Abstract
Metagenomics is the study of the genomic content of a sample of organisms obtained from a common habitat using targeted or random sequencing. Goals include understanding the extent and role of microbial diversity. The taxonomical content of such a sample is usually estimated by comparison against sequence databases of known sequences. Most published studies use the analysis of paired-end reads, complete sequences of environmental fosmid and BAC clones, or environmental assemblies. Emerging sequencing-by-synthesis technologies with very high throughput are paving the way to low-cost random “shotgun” approaches. This paper introduces MEGAN, a new computer program that allows laptop analysis of large metagenomic data sets. In a preprocessing step, the set of DNA sequences is compared against databases of known sequences using BLAST or another comparison tool. MEGAN is then used to compute and explore the taxonomical content of the data set, employing the NCBI taxonomy to summarize and order the results. A simple lowest common ancestor algorithm assigns reads to taxa such that the taxonomical level of the assigned taxon reflects the level of conservation of the sequence. The software allows large data sets to be dissected without the need for assembly or the targeting of specific phylogenetic markers. It provides graphical and statistical output for comparing different data sets. The approach is applied to several data sets, including the Sargasso Sea data set, a recently published metagenomic data set sampled from a mammoth bone, and several complete microbial genomes. Also, simulations that evaluate the performance of the approach for different read lengths are presented.
The genomic revolution of the early 1990s targeted the study of individual genomes of microorganisms, plants, and animals. While this type of analysis has almost become routine, the genomic analysis of complex mixtures of organisms remains challenging. Metagenomics has been defined as “the genomic analysis of microorganisms by direct extraction and cloning of DNA from an assemblage of microorganisms” (Handelsman 2004), and its importance stems from the fact that 99% or more of all microbes are deemed to be unculturable. Goals of metagenomic studies include assessing the coding potential of environmental organisms, quantifying the relative abundances of (known) species, and estimating the amount of unknown sequence information (environmental sequences) for which no species, or only distant relatives, have yet been described. It is useful to extend Handelsman’s definition to also include sequences from higher organisms as well as just microorganisms, thus opening the door to “environmental forensics.” By vastly extending the currently available sequences in databases, metagenomics promises to lead to the discovery of new genes that have useful applications in biotechnology and medicine (Steele and Streit 2005).
Early metagenomics projects (Béja et al. 2000, 2001) were plagued by potential biases that are due to DNA extraction and cloning methods (Martiny et al. 2006). Clone libraries were constructed from environmental DNA using fosmid and BAC vectors as vehicles for DNA propagation and amplification. The libraries were subsequently screened for specific phylogenetic markers, and paired-end sequencing was undertaken on clones of interest. Overlapping clones, sequenced in their entity, were scaffolded into super-contigs, giving a snapshot of an organism’s genomic features, such as GC content, codon usage, or coding density. This strategy was soon complemented by whole (meta)-genome sequencing using a “shotgun” approach (Venter et al. 2004) that employs cloning and paired-end sequencing of plasmid libraries. Recent projects based on these methodologies include data sets from an acid mine biofilm (Tyson et al. 2004), seawater samples (Venter et al. 2004; DeLong et al. 2006), deep-sea sediment (Hallam et al. 2004), or soil and whale falls (Tringe et al. 2005).
These projects all use “Sanger sequencing,” based on cloning, fluorescent dideoxynucleotides, and capillary electrophoresis (Meldrum 2000a, b). Recently, a new “sequencing-by-synthesis” strategy was published (Margulies et al. 2005; Zhang et al. 2006). This approach uses emulsion-based PCR amplification of a large number of DNA fragments and parallel pyro-sequencing with high throughput. In a single sequencing run, >20 million base pairs of sequence can be generated, at a lower price per base than Sanger-based methods. The current drawbacks of the method are short read lengths of ∼100 bp, in contrast to ∼800 bp using Sanger sequencing, a slightly higher sequencing error rate due to difficulties determining base pair counts in homopolymer stretches, and a substantial reduction of read length when sequencing pair-ended reads. The most important advantage of the new sequencing approach for metagenomics is that it does not require cloning of the target DNA fragments and therefore avoids cloning biases resulting from toxic sequences killing their cloning hosts.
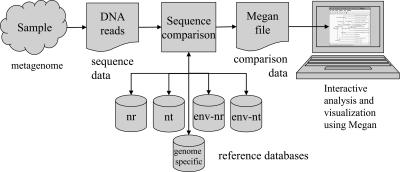
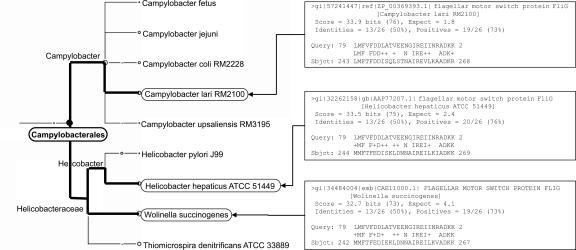
In this study, we present a new approach to the initial analysis of a metagenomic data set that avoids the problems associated with environmental assemblies or the use of a limited number of phylogenetic markers. Our strategy can be applied to DNA reads collected within the framework of any metagenomics project, regardless of the sequencing technology used, and thus provides an easily deployable alternative to other types of analysis. We provide a new computer program called MEGAN (Metagenome Analyzer) that allows analysis of large data sets by a single scientist. In a pre-processing step, the set of DNA reads (or contigs) is compared against databases of known sequences using a comparison tool such as BLAST (see Fig. 1). MEGAN is then used to estimate and interactively explore the taxonomical content of the data set, using the NCBI taxonomy to summarize and order the results. The program uses a simple algorithm that assigns each read to the lowest common ancestor (LCA) of the set of taxa that it hit in the comparison (see Fig. 2). As a result, species-specific sequences are assigned to taxa near the leaves of the NCBI tree, whereas widely conserved sequences are assigned to high-order taxa closer to the root.
Figure 1.
For a given sample of organisms, a randomly selected collection of DNA fragments is sequenced. The resulting reads are then compared with one or more reference databases using an appropriate sequence comparison program such as BLAST (Altschul et al. 1990). The resulting data are processed by MEGAN to produce an interactive analysis of the taxonomical content of the sample.
Figure 2.
On the right, we list the three BLASTX matches obtained for a specific read r from the mammoth data set, to sequences representing Campylobacter lari, Helicobacter hepaticus, and Wolinella, respectively. The LCA-assignment algorithm assigns r to the taxon Campylobacterales, shown on the left, as it is the lowest-common taxonomical ancestor of the three matched species.
We first illustrate this approach by applying it to a subset of the Sargasso Sea data set (Venter et al. 2004), which was obtained by Sanger sequencing. We then apply it to a set of ∼300,000 reads obtained from a sample of mammoth bone (Poinar et al. 2006), which used the “sequencing-by-synthesis” approach. Finally, we address the question of whether species can be identified with confidence from individual short reads, using the genome sequences of Escherichia coli and Bdellovibrio bacteriovorus.
Ease of use is a main design criterion of MEGAN. An analysis is initiated by simply opening the output file of any member of the BLAST family of programs, or from some other sequence comparison tool, and is then performed interactively via a graphical user interface. The program was carefully engineered to run quickly and responsively on a laptop, even when processing large data sets. For maximum portability, the program is written in Java, and installers for Linux/Unix, MacOS and Windows are freely available to the academic community from http://www-ab.informatik.uni-tuebingen.de/software/megan.
Results
The MEGAN processing pipeline
Figure 1 illustrates a typical processing pipeline in which MEGAN is used to perform the initial analysis of a metagenomic sample. Firstly, reads are collected from the sample using any random shotgun protocol. Secondly, a sequence comparison of all reads against one or more databases of known reads is performed, using BLAST or a similar comparison tool. Thirdly, MEGAN processes the results of the comparison to collect all hits of reads against known sequences and assigns a taxon ID to each sequence based on the NCBI taxonomy. This produces a MEGAN file that contains all information needed for analyzing and generating graphical and statistical output. Fourthly, the user interacts with the program to run the lowest common ancestor (LCA) algorithm (see Fig. 2), to analyze the data, to inspect the assignment of individual reads to taxa based on their hits, and to produce summaries of the results at different levels of the NCBI taxonomy (see Figs. 3 and 5–8 below).
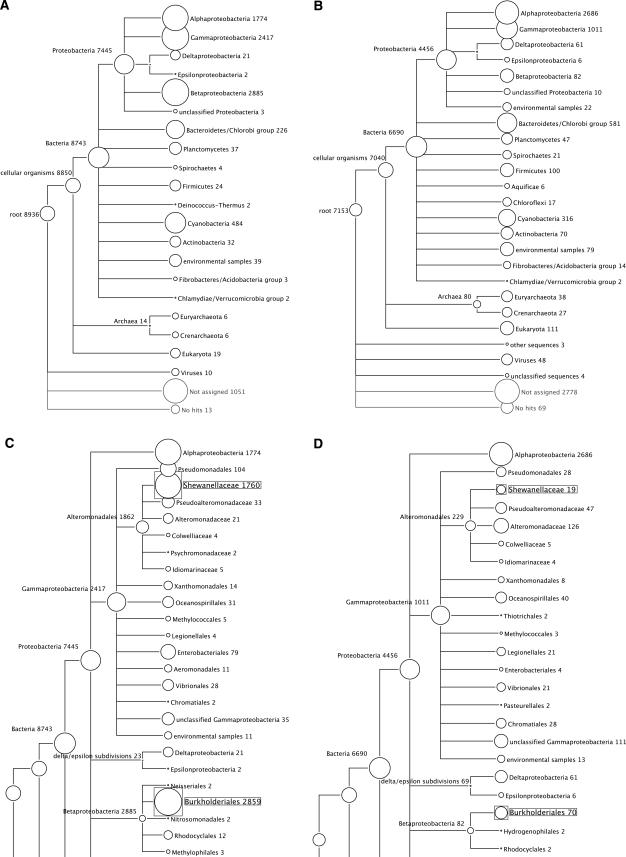
Figure 3.
Phylogenetic diversity of the Sargasso Sea sequences computed by MEGAN. The microheterogeneity of Sample 1 was investigated by comparing it to pooled Samples 2, 3, and 4 (Venter et al. 2004). (A) Analysis of 10,000 reads randomly chosen from Sample 1. (B) Analysis of 10,000 reads randomly chosen from Sample 2. (C,D) A more detailed view of Sample 1 and Samples 2–4, respectively, illustrating a significant difference of relative frequencies of Shewanella and Burkholderia species in the two data sets. In all such figures, each circle represents a taxon in the NCBI taxonomy and is labeled by its name and the number of reads that are assigned either directly to the taxon, or indirectly via one of its subtaxa. The size of the circle is scaled logarithmically to represent the number of reads assigned directly to the taxon.
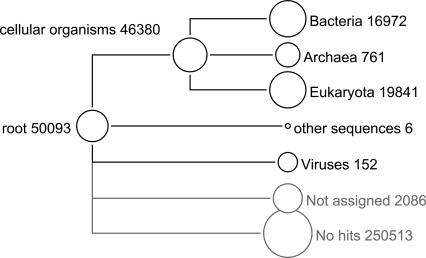
Figure 5.
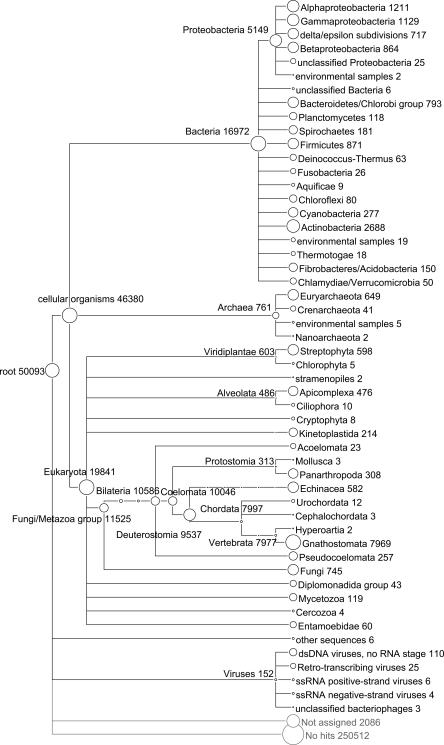
High-level summary of a MEGAN analysis of the mammoth data set, based on a BLASTX comparison of the 302,692 reads against the NCBI-NR database.
Figure 8.
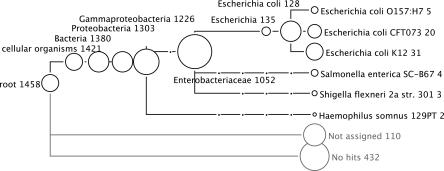
MEGAN analysis of 2000 reads collected from B. bacteriovorus HD100 using Roche GS20 sequencing. (A) Analysis based on a BLASTX comparison with NCBI-NR. (B) The same analysis, but with all hits matching database sequences representing the B. bacteriovorus HD100 genome removed, mimicking the situation in which the reads originate from a genome that is not represented in NCBI-NR.
As different metagenomics projects need to use different alignment tools and databases, we have designed MEGAN in such a way that gives users unrestricted choice in this matter. In our studies, we used BLAST comparisons (Altschul et al. 1990) against the NCBI-NR, NCBI-NT, NCBI-ENV-NR, and NCBI-ENV-NT databases (Benson et al. 2006), and additional genome-specific databases, where appropriate.
Although well established and trivial to carry out, sequence comparison is the main computational bottleneck in metagenomic analysis and will become increasingly critical, as the size of data sets and databases continues to grow. There is a tradeoff to be considered: Whole-genome approaches are easier to execute and potentially provide better taxonomical resolution than projects that target specific phylogenetic markers, but the additional computational burden can be immense.
Re-analysis of the Sargasso Sea data set
In the Sargasso Sea project (Venter et al. 2004), samples of seawater were collected, and organisms of size 0.1–3 μm were extracted to produce a metagenomic data set. From four individual sampling sites, ∼1.66 million reads of average length 818 bp were determined using Sanger sequencing. The biological diversity and species richness was measured using environmental assemblies, and also by analyzing six specific phylogenetic markers (rRNA, RecA/RadA, HSP70, RpoB, EF-Tu, and Ef-G). The species profile of 16 taxonomical groups generated by this approach shows a prevalence of Alphaproteobacteria and Gammaproteobacteria by a factor of 2–4 over the remaining 14 taxonomic groups, with only the Cyanobacteria being notably more frequent than the remaining taxa.
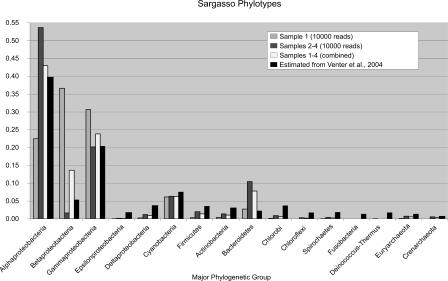
The Venter et al. (2004) study pioneered random genome sequencing of environmental samples. Their analysis of the data relies on the frequency of individual species to their contribution of scaffolds and contigs or matches to six established phylogenetic markers. The analysis performed by MEGAN uses an independent statistical approach, arriving at a very similar result for the species distribution. In Figure 3, A and B, we present the result of a MEGAN analysis for Sample 1 and pooled Samples 2–4, respectively, based on two subsets of 10,000 reads per data set. These results closely resemble the species distribution reported in Venter et al. (2004). In Figure 4, we report the averaged weighted percentage of the six phylogenetic markers for each of the 16 taxonomic groups, as estimated from Venter et al. (2004), and compare the result to the corresponding values produced by MEGAN.
Figure 4.
The distribution of reads from Sample 1, pooled Samples 2–4, and the weighted average of these two data sets, over 16 major phylogenetic groups, as computed by MEGAN. For the sake of comparison, the diagram also shows the relative contribution of organisms to these groups, as estimated from Venter et al. (2004) by averaging over the values for all six genes that are reported there.
Figure 3 demonstrates that MEGAN can easily detect a sampling bias between Sample 1 and pooled Samples 2–4, despite the fact that only a small fraction (20,000 reads, ∼1% of the total data set) was analyzed. This discrepancy, referred to as “microheterogeneity” by Venter et al. (2004), concerns an over-representation of members of the proteobacteria groups Shewanella and Burkholderia in Sample 1 (DeLong 2005). Both bacteria are not expected to be present in pelagic marine samples, as they live either in aquatic, nutrient-rich environments (Shewanella) or are found in terrestrial settings (Burkholderia) (Hicks et al. 2000; Nealson and Scott 2003; DeLong 2005).
To describe our process in more detail, firstly, we downloaded the complete set of Sargasso Sea Samples 1–4 from DDBJ/EMBL/GenBank (accession no. AACY0100000). We then selected the first 10,000 reads from Sample 1 and randomly selected a pooled set of 10,000 reads from Samples 2–4. On both data sets, we ran a BLASTX comparison against the NCBI-NR database, using default parameters. For the Sample 1 data set, only 1% of the reads had no hits (13) or remained unassigned (1051). Similarly, for the Sample 2–4 data set, <3% of the reads had no hits (69) or remained unassigned (2778).
We performed a MEGAN analysis of both data sets using a bit-score threshold of 100 (min-score filter; see Methods for more details on these parameters) and retaining only those hits whose bit scores lie within 5% of the best score (top-percent filter). In addition, all isolated assignments (that is, taxa that were hit by only one read) were discarded (min-support filter). For Sample 1, ∼83% (8336) of all reads were assigned to taxa that were more specific than the kingdom level, a majority of which (8298) were assigned to bacterial groups. For Samples 2–4, ∼59% (5195) of all reads were assigned to taxa that are more specific than the kingdom level, a majority of which (5709) were assigned to bacterial groups. In both cases, the numbers of reads assigned to eukaryotes and viruses are very small, which is readily explained by the size filtering used. However, size filtering does not explain why the number of Archaea is 100 times smaller than the number of Bacteria in the pelagic environment sampled. The observed difference in frequency may in part be explained by the fact that there is at least 10 times as much bacterial sequence information in the public databases as there is archaeal. Whether the remaining 10-fold difference reflects the true situation in the environment is currently an open question.
The analysis of the 16 taxonomic groups performed in Venter et al. (2004) does not provide an estimation of the absolute numbers of reads allowing assignment to a taxonomic group. MEGAN can readily produce such statistics because the LCA algorithm explicitly assigns every individual read, for which database hits are available, to some taxon in the NCBI taxonomy, regardless of the read’s suitability as a phylogenetic marker. As an example of the quantification of assigned reads, out of the 10,000 reads of Sample 1, a total of 8743 reads are assigned to the node labeled “Bacteria,” or to one of the descendants of this node. Furthermore, 7445 reads are assigned to Proteobacteria, of which 1774, 2885, 2417, 21, 2, and 3 are more specifically assigned to Alpha-, Beta-, Gamma-, Delta-, Epsilon-, and unclassified Proteobacteria, respectively (see Fig. 3A).
Analysis of the mammoth data set
In (Poinar et al. 2006), we used Roche GS20 sequencing technology (Margulies et al. 2005) to randomly sequence DNA from a sample of 1 g of bone taken from a mammoth that was preserved in permafrost for 28,000 yr. We obtained 302,692 reads of mean length 95 bp. We refer to this as the “mammoth data set.” As similar specimens were shown to contain large amounts of environmental sequences in addition to host DNA, the study was designed as a metagenomics project.
To identify those reads that come from the mammoth genome, we performed BLASTZ (Schwartz et al. 2003) comparisons against genome sequences for elephant, human, and dog, downloaded from http://www.genome.ucsc.edu. As a result of this computation, we estimate that at least 45.4% of the reads represent mammoth DNA (Poinar et al. 2006). The remaining portion of reads was likely to be derived from environmental organisms, such as bacteria, fungi, amoeba, and nematodes. These organisms are likely to have lived on the carcass of the mammoth and may have contributed to the putrification process.
To determine the distribution of environmental sequences in the sample, we first used BLASTX to compare all reads against the NCBI-NR (“non-redundant”) protein database (Benson et al. 2006), which does not contain any sequence information from the elephant genome project. This computation resulted in a file of size 1.4 GB containing 2,911,587 local alignments of reads to sequences in the database. Of the 302,692 reads, 52,179 resulted in one or more alignments (17.2%). We then loaded the results of the BLASTX search into a prelimary version of MEGAN and applied the LCA algorithm to compute an assignment of reads to taxa, thus obtaining an estimation of the taxonomical content of the sample.
Here we provide details of the MEGAN analysis, using a bit-score threshold of 30 and discarding any isolated assignments, that is, any taxon that has only a single read assigned to it. The LCA algorithm assigned 50,093 reads to taxa, and 2086 remained unassigned either because the bit-score of their matches fell below the threshold or because they gave rise to an isolated hit.
A total of 19,841 reads were assigned to Eukaryota, of which 7969 were assigned to Gnathostomata (jawed vertebrates) and thus presumably derive from mammoth sequences. Furthermore, a total of 16,972 reads were assigned to Bacteria, 761 to Archea, and 152 to Viruses, respectively. These numbers are marginally lower than those reported in Poinar et al. (2006) because of our new filters, thus underlining the intrinsic robustness of the LCA approach.
Figures 5 and 6 demonstrate the ability of MEGAN to summarize results at different levels of the NCBI taxonomy. A distinctive feature of the program is that such summaries are computed dynamically on-the-fly, as the user changes parameters of the LCA algorithm or expands or collapses parts of the taxonomy. The relative abundance of reads at a certain node or leaf is indicated visually by the size of the circle representing the node, or by numerical labels. The cladograms produced by MEGAN can be considered “species profiles” and can be produced as tables, for example, for side-by-side comparisons of series of samples (see Fig. 4).
Figure 6.
A low level view of the MEGAN analysis of the mammoth data set.
Species identification from short reads
Several companies are developing new sequencing technologies that promise to produce high-throughput sequencing at substantially reduced cost, albeit with reads as short as 35 bp. The average length of reads produced using current Roche GS20 sequencing technology, introduced last year (Margulies et al. 2005), is ∼100 bp, and reads obtainable by current Sanger sequencing are ∼800 bp in length (Franca et al. 2002). The question therefore arises what read length is required to identify species in a metagenomic sample reliably.
A simple approach to addressing this is to collect a set of reads from a known genome, to process the data as a metagenomic data set (as described above), and then to evaluate the accuracy of the assignments. For this purpose, the genome sequence of the two organisms E. coli K12 and B. bacteriovorus HD100 were used. We chose E. coli as it is used as a cloning host in most clone-based sequencing projects and is thus likely to occur in several different database sequences by mistake. The second test organism, B. bacteriovorus, is very distinctive in its sequence from other Proteobacteria and has no close relatives that are currently represented in the sequence databases. Its metagenomic analysis should therefore result in a much better signal/noise ratio than for E. coli.
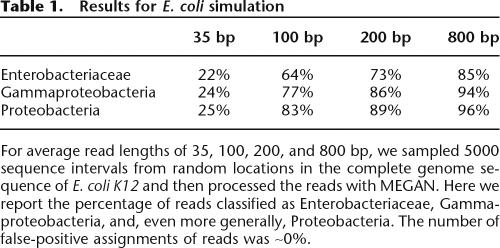
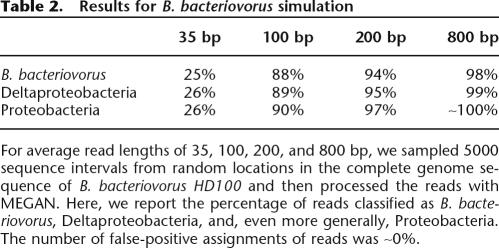
We show the results of simulation studies for the two genomes in Tables 1 (E. coli) (Blattner et al. 1997) and 2 (B. bacteriovorus) (Rendulic et al. 2004). For each genome, we use sequence intervals of length 35 bp, 100 bp, 200 bp, and 800 bp, as these lengths correspond to upcoming or existing sequencing technology. We simulated 5000 random shotgun reads for each datapoint, compared them to the NCBI-NR database using BLASTX, and then processed the reads with MEGAN, using a bit-score threshold of 35, retaining only those hits that are within 20% of the best hit for a read, and discarding all isolated assignments. The percentage of reads classified as Enterobacteriaceae ranged from 22% to 85%, Gammaproteobacteria from 24% to 94%, and Proteobacteria from 25% to 96% in the case of E. coli. The number of false-positive assignments of reads was 0%. In the case of B. bacteriovorus, the percentage of reads classified as B. bacteriovorus ranges from 25% to 98%, Deltaproteobacteria from 26% to 99%, and Proteobacteria from 26% to ∼100%. No false-positive hits were detected. The result demonstrates that short reads in general can be used for metagenomic analysis, albeit at the cost of a high rate of under-prediction.
Table 1.
Results for E. coli simulation
For average read lengths of 35, 100, 200, and 800 bp, we sampled 5000 sequence intervals from random locations in the complete genome sequence of E. coli K12 and then processed the reads with MEGAN. Here we report the percentage of reads classified as Enterobacteriaceae, Gammaproteobacteria, and, even more generally, Proteobacteria. The number of false-positive assignments of reads was ∼0%.
Table 2.
Results for B. bacteriovorus simulation
For average read lengths of 35, 100, 200, and 800 bp, we sampled 5000 sequence intervals from random locations in the complete genome sequence of B. bacteriovorus HD100 and then processed the reads with MEGAN. Here, we report the percentage of reads classified as B. bacteriovorus, Deltaproteobacteria, and, even more generally, Proteobacteria. The number of false-positive assignments of reads was ∼0%.
Using Roche GS20 sequencing technology, we sequenced a test set of 2000 reads from random positions in the E. coli K12 genome of length ∼100 bp. Figure 7 shows the details of a MEGAN analysis of these data, which is based on a BLASTX comparison of the reads against the NCBI-NR database, using the same parameters as above. Of the 2000 reads, ∼25% (432) have no hits, and 110 reads are not assigned. Of the remaining 1458 reads, ∼75% (1052) are assigned to Enterobacteriaceae, thus making a correct assignment up to the taxonomic level of family. All other reads, except two, are assigned to super-taxa, thus producing correct, if increasingly weak, predictions.
Figure 7.
MEGAN analysis of 2000 reads collected from E. coli K12 using Roche GS20 sequencing, based on a BLASTX comparison with the NCBI-NR database.
The two false-positive assignments to Haemophilus somnus appear to be due to false entries in the NCBI-NR database: the two database sequences are labeled “hypothetical proteins”; however, one is identical to the 16S rRNA sequence in E. coli, and the other is identical to the 23S rRNA sequence in E. coli.
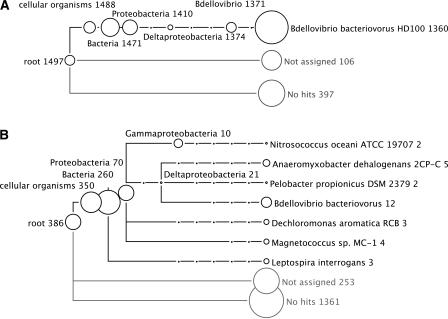
In a second experiment, we considered 2000 reads of length ∼100 bp randomly collected from B. bacteriovorus HD100 using the same sequencing technology. In Figure 8A, we show the resulting MEGAN analysis, which is based on a BLASTX comparison of the reads against the NCBI-NR database, using the same parameters as above. Of the 2000 reads, ∼20% (397) have no hits, and 5% (106) are not assigned. Of the remaining 1498 reads, ∼70% (1360) are assigned to B. bacteriovorus HD100. All other reads are assigned to super-taxa, once again producing correct, if increasingly weak, predictions. There are no false-positive predictions.
In Figure 8B, we show a similar MEGAN analysis obtained when using a copy of the NCBI-NR database from which all sequences representing the B. bacteriovorus HD100 genome have been removed. This mimics the case in which reads are obtained from a genome that is not yet represented in the database. Of the 2000 reads, ∼65% (1361) have no hits, and ∼13% (253) are not assigned. A small number of false positives occur up to the level of Bacteria.
While these two experiments conducted with organisms of known phylogenetic distance demonstrate the robustness of the LCA algorithm, its performance on unknown, more distantly related sequences can only be estimated. Given the logical structure of the LCA algorithm, however, we predict a low rate of false-positive assignments at the price of producing fairly large numbers of unspecific assignments or no hits. Independent of MEGAN’s design, the outcome of each analysis will be biased by the content of the database used and will only improve as sequence databases become more complete. In addition to the generation of more sequence data, new algorithms will be required to structure databases of environmental content, as currently the taxon frequencies of unknown organisms cannot be assessed.
Species and strain identification through species–specific genes
For in-depth metagenomic analysis, it is of particular interest to resolve the taxonomical tree down to the species level, as illustrated in Figure 7. The analysis of random reads allows one to distinguish between closely related species and strains, and thus to obtain a level of resolution that is not possible using phylogenetic markers. This is due to the fact that random sequencing also targets species- and strain-specific genes that are not usually used in a phylogenetic analysis. Furthermore, in many cases the diffentiation between a pathogenic and a nonpathogenic strain can only be based on gene content and not on the similarity of shared genes. The presence of reads that clearly distinguish pathogenic variants from mutualistic ones will contribute toward the understanding of potential pathogens in the environment. To this end, species or taxa of interest can be searched for using a Find tool (Fig. 9A), and the distribution of reads over known strains of a species can be viewed (Fig. 9B). Underlying sequence alignments can be manually inspected (Fig. 9C), and individual sequences can be extracted for evaluation with other tools.
Figure 9.
(A) MEGAN provides a Find tool to search for specific taxa of interest. (B) The result of a search is highlighted in a detailed summary of the analysis. (C) MEGAN provides an Inspector tool to view the individual sequence comparisons upon which the assignment of a particular read to a particular taxon is based.
Discussion
Early metagenomic studies resorted to screening of environmental libraries for the presence of known phylogenetic markers and subsequent sequencing of clones of interest (Béja et al. 2000, 2001; Rondon et al. 2000; Quaiser et al. 2003; Treusch et al. 2004). Venter et al. (2004) pioneered random genome sequencing of environmental samples, producing data on a much larger scale, and shifted the focus from short scaffolds to high coverage contigs of dozens of kilobases long. Sequence information of this type allows for a rough annotation of the metabolic capacity of a microbial community of interest, and the statistics of such assemblies can be used in a population genetics context to distinguish between discrete species and populations of closely related biotypes.
The problem of species identification in a mixture of organisms has been addressed using proven phylogenetic markers, such as the ribosomal genes (16S, 18S, and 23S rRNA) or coding sequences of genes involved in the transcription or translation machinery of the cell (e.g., recA/radA, hsp70, EF-Tu, Ef-G, rpoB). By definition, such markers are based on slow-evolving genes and aim at distinguishing between species at large evolutionary distances, and are thus unsuitable for resolving closely related organisms.
MEGAN deviates from the analytical pattern of previous metagenomic analysis pipelines and builds on the statistical power of comparing random sequence intervals with unspecified phylogenetic properties against databases of known sequences. This study demonstrates that even given the current incomplete and biased state of the DNA-, protein-, and environmental databases, a meaningful categorization of random reads is possible as a useful first phylogenetic analysis of metagenomic data. The ability to identify species depends, of course, on the presence or absence of closely related sequences in the databases, as demonstrated in Figure 8. Removal of the source genome B. bacteriovorus HD100 from the database results in a threefold increase of completely unassigned reads, while producing only a small number of false-positive identifications above the level of Proteobacteria. This underlines the fact that MEGAN takes a conservative approach to taxon identification. Lack of data may result in severe under-prediction or large numbers of unassigned reads, but will not result in a significant amount of over-prediction.
Laptop analysis
Early approaches to metagenomic analysis frequently involved large teams of bioinformaticians who generated intricate analysis pipelines with complex outputs.
MEGAN can be used to analyze DNA reads collected within the framework of any metagenomics project, regardless of the sequencing technology used. In a pre-processing step, the set of DNA reads (or contigs) is compared against databases of known sequences using BLAST or other comparison tools. This computationally demanding task will usually be performed on a high-performance computer cluster. Once completed, the resulting files can be downloaded onto a laptop or workstation and then interactively analyzed using MEGAN.
Assuming that the reads are randomly selected from the metagenomic sample, MEGAN analysis can be viewed as a statistical approach with several attractive features. Because the reads are independently sampled from random regions of the genomes that can have very different levels of conservation, this type of analysis will show better resolution at all levels of the taxonomy, and particularly at the species and strain level, than an analysis based on a small set of phylogenetic markers, as their rate of evolution is slower than average. Because the analysis does not require an assembly of the reads into contigs, all problems associated with assembling data from a mixture of potentially very similar genomes are avoided.
The software is easy to deploy as it operates on data produced by existing and widely available bioinformatics software tools for alignments (such as BLAST, BLASTZ, and other comparison tools) and publicly accessible data resources (sequence databases and the NCBI taxonomy). As sequence comparisons are computationally intensive and time-consuming, they should be performed only once with sufficiently relaxed alignment parameters. MEGAN provides filters to adjust the level of stringency later to an appropriate level. An investigator can perform a detailed analysis of a large metagenomic data set and manually inspect the correctness of each classification without needing to rerun the sequence comparison at various cutoff levels.
Intrinsic biases of current metagenomic analysis
The three key elements in the analysis pipeline are the sequence database, the alignment software for sequence comparison, and a generally accepted taxonomy of known organisms. The first element consists of public sequence databases, which are curated by NCBI, EBI, and DDBJ. The content of such databases is heavily biased by an anthropocentric research focus, and only poorly reflects the biological diversity of this planet. This fact introduces the largest bias in any metagenomic analysis, which presently cannot be circumvented. The second component, the sequence alignment tool, is the most critical with regard to the computational cost of an analysis. As sequence databases continue to grow and metagenomic projects increase in size, the computational cost will also increase. However, as databases begin to provide a better coverage of the diversity of life, the computational cost of performing these analyses may actually begin to sink again, as more stringent global alignments will begin to replace less stringent (and thus more costly) local comparisons.
The third component is the taxonomical classification of species used. Our approach is based on the NCBI taxonomic system, which is maintained and updated by a team of taxonomy experts, who incorporate both sequence-based and non-sequence-based taxonomic information. However, MEGAN allows for the integration of other taxonomic systems as well.
Current issues and future extensions
MEGAN is designed to post-process the results of a set of sequence comparisons against one or more databases and places no explicit restrictions on the type of reads, the sequence comparison method, or databases used. Hence, we anticipate that our approach will remain valid even when innovations are introduced in any of these areas.
The current LCA assignment algorithm bases its decision solely on the presence or absence of hits between reads and taxa. We are currently contemplating a more sophisticated approach that will not only take the presence or absence of hits into account, but will also make use of the quality of the matches and the levels of similarity that are typical for given genes in a given clade of sequences.
It is intriguing to see how robust and correct the taxonomical assignments based on local alignments performed with either BLASTN or BLASTX can be. While these tools create alignments of variable length from sequence intervals of unspecified phylogenetic relevance, potential problems are overcome by the power of statistics. By default, MEGAN requires that at least two reads are assigned to a taxon before that taxon is deemed to be present, and this helps to prevent false positives. Moreover, by design, short, highly conserved domains will lead to an unspecific assignment, rather than to a false one.
The analysis of any metagenomic data set will produce a significant set of sequences that cannot be assigned to any known taxon, and the question arises how to estimate the number of unknown species. In our experience (data not shown), anywhere between 10% and 90% of all reads may fail to produce any hits when compared with BLASTX against NCBI-NR. To estimate how many of these reads actually come from unknown species, one must take into account that most known species are only partially represented in current databases. If, for example, only 10% of the genome of a species is present in the databases, then for every correctly identified read, there will be as many as nine that do not produce a hit. As there is insufficient information on the size of genomes to make such estimations in a precise way, such calculations have not yet been implemented in MEGAN.
Can short sequence intervals identify a species?
The currently available sequencing technologies provide sequencing reads from 35 bp (upcoming sequencing-by-synthesis approaches) to ∼800 bp (Sanger sequencing). Assignments based on very short reads of less than ∼50 bp will suffer from low confidence values (such as bit scores in the case of BLAST), whereas reads of length ∼100 bp can be assigned with a reasonable level of confidence (BLASTX bit-scores of 30 and higher). As shown in Tables 1 and 2, MEGAN analysis correctly assigns fragments as short as 35 bp. However, short read lengths result in severe under-prediction, which will reduce the cost efficiency of the new technologies. While our work indicates that reads of length 35 bp and 100 bp are long enough to identify a species, the hit statistics from Tables 1 and 2 suggest that 200 bp might constitute an optimal tradeoff between the rate of under-prediction and the production cost of such reads.
While new developments in sequencing technology will continue to impact metagenomic projects in terms of cost and throughput, we believe that MEGAN analysis will remain a valuable tool for analyzing the new data and will help scientists to dissect the sequence information of their environmental samples.
Methods
Sequence comparisons
In our studies, we performed sequence comparisons against the NCBI-NR database of nonredundant protein sequences using BLASTX with the default settings, the NCBI-NT database on nucleotide sequences using BLASTN with the default settings, and against whole-genome sequences obtained from dog, elephant, and human, using BLASTZ. Sequence comparison is a computationally challenging task that is likely to grow even more demanding as databases continue to grow and larger metagenome data sets are analyzed. For example, comparing the mammoth data set against NCBI-NR took almost 180 h real time on a cluster of 64 CPUs. We estimate that performing the same computation on the 1.6 million reads of the complete Sargasso Sea data set would require ∼1000 h real time on our system.
Analysis using MEGAN
At the startup, MEGAN loads the complete NCBI taxonomy, currently containing >280,000 taxa, which can then be interactively explored using customized tree-navigation features. However, the main application of MEGAN is to process the results of a comparison of reads against a database of known sequences. The program parses files generated by BLASTX, BLASTN, or BLASTZ, and saves the results as a series of read–taxon matches in a program-specific metafile. (Additional parsers may be added to process the results generated by other sequence comparison methods.)
The program assigns reads to taxa using the LCA algorithm and then displays the induced taxonomy. Nodes in the taxonomy can be collapsed or expanded to produce summaries at different levels of the taxonomy. Additionally, the program provides a search tool to search for specific taxa, and an Inspector tool to view individual BLAST matches (see Fig. 9).
The approach uses several thresholds. First, the min-score filter sets a threshold for the score that an alignment must achieve to be considered in the calculations. For reads of length ∼100 bp and using BLASTX to compare against NCBI-NR, a min-score of 35 or higher is recommended; while for reads of length ∼800 bp, a min-score of 100 is more suitable. Second, to help distinguish between hits due to sequence identity and those due to homology, the top-percent filter is used to retain only those hits for a given read r whose scores lie within a given percentage of the highest score involving r. (Note that this is not the same as keeping a certain percentage of the hits.) The smaller the set value is, the more specific a calculated assignment will be, but also the greater the chance of producing an over-prediction, that is, a false prediction due to the absence of the true taxon in the database. A useful range of values is 10%–20%. Third, a win-score threshold can be set such that, for any given read, if any match scores above the threshold, then for that read, only those matches are considered that score above the threshold. Fourth, to help reduce false positives, the min-support filter is used to set a threshold for the minimum number of reads that must be assigned to a taxon t, or to any of its descendants in the taxonomical tree. After the main computation, all reads that are assigned to a taxon that does not meet this requirement are reassigned to the special taxon “Not Assigned.” By default, this parameter is set to 2.
The result of the LCA algorithm is presented to the user as the partial taxonomy T that is induced by the set of taxa that have been identified (see Fig. 5). The program allows the user to explore the results at many different taxonomical levels, by providing methods for collapsing and expanding different parts of the taxonomy T. Each node in T represents a taxon t and can be queried to determine which reads have been assigned directly to t, and how many reads have been assigned to taxa below t. Additionally, the program allows the user to view the sequence alignments upon which specific assignments are based (see Fig. 9).
Acknowledgments
D.H. thanks the DFG for funding and Ramona Schmid and Mike Steel for helpful discussions. S.S. thanks The Gordon and Betty Moore Foundation for supporting a part of this project. S.S. and D.H. thank Webb Miller and Francesca Chiaromonte for stimulating discussions and comments on the computational approach.
Footnotes
[MEGAN is freely available at http://www-ab.informatik.uni-tuebingen.de/software/megan.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5969107
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Béja O., Aravind L., Koonin E.V., Suzuki M.T., Hadd A., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Aravind L., Koonin E.V., Suzuki M.T., Hadd A., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Koonin E.V., Suzuki M.T., Hadd A., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Suzuki M.T., Hadd A., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Hadd A., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Nguyen L.P., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Jovanovich S.B., Gates C.M., Feldman R.A., Spudich J.L., Gates C.M., Feldman R.A., Spudich J.L., Feldman R.A., Spudich J.L., Spudich J.L., et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béja O., Spudich E.N., Spudich J.L., Leclerc M., DeLong E.F., Spudich E.N., Spudich J.L., Leclerc M., DeLong E.F., Spudich J.L., Leclerc M., DeLong E.F., Leclerc M., DeLong E.F., DeLong E.F. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Benson D., Karsch-Mizrachi I., Lipman D., Ostell J., Wheeler D., Karsch-Mizrachi I., Lipman D., Ostell J., Wheeler D., Lipman D., Ostell J., Wheeler D., Ostell J., Wheeler D., Wheeler D. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Glasner J.D., Rode C.K., Mayhew G.F., Rode C.K., Mayhew G.F., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- DeLong E.F. Microbial community genomics in the ocean. Nat. Rev. Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- DeLong E.F., Preston C.M., Mincer T., Rich V., Hallam S.J., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Preston C.M., Mincer T., Rich V., Hallam S.J., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Mincer T., Rich V., Hallam S.J., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Rich V., Hallam S.J., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Hallam S.J., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Frigaard N.-U., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Martinez A., Sullivan M.B., Edwards R., Rodriguez Brito B., Sullivan M.B., Edwards R., Rodriguez Brito B., Edwards R., Rodriguez Brito B., Rodriguez Brito B., et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;27:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Franca L.T., Carrilho E., Kist T.B., Carrilho E., Kist T.B., Kist T.B. A review of DNA sequencing techniques. Q. Rev. Biophys. 2002;35:169–200. doi: 10.1017/s0033583502003797. [DOI] [PubMed] [Google Scholar]
- Hallam S.J., Putnam N., Preston C., Detter J., Rokhsar D., Putnam N., Preston C., Detter J., Rokhsar D., Preston C., Detter J., Rokhsar D., Detter J., Rokhsar D., Rokhsar D. Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C.L., Kinoshita R., Ladds P.W., Kinoshita R., Ladds P.W., Ladds P.W. Pathology of melioidosis in captive marine mammals. Aust. Vet. J. 2000;78:193–195. doi: 10.1111/j.1751-0813.2000.tb10593.x. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W., Attiya S., Bader J., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Egholm M., Altman W., Attiya S., Bader J., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Altman W., Attiya S., Bader J., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Attiya S., Bader J., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Bader J., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Bemben L., Berka J., Braverman M., Chen Y.-J., Chen Z., Berka J., Braverman M., Chen Y.-J., Chen Z., Braverman M., Chen Y.-J., Chen Z., Chen Y.-J., Chen Z., Chen Z., et al. Genome sequencing in micro-fabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny J.B., Bohannan B.J., Brown J.H., Colwell R.K., Fuhrman J.A., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Bohannan B.J., Brown J.H., Colwell R.K., Fuhrman J.A., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Brown J.H., Colwell R.K., Fuhrman J.A., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Colwell R.K., Fuhrman J.A., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Fuhrman J.A., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Green J.L., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Horner-Devine M.C., Kane M., Krumins J.A., Kuske C.R., Kane M., Krumins J.A., Kuske C.R., Krumins J.A., Kuske C.R., Kuske C.R., et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Meldrum D. Automation for genomics, part one: Preparation for sequencing. Genome Res. 2000a;10:1081–1092. doi: 10.1101/gr.101400. [DOI] [PubMed] [Google Scholar]
- Meldrum D. Automation for genomics, part two: Sequencers, microarrays, and future trends. Genome Res. 2000b;10:1288–1303. doi: 10.1101/gr.157400. [DOI] [PubMed] [Google Scholar]
- Nealson K.H., Scott J., Scott J. In: The prokaryotes: An evolving electronic resource for the microbiological community. Dworkin E.A., editor. Springer-Verlag; New York: 2003. [Google Scholar]
- Poinar H.N., Schwarz C., Qi J., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., Schwarz C., Qi J., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., Qi J., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., Buigues B., Tikhonov A., Huson D., Tomsho L.P., Auch A., Tikhonov A., Huson D., Tomsho L.P., Auch A., Huson D., Tomsho L.P., Auch A., Tomsho L.P., Auch A., Auch A., et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;331:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Quaiser A., Ochsenreiter T., Lanz C., Schuster S.C., Treusch A.H., Eck J., Schleper C., Ochsenreiter T., Lanz C., Schuster S.C., Treusch A.H., Eck J., Schleper C., Lanz C., Schuster S.C., Treusch A.H., Eck J., Schleper C., Schuster S.C., Treusch A.H., Eck J., Schleper C., Treusch A.H., Eck J., Schleper C., Eck J., Schleper C., Schleper C. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 2003;50:563–575. doi: 10.1046/j.1365-2958.2003.03707.x. [DOI] [PubMed] [Google Scholar]
- Rendulic S., Jagtap P., Rosinus A., Eppinger M., Baar C., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Jagtap P., Rosinus A., Eppinger M., Baar C., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Rosinus A., Eppinger M., Baar C., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Eppinger M., Baar C., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Baar C., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Lanz C., Keller H., Lambert C., Evans K.J., Goesmann A., Keller H., Lambert C., Evans K.J., Goesmann A., Lambert C., Evans K.J., Goesmann A., Evans K.J., Goesmann A., Goesmann A., et al. A predator unmasked: Life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Rondon M.R., August P.R., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., August P.R., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Lynch B.A., MacNeil I.A., Minor C., MacNeil I.A., Minor C., Minor C., et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele H., Streit W., Streit W. Metagenomics: Advances in ecology and biotechnology. FEMS Microbiol. Lett. 2005;247:105–111. doi: 10.1016/j.femsle.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Treusch A.H., Kletzin A., Raddatz G., Ochsenreiter T., Quaiser A., Meurer G., Schuster S.C., Schleper C., Kletzin A., Raddatz G., Ochsenreiter T., Quaiser A., Meurer G., Schuster S.C., Schleper C., Raddatz G., Ochsenreiter T., Quaiser A., Meurer G., Schuster S.C., Schleper C., Ochsenreiter T., Quaiser A., Meurer G., Schuster S.C., Schleper C., Quaiser A., Meurer G., Schuster S.C., Schleper C., Meurer G., Schuster S.C., Schleper C., Schuster S.C., Schleper C., Schleper C. Characterization of large-insert DNA libraries from soil for environmental genomic studies of Archaea. Environ. Microbiol. 2004;6:970–980. doi: 10.1111/j.1462-2920.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- Tringe S.G., von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Podar M., Short J.M., Mathur E.J., Detter J.C., Short J.M., Mathur E.J., Detter J.C., Mathur E.J., Detter J.C., Detter J.C., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Raml R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Chapman J., Hugenholtz P., Allen E.E., Raml R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Hugenholtz P., Allen E.E., Raml R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Allen E.E., Raml R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Raml R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F., Rubin E.M., Rokhsar D.S., Banfield J.F., Rokhsar D.S., Banfield J.F., Banfield J.F., et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Wu D., Paulsen I., Nelson K.E., Nelson W., Paulsen I., Nelson K.E., Nelson W., Nelson K.E., Nelson W., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Zhang K., Martiny A.C., Reppas N.B., Barry K.W., Malek J., Chisholm S.W., Church G.M., Martiny A.C., Reppas N.B., Barry K.W., Malek J., Chisholm S.W., Church G.M., Reppas N.B., Barry K.W., Malek J., Chisholm S.W., Church G.M., Barry K.W., Malek J., Chisholm S.W., Church G.M., Malek J., Chisholm S.W., Church G.M., Chisholm S.W., Church G.M., Church G.M. Sequencing genomes from single cells via polymerase clones. Nat. Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]