If these are split into 12 normal and 12 affected samples, this yields, on average, four heterozygous SNPs (the SNPs on the array display an average heterozygosity of 30%–40%) per probe, which is insufficient to follow an approach similar to that used for sample normalization. Therefore, we summed the intensities of both alleles from normal (nontumorous) samples within the experiment to perform per SNP normalizations, where we assumed that these samples are unaffected by CNVs and had a copy number of 2.
Selection criteria for best settings
In order to determine the proper way to process the data, we tested combinations of background correction methods while using either all or just the heterozygous loci, varying the cutoff value for the proportion of loci with the highest rGCS to use in sample normalization, and using quantile normalization (Qnt) between the channels of a sample. The selection goals were (1) low variability in regions with the same copy number, (2) a good approximation to two for normal chromosomal regions, and (3) a high amplitude in regions with CNVs compared with normal regions.
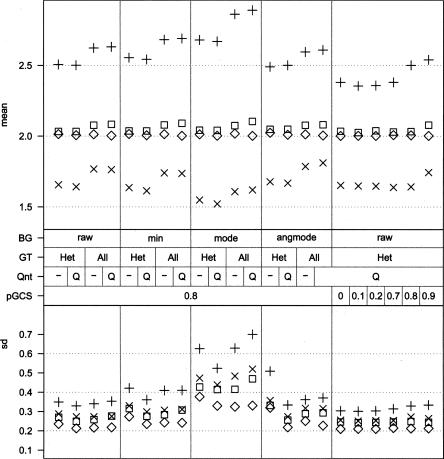
Figure 2 shows the effects of the normalization settings on the amplitude and variability of the signal. Background subtraction (BG) shows a clear increase in the amplitude of the signal, especially for the “mode” method. However, all methods substantially increase variability (e.g., the mode method increases the amplitude but nearly doubles the standard deviation).
Figure 2.
Effects of background estimation and sample normalization. The effects on amplitude (top panel) and variability (bottom panel) of different preprocessing and normalization strategies on the same data set are shown. The symbols indicate different copy number states: ◇, blood; ◽, tumor, normal; ×, tumor, loss; +, tumor, gain. The middle panel indicates the settings. BG, background estimation method: raw, no background estimation; min, minimum intensity in sample; mode, mode of intensities in sample; angmode, mode of angle in polar coordinates near the quadrants. Qnt: Q indicates quantile normalization between channels of a sample. GT: All, use all loci for normalization; Het, use heterozygous loci to calculate normalization factor. pGCS, proportion of relative gene call score. Use only loci with GCS higher than value to calculate normalization factor.
Selecting only heterozygous loci (GT) for use as an invariant set improves the normalization since the resulting copy number for unaffected regions in tumor samples is close to 2, with low variability.
Qnt between the channels of a sample decreases variability with little or no impact on the other goals.
Selection of high-quality heterozygous loci (pGCS) improves normalization when the cutoff is around the 80th percentile. At that point the amplitude of the signal for affected regions is the highest, with only a small effect on variability. Consequently, we chose the following settings for pre-processing and normalization (Table 1): (1) Use Qnt between the red and green signals for each sample; (2) no background estimation and correction; (3) use only the top 20% of heterozygous loci in each sample on the rGCS scale.
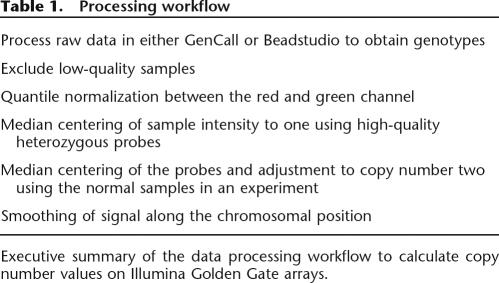
Table 1.
Processing workflow
Executive summary of the data processing workflow to calculate copy number values on Illumina Golden Gate arrays.
Validation
We validated our processing strategy by evaluating sex chromosomes, comparing the results to other methods, and assessing the reproducibility of samples in different experiments.
Performance with known CNV
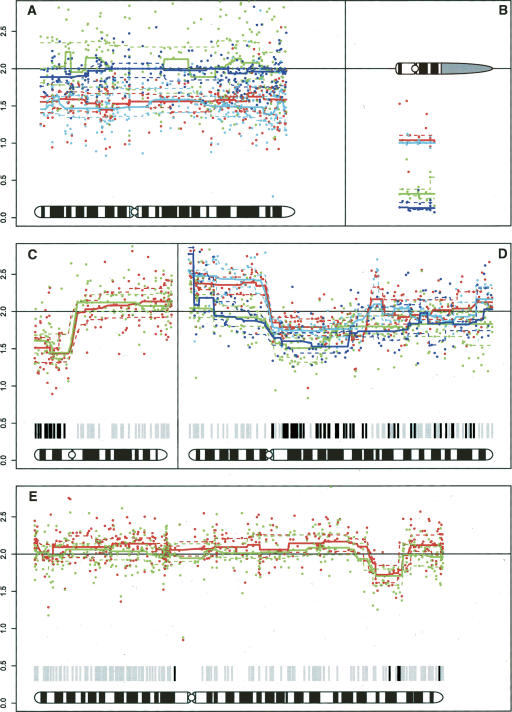
In normal samples, the sex chromosomes can provide insight into the behavior of regions with physical loss. For the X-chromosome, we used female samples for locus normalization. The X-chromosome of normal male samples showed calculated copy number values between 1.5 and 1.6 (see Fig. 4A, below). The loci on the Y-chromosome were normalized using the male subjects, with an assumed copy number of one. We found that the signal intensity for Y-chromosomal loci in females was close to zero (see Fig. 4B, below).
Figure 4.
Chromosomal plots from BeadArrays. Each panel depicts the smoothed copy number as a continuous line and the 10th and 90th percentiles as dashed lines. The unsmoothed copy number values are shown as dots. (A,B) X- and Y-chromosome from leukocyte DNA. Red, 44 male; blue, 106 female; cyan, 108 male; green, 514 female. (C–E) Comparisons of copy numbers. The green lines depict FFPE tumor tissue, and the red lines depict fresh frozen tumor tissue. Bars below the plot indicate heterozygous SNPs in the corresponding normal sample. At black bars, the SNP has switched to homozygosity in the tumor. Physical loss is called when the upper percentile line drops below 2; gain is called when the lower percentile line exceeds 2. (C) Chromosome 17 in tumor 106. (D) Chromosome 5 in tumor 44. Blue line, FFPE BAC array; cyan line, fresh frozen BAC array. (E) Chromosome 2 in tumor 514.
Comparison to other methods
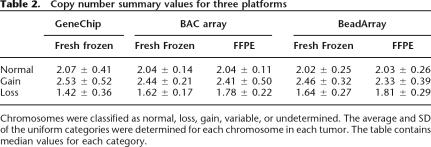
Next, we compared four colorectal tumors using BeadArrays, GeneChip arrays, and BAC arrays. All autosomes of these tumors were divided into five categories based on GeneChip and BAC array analysis: normal, gain, loss, variable, and undetermined. Averages and standard deviations for the first three, or uniform, categories are shown in Table 2. There is a slight overestimation of the copy number for normal chromosomes for all methods. The values for chromosomes with a gain are comparable, while the GeneChip copy numbers for chromosomes with a loss are somewhat lower than for the other two methods. The variability of the signal in GeneChips is larger than for BeadArrays, with BAC arrays showing the smallest amount of variability.
Table 2.
Copy number summary values for three platforms
Chromosomes were classified as normal, loss, gain, variable, or undetermined. The average and SD of the uniform categories were determined for each chromosome in each tumor. The table contains median values for each category.
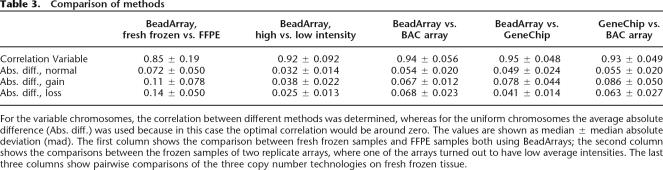
The basic pattern of CNVs is comparable between these platforms, with a correlation of >0.9 between the same tumors on different platforms and low correlations between the experimental samples (Table 3). Moreover, the visual resemblance between the smoothed signals from these methods is remarkable (Fig. 3).
Table 3.
Comparison of methods
For the variable chromosomes, the correlation between different methods was determined, whereas for the uniform chromosomes the average absolute difference (Abs. diff.) was used because in this case the optimal correlation would be around zero. The values are shown as median ± median absolute deviation (mad). The first column shows the comparison between fresh frozen samples and FFPE samples both using BeadArrays; the second column shows the comparisons between the frozen samples of two replicate arrays, where one of the arrays turned out to have low average intensities. The last three columns show pairwise comparisons of the three copy number technologies on fresh frozen tissue.
Figure 3.
Comparison of platforms for calculation of copy numbers for tumor T106. Smoothed values are plotted along idiograms. Red, GeneChip; green, BAC array; blue, BeadArray. Below each idiogram, gray bars indicate heterozygous SNPs in the corresponding normal sample. Loci that are not heterozygous in the tumor are shown as black bars.
Reproducibility
The set of samples were hybridized twice to separate Illumina Sentrix arrays. Despite the far lower intensities in one of these arrays (3900 vs. 820), the data show a very good concordance (Table 3)
Combined analysis of LOH and copy number
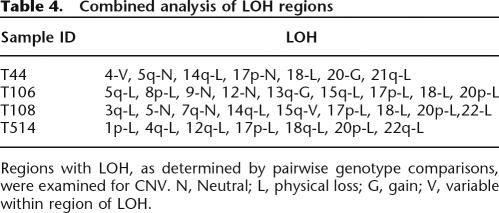
This method allows us to extend the LOH analysis from our previous article (Lips et al. 2005). Besides the copy number profiles, Figure 3 also shows the loci that show heterozygosity in the paired normal sample and homozygosity in the tumor sample. Although most regions with LOH show physical loss for this sample, chromosomes 9 and 12 show copy neutral LOH. The findings for all tumors in the study are summarized in Table 4.
Table 4.
Combined analysis of LOH regions
Regions with LOH, as determined by pairwise genotype comparisons, were examined for CNV. N, Neutral; L, physical loss; G, gain; V, variable within region of LOH.
Comparison of CNV in frozen and FFPE tumor tissue
The variability of the unsmoothed copy numbers at different levels of CNV was comparable between frozen tissue and FFPE samples (Table 2). The correlation between the variable chromosomes in FFPE and frozen samples from the same patient was less than between different methods with frozen samples or between replicates (Table 3). This was mainly due to sample T44. When this tumor was excluded, 50% of the values were >0.96. In order to test the origin of the differences, we also performed BAC arrays on the FFPE-extracted DNA. There was insufficient material left to process T108, but the patterns of CNVs in the other three samples, and especially the differences between frozen and FFPE material, were comparable between BAC arrays and BeadArrays (Fig. 4D). Also, for each, tumor chromosomes can be selected that show perfect concordance, while other chromosomes perform less well (Fig. 4C–E). The average absolute distances between frozen and FFPE samples from both normal and affected chromosomes show excellent concordance.
Application in FFPE tissue
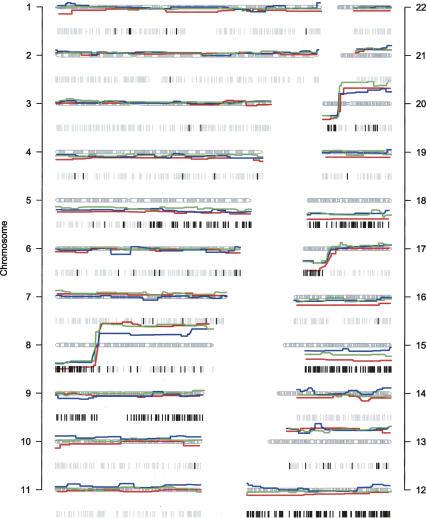
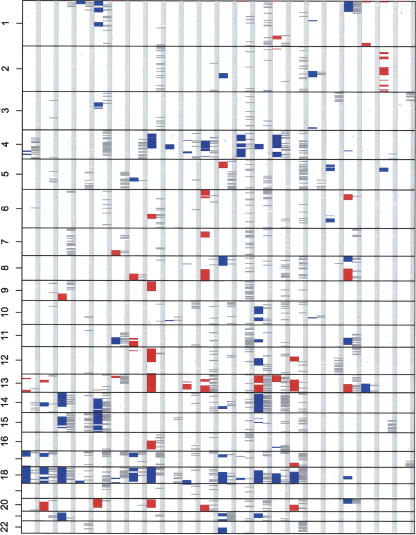
We have applied this method to a series of 22 colorectal tumors that have been stored in the paraffin archive for up to 10 yr. Several characteristic patterns of LOH and CNVs can be identified in this series (Fig. 5) (Diep et al. 2006). Frequent events include physical loss on chromosomes 4 and 18, gain of chromosomes 13 and 20, and copy neutral LOH on chromosomes 4, 5, 10, and 17.
Figure 5.
Analysis of 22 colorectal carcinomas. Validation of the application was demonstrated on a series of 22 colorectal carcinomas. Each column in the plot shows a genome-wide overview of the numerical changes in red (gain) and blue (loss) on the left and probes with LOH on the right. The common patterns of chromosomal instability (Diep et al. 2006), such as physical loss on chromosomes 4 and 18, gain of chromosomes 13 and 20, and copy neutral LOH on chromosomes 4, 5, 10, and 17 can all be identified from this series.
Discussion
We have developed a method to determine CNVs using an Illumina BeadArray in combination with the GoldenGate assay and validated it using established copy number methodologies. The technical properties of the GoldenGate assay allow the analysis of partly degraded DNA, and we have shown that copy number analysis of paraffin embedded tissue shows comparable results to the analysis of fresh frozen tissue.
Copy number analysis using SNP microarrays enables the combined analysis of CNV and LOH in one assay (Bignell et al. 2004; Zhou et al. 2004b), and makes it possible to distinguish several forms of LOH: physical loss of one or both alleles and homologous mitotic recombination.
Normalization procedures usually assume that the cell population is diploid or aneuploid when no further information is available. The DNA index can be used to improve estimation of chromosomal copy numbers in cases of aneuploid or multiploid tumors. For the GoldenGate assay, this is not immediately feasible because the four linkage panels each contain a different set of chromosomes and any CNV will not usually be distributed evenly among the chromosomes. To cope with this situation, linkage panels with SNPs distributed evenly across the genome are required.
The absence of a single chromosome, e.g., the X-chromosome in males, shows a signal of 1.5 rather than the theoretically expected one. This reduced linearity is also found for BAC arrays and GeneChips and is probably a consequence of the complexity of the process. However, it is also the case that the design of the BeadArrays is optimized to discriminate heterozygous from homozygous loci, rather than measure copy number. Consequently, the allele-specific PCR likely approaches saturation, leading to reduced linearity. Reducing the number of PCR cycles could potentially improve the linearity, although the effect of this remains to be tested.
In general, calculated copy numbers from SNP and BAC arrays show too much variability to assign discrete copy number values to individual probes. Usually, analysis methods use information from flanking SNPs to calculate copy number under the assumption that genomic events are not restricted to single SNPs. The effect is that a smoothed signal is calculated along the physical position on the genome. The extent of smoothing has an effect on the spatial resolution of the measurement. For noisy data, stronger smoothing is required, thus increasing the minimum size of detectable CNVs.
Of the various smoothing algorithms (Fridlyand et al. 2004; Jong et al. 2004; Eilers and de Menezes 2005; Lai et al. 2005), we utilize quantile smoothing, mainly for visualization. Which analysis method optimally deals with the statistical properties of genomic arrays is currently unclear (Lai et al. 2005). Besides median smoothing, which calculates the smoothed copy number value, quantile smoothing can also calculate smoothed signals to indicate the bandwidth of the raw data (Fig. 4).
The comparison of copy number analysis from fresh frozen tissue and FFPE tissue from the same tumor showed varying degrees of similarity. Three of the tested tumors showed a median correlation at the same level as comparisons between the methods in the validation section. The limited concordance between frozen and FFPE samples from the fourth tumor could likely be explained by tumor heterogeneity (Fukunari et al. 2003), because the same CNVs can be identified on BAC arrays performed on the FFPE material. BAC array, GeneChip, and BeadArray profiles were derived from the same DNA isolate of (fresh frozen) tumor material, while the comparison between frozen and FFPE tissue was necessarily sampled from different locations within the tumor.
Taking into account that the PCR amplification protocol was essentially the same for both types of tissue and that the variability of the signal is comparable for both, these findings show that the Illumina GoldenGate array can reliably determine copy number changes from FFPE tissue. A previous study (Thompson et al. 2005) has shown copy number analysis from FFPE tissue with GeneChips, but in that study the number of amplification cycles was higher in FFPE samples and the apparent variability of the copy number signal was higher. Copy number analysis can readily be performed on FFPE material using array-CGH, but this platform lacks the ability to additionally determine LOH and cannot identify copy neutral genomic events, like chromosomes 9 and 12 in Figure 3 and chromosomes 4, 5, 10, and 17 in Figure 5.
We have previously shown that BeadArrays can be used to reliably genotype and detect LOH on FFPE tissue (Lips et al. 2005). We now show that this platform allows copy number analysis that performs equally well on FFPE and frozen material, and that results from this approach show high agreement with other copy number methodologies. These findings open the large pathological archives for genomic analysis, and are especially relevant in hereditary disease because there are often problems in obtaining fresh material from affected relatives.
Methods
Subjects/material
Colorectal tumor tissue that was known to have genomic aberrations and corresponding normal tissue from four patients was used, following medical ethical guidelines. Ploidy status of the tumors was previously assessed by flow cytometry. A pathologist (H. Morreau) assessed the normal and tumor areas and the percentage of tumor cells based on H&E slides. The samples included a rectal adenoma (T514, 60% tumor, aneuploid), one right-sided Dukes B (T44, 50% tumor, aneuploid), and two Dukes C carcinomas (T106, 90% tumor, multiploid; T108, 80% tumor, multiploid). From the departmental FFPE archives, we collected 22 colorectal carcinomas, for which normal DNA from either leukocytes or histologically normal FFPE tissue was also available.
DNA isolation
From fresh frozen tissue, twenty 30-μm-thick sections were cut from each tumor. DNA was isolated with the Genomic Wizard kit according to the manufacturer’s protocol (Promega). Leukocyte DNA was obtained by salting out precipitation as described previously (Miller et al. 1988). DNA from FFPE tissue was extracted using the Chelex extraction method (De Jong et al. 2004). Briefly, three tissue punches (diameter, 0.6 mm) were obtained by a tissue microarrayer (Beecher Instruments) from both paraffin blocks containing tumor and blocks from the same patient that did not contain tumor tissue. DNA was isolated with Chelex and proteinase K. FFPE DNA was subsequently cleaned up using the Genomic Wizard kit (Promega). DNA concentrations were measured by the PicoGreen method (Invitrogen-Molecular Probes) and DNA quality was checked on a 1% agarose gel. For each cell isolate, 1 μg of DNA obtained from either FFPE or frozen samples was used for the BeadArrays, whereas 250 ng DNA was used for the GeneChip arrays and 450 ng from frozen samples was used for the BAC arrays according to the manufacturer’s instructions.
Array platforms
Illumina Sentrix BeadArrays were used with the linkage mapping panel version IV (Illumina). This platform contains 5861 SNP markers distributed evenly over the genome with a physical distance of, on average, 482 kb. The probes are divided among four OPA panels because the method is restricted to ∼1500 different SNPs per hybridization. Samples were prepared according to the GoldenGate assay (Fan et al. 2003).
Affymetrix GeneChip Mapping 10K Xba1 2.0 arrays (Affymetrix) contain 10,204 markers with a mean intermarker distance of 258 kb.
BAC array slides were produced at the LUMC department of Molecular Cell Biology. This platform contains 3700 probes spotted in triplicate and uses 1-Mb-spaced BACs distributed by the Wellcome Trust Sanger Institute (Knijnenburg et al. 2005). All laboratory processing and hybridizations were performed according to manufacturer’s protocols.
Data analysis
Throughout our analysis, we have chosen to use data on a linear scale. In the case of CNVs, this is the more intuitive alternative compared with logarithmic scaling. A linear scale includes zero to indicate complete loss of a chromosomal region, and losing and acquiring an allele show the same impact.
For BeadArrays, gene calls were extracted using the gene calling program GenCall version 6.0.7 (Illumina). The software provides two quality scores per locus, an experiment-wide gene train score (GTS) and a sample-specific gene call score (GCS). From these, we computed the relative gene call score (rGCS) as GCS/GTS. In order to retrieve intensity measurements, the Settings.xml file for the BeadArray software has to be adapted: line <SaveTextFiles>false</SaveTextFiles> has to be changed to <SaveTextFiles>true</SaveTextFiles>. Samples were excluded when the raw median intensity in either of the channels was <1250. Normalization procedures for Illumina arrays are discussed in the Results section.
There are a number of methods available to calculate copy numbers from Affymetrix SNP arrays. We have evaluated CNAT, CNAG, and dChip. From these methods, we chose dChip version 1.3 because of its performance with regard to variability and amplitude of the signal for changed chromosomes.
The scanned images for the BAC arrays were processed using GenePix 4.1 software. The BioConductor package Limma (Smyth and Speed 2003) version 2.6.0 was used to perform background correction and normalization. After normalization, the replicate spots were averaged.
LOH was determined by comparing the genotypes from frozen tumor tissue and blood leukocytes from the same patient. LOH was called for stretches of two or more SNPs within 500,000 bp that were heterozygous in normal tissue and homozygous in tumor tissue
Basic properties of the methods, such as average and variability, were calculated from the normalized copy numbers. Comparisons between methods and samples were calculated from binned, smoothed copy numbers with a bin size of 2500 kb. A genomic smoother was used as described in Eilers and de Menezes (2005). The smoothing parameter was empirically chosen as the number of autosomal genomic markers on an assay divided by 1500.
In order to promote the concept of reproducible research, the R-scripts to create the figures and tables from the raw data are bundled together with the data sets.
Acknowledgments
We thank A. Middeldorp and M. van Puijenbroek for discussions, J.W.F. Dierssen for providing colorectal tumor samples, and R. van’t Slot for sample and array processing.
Footnotes
[Supplemental material is available online at www.genome.org. The R-package BeadArray SNP used to perform the analysis is available from http://www.bioconductor.org. The data sets are available from the Gene Expression Omnibus with accession number GSE5347 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5347).]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5686107
References
- Bignell G.R., Huang J., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Huang J., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Greshock J., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Watt S., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Butler A., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., West S., Grigorova M., Jones K.W., Wei W., Stratton M.R., Grigorova M., Jones K.W., Wei W., Stratton M.R., Jones K.W., Wei W., Stratton M.R., Wei W., Stratton M.R., Stratton M.R., et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong A.E., van Puijenbroek M., Hendriks Y., Tops C., Wijnen J., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., van Puijenbroek M., Hendriks Y., Tops C., Wijnen J., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Hendriks Y., Tops C., Wijnen J., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Tops C., Wijnen J., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Wijnen J., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Ausems M.G., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Meijers-Heijboer H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Wagner A., Van Os T.A., Brocker-Vriends A.H., Van Os T.A., Brocker-Vriends A.H., Brocker-Vriends A.H., et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin. Cancer Res. 2004;10:972–980. doi: 10.1158/1078-0432.ccr-0956-3. [DOI] [PubMed] [Google Scholar]
- Diep C.B., Kleivi K., Ribeiro F.R., Teixeira M.R., Lindgjaerde O.C., Lothe R.A., Kleivi K., Ribeiro F.R., Teixeira M.R., Lindgjaerde O.C., Lothe R.A., Ribeiro F.R., Teixeira M.R., Lindgjaerde O.C., Lothe R.A., Teixeira M.R., Lindgjaerde O.C., Lothe R.A., Lindgjaerde O.C., Lothe R.A., Lothe R.A. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- Dumur C.I., Dechsukhum C., Ware J.L., Cofield S.S., Best A.M., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Dechsukhum C., Ware J.L., Cofield S.S., Best A.M., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Ware J.L., Cofield S.S., Best A.M., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Cofield S.S., Best A.M., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Best A.M., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Wilkinson D.S., Garrett C.T., Ferreira-Gonzalez A., Garrett C.T., Ferreira-Gonzalez A., Ferreira-Gonzalez A. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- Eilers P.H., de Menezes R.X., de Menezes R.X. Quantile smoothing of array CGH data. Bioinformatics. 2005;21:1146–1153. doi: 10.1093/bioinformatics/bti148. [DOI] [PubMed] [Google Scholar]
- Fan J.B., Oliphant A., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Oliphant A., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Hansen M., Steemers F., Butler S.L., Deloukas P., Steemers F., Butler S.L., Deloukas P., Butler S.L., Deloukas P., Deloukas P., et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Fridlyand J., Snijders A.M., Pinkel D., Albertson D.G., Jain A.N., Snijders A.M., Pinkel D., Albertson D.G., Jain A.N., Pinkel D., Albertson D.G., Jain A.N., Albertson D.G., Jain A.N., Jain A.N. Hidden Markov models approach to the analysis of array CGH data. J. Multivariate Anal. 2004;90:132–153. [Google Scholar]
- Fukunari H., Iwama T., Sugihara K., Miyaki M., Iwama T., Sugihara K., Miyaki M., Sugihara K., Miyaki M., Miyaki M. Intratumoral heterogeneity of genetic changes in primary colorectal carcinomas with metastasis. Surg. Today. 2003;33:408–413. doi: 10.1007/s10595-002-2520-1. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Beroukhim R., Milner D.A., Granter S.R., Du J., Milner D.A., Granter S.R., Du J., Granter S.R., Du J., Du J., et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Herr A., Grutzmann R., Matthaei A., Artelt J., Schrock E., Rump A., Pilarsky C., Grutzmann R., Matthaei A., Artelt J., Schrock E., Rump A., Pilarsky C., Matthaei A., Artelt J., Schrock E., Rump A., Pilarsky C., Artelt J., Schrock E., Rump A., Pilarsky C., Schrock E., Rump A., Pilarsky C., Rump A., Pilarsky C., Pilarsky C. High-resolution analysis of chromosomal imbalances using the Affymetrix 10K SNP genotyping chip. Genomics. 2005;85:392–400. doi: 10.1016/j.ygeno.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Irving J.A., Bloodworth L., Bown N.P., Case M.C., Hogarth L.A., Hall A.G., Bloodworth L., Bown N.P., Case M.C., Hogarth L.A., Hall A.G., Bown N.P., Case M.C., Hogarth L.A., Hall A.G., Case M.C., Hogarth L.A., Hall A.G., Hogarth L.A., Hall A.G., Hall A.G. Loss of heterozygosity in childhood acute lymphoblastic leukemia detected by genome-wide microarray single nucleotide polymorphism analysis. Cancer Res. 2005;65:3053–3058. doi: 10.1158/0008-5472.CAN-04-2604. [DOI] [PubMed] [Google Scholar]
- Ishikawa S., Komura D., Tsuji S., Nishimura K., Yamamoto S., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Komura D., Tsuji S., Nishimura K., Yamamoto S., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Tsuji S., Nishimura K., Yamamoto S., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Nishimura K., Yamamoto S., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Yamamoto S., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Panda B., Huang J., Fukayama M., Jones K.W., Aburatani H., Huang J., Fukayama M., Jones K.W., Aburatani H., Fukayama M., Jones K.W., Aburatani H., Jones K.W., Aburatani H., Aburatani H. Allelic dosage analysis with genotyping microarrays. Biochem. Biophys. Res. Commun. 2005;333:1309–1314. doi: 10.1016/j.bbrc.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Janne P.A., Li C., Zhao X., Girard L., Chen T.H., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Li C., Zhao X., Girard L., Chen T.H., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Zhao X., Girard L., Chen T.H., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Girard L., Chen T.H., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Chen T.H., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Minna J., Christiani D.C., Johnson B.E., Meyerson M., Christiani D.C., Johnson B.E., Meyerson M., Johnson B.E., Meyerson M., Meyerson M. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- Jong K., Marchiori E., Meijer G., Vaart A.V., Ylstra B., Marchiori E., Meijer G., Vaart A.V., Ylstra B., Meijer G., Vaart A.V., Ylstra B., Vaart A.V., Ylstra B., Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–3637. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- Kallioniemi O.P., Kallioniemi A., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Kallioniemi A., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Gray J.W., Waldman F., Pinkel D., Waldman F., Pinkel D., Pinkel D. Comparative genomic hybridization: A rapid new method for detecting and mapping DNA amplification in tumors. Semin. Cancer Biol. 1993;4:41–46. [PubMed] [Google Scholar]
- Kennedy G.C., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Liu G., Su X., Cao M., Chen W., Zhang J., Su X., Cao M., Chen W., Zhang J., Cao M., Chen W., Zhang J., Chen W., Zhang J., Zhang J., et al. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- Knijnenburg J., Szuhai K., Giltay J., Molenaar L., Sloos W., Poot M., Tanke H.J., Rosenberg C., Szuhai K., Giltay J., Molenaar L., Sloos W., Poot M., Tanke H.J., Rosenberg C., Giltay J., Molenaar L., Sloos W., Poot M., Tanke H.J., Rosenberg C., Molenaar L., Sloos W., Poot M., Tanke H.J., Rosenberg C., Sloos W., Poot M., Tanke H.J., Rosenberg C., Poot M., Tanke H.J., Rosenberg C., Tanke H.J., Rosenberg C., Rosenberg C. Insights from genomic microarrays into structural chromosome rearrangements. Am. J. Med. Genet. A. 2005;132:36–40. doi: 10.1002/ajmg.a.30378. [DOI] [PubMed] [Google Scholar]
- Lai W.R., Johnson M.D., Kucherlapati R., Park P.J., Johnson M.D., Kucherlapati R., Park P.J., Kucherlapati R., Park P.J., Park P.J. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberfarb M.E., Lin M., Lechpammer M., Li C., Tanenbaum D.M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Lin M., Lechpammer M., Li C., Tanenbaum D.M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Lechpammer M., Li C., Tanenbaum D.M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Li C., Tanenbaum D.M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Tanenbaum D.M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Febbo P.G., Wright R.L., Shim J., Kantoff P.W., Loda M., Wright R.L., Shim J., Kantoff P.W., Loda M., Shim J., Kantoff P.W., Loda M., Kantoff P.W., Loda M., Loda M., et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. [PubMed] [Google Scholar]
- Lin M., Wei L.J., Sellers W.R., Lieberfarb M., Wong W.H., Li C., Wei L.J., Sellers W.R., Lieberfarb M., Wong W.H., Li C., Sellers W.R., Lieberfarb M., Wong W.H., Li C., Lieberfarb M., Wong W.H., Li C., Wong W.H., Li C., Li C. dChipSNP: Significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Tanenbaum D.M., Daly M.J., Winchester E., Lui W.O., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Tanenbaum D.M., Daly M.J., Winchester E., Lui W.O., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Daly M.J., Winchester E., Lui W.O., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Winchester E., Lui W.O., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Lui W.O., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Villapakkam A., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Stanton S.E., Larsson C., Hudson T.J., Johnson B.E., Larsson C., Hudson T.J., Johnson B.E., Hudson T.J., Johnson B.E., Johnson B.E., et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- Lips E.H., Dierssen J.W.F., van Eijk R., Oosting J., Eilers P.H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., Dierssen J.W.F., van Eijk R., Oosting J., Eilers P.H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., van Eijk R., Oosting J., Eilers P.H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., Oosting J., Eilers P.H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., Eilers P.H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., Tollenaar R.A., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., de Graaff E.J., Wijmenga C., van’t Slot R., Morreau H., Wijmenga C., van’t Slot R., Morreau H., van’t Slot R., Morreau H., Morreau H., et al. Reliable high-throughput genotyping and loss of heterozygosity detection in formalin-fixed paraffin-embedded tumors using single nucleotide polymorphism arrays. Cancer Res. 2005;65:10188–10191. doi: 10.1158/0008-5472.CAN-05-2486. [DOI] [PubMed] [Google Scholar]
- Mao X., Barfoot R., Hamoudi R.A., Easton D.F., Flanagan A.M., Stratton M.R., Barfoot R., Hamoudi R.A., Easton D.F., Flanagan A.M., Stratton M.R., Hamoudi R.A., Easton D.F., Flanagan A.M., Stratton M.R., Easton D.F., Flanagan A.M., Stratton M.R., Flanagan A.M., Stratton M.R., Stratton M.R. Allelotype of uterine leiomyomas. Cancer Genet. Cytogenet. 1999;114:89–95. doi: 10.1016/s0165-4608(99)00053-9. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Loi H., Dong S., Tsai Y.Y., Fang J., Law J., Di X., Liu W.M., Yang G., Liu G., Loi H., Dong S., Tsai Y.Y., Fang J., Law J., Di X., Liu W.M., Yang G., Liu G., Dong S., Tsai Y.Y., Fang J., Law J., Di X., Liu W.M., Yang G., Liu G., Tsai Y.Y., Fang J., Law J., Di X., Liu W.M., Yang G., Liu G., Fang J., Law J., Di X., Liu W.M., Yang G., Liu G., Law J., Di X., Liu W.M., Yang G., Liu G., Di X., Liu W.M., Yang G., Liu G., Liu W.M., Yang G., Liu G., Yang G., Liu G., Liu G., et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.A., Dykes D.D., Polesky H.F., Dykes D.D., Polesky H.F., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannya Y., Sanada M., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Sanada M., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Nakazaki K., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Hosoya N., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Wang L., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Hangaishi A., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Kurokawa M., Chiba S., Bailey D.K., Kennedy G.C., Chiba S., Bailey D.K., Kennedy G.C., Bailey D.K., Kennedy G.C., Kennedy G.C., et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Oliphant A., Barker D.L., Stuelpnagel J.R., Chee M.S., Barker D.L., Stuelpnagel J.R., Chee M.S., Stuelpnagel J.R., Chee M.S., Chee M.S. BeadArray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;56 (Suppl 8):60–61. [PubMed] [Google Scholar]
- Pinkel D., Albertson D.G., Albertson D.G. Comparative genomic hybridization. Annu. Rev. Genomics Hum. Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Collins C., Kuo W.L., Chen C., Zhai Y., Kuo W.L., Chen C., Zhai Y., Chen C., Zhai Y., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Jeffrey S.S., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Primdahl H., Wikman F.P., von der Masse H., Zhou X.G., Wolf H., Orntoft T.F., Wikman F.P., von der Masse H., Zhou X.G., Wolf H., Orntoft T.F., von der Masse H., Zhou X.G., Wolf H., Orntoft T.F., Zhou X.G., Wolf H., Orntoft T.F., Wolf H., Orntoft T.F., Orntoft T.F. Allelic imbalances in human bladder cancer: Genome-wide detection with high-density single-nucleotide polymorphism arrays. J. Natl. Cancer Inst. 2002;94:216–223. doi: 10.1093/jnci/94.3.216. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H., Lengauer C., Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Shen R., Fan J.B., Campbell D., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Fan J.B., Campbell D., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Campbell D., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Doucet D., Yeakley J., Bibikova M., Wickham G.E., McBride C., Yeakley J., Bibikova M., Wickham G.E., McBride C., Bibikova M., Wickham G.E., McBride C., Wickham G.E., McBride C., McBride C., et al. High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Speed T., Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Thompson E.R., Herbert S.C., Forrest S.M., Campbell I.G., Herbert S.C., Forrest S.M., Campbell I.G., Forrest S.M., Campbell I.G., Campbell I.G. Whole genome SNP arrays using DNA derived from formalin-fixed, paraffin-embedded ovarian tumor tissue. Hum. Mutat. 2005;26:384–389. doi: 10.1002/humu.20220. [DOI] [PubMed] [Google Scholar]
- Tomlinson I., Rahman N., Frayling I., Mangion J., Barfoot R., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Rahman N., Frayling I., Mangion J., Barfoot R., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Frayling I., Mangion J., Barfoot R., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Mangion J., Barfoot R., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Barfoot R., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Hamoudi R., Seal S., Northover J., Thomas H.J., Neale K., Seal S., Northover J., Thomas H.J., Neale K., Northover J., Thomas H.J., Neale K., Thomas H.J., Neale K., Neale K., et al. Inherited susceptibility to colorectal adenomas and carcinomas: Evidence for a new predisposition gene on 15q14-q22. Gastroenterology. 1999;116:789–795. doi: 10.1016/s0016-5085(99)70061-2. [DOI] [PubMed] [Google Scholar]
- Zhao X., Li C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Li C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Chen T.H., Girard L., Minna J., Christiani D., Leo C., Girard L., Minna J., Christiani D., Leo C., Minna J., Christiani D., Leo C., Christiani D., Leo C., Leo C., et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- Zhou X., Li C., Mok S.C., Chen Z., Wong D.T., Li C., Mok S.C., Chen Z., Wong D.T., Mok S.C., Chen Z., Wong D.T., Chen Z., Wong D.T., Wong D.T. Whole genome loss of heterozygosity profiling on oral squamous cell carcinoma by high-density single nucleotide polymorphic allele (SNP) array. Cancer Genet. Cytogenet. 2004a;151:82–84. doi: 10.1016/j.cancergencyto.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Zhou X., Mok S.C., Chen Z., Li Y., Wong D.T., Mok S.C., Chen Z., Li Y., Wong D.T., Chen Z., Li Y., Wong D.T., Li Y., Wong D.T., Wong D.T. Concurrent analysis of loss of heterozygosity (LOH) and copy number abnormality (CNA) for oral premalignancy progression using the Affymetrix 10K SNP mapping array. Hum. Genet. 2004b;115:327–330. doi: 10.1007/s00439-004-1163-1. [DOI] [PubMed] [Google Scholar]