Abstract
A meiotic linkage map is essential for mapping traits of interest and is often the first step toward understanding a cryptic genome. Specific strains of silver fox (a variant of the red fox, Vulpes vulpes), which segregate behavioral and morphological phenotypes, create a need for such a map. One such strain, selected for docility, exhibits friendly dog-like responses to humans, in contrast to another strain selected for aggression. Development of a fox map is facilitated by the known cytogenetic homologies between the dog and fox, and by the availability of high resolution canine genome maps and sequence data. Furthermore, the high genomic sequence identity between dog and fox allows adaptation of canine microsatellites for genotyping and meiotic mapping in foxes. Using 320 such markers, we have constructed the first meiotic linkage map of the fox genome. The resulting sex-averaged map covers 16 fox autosomes and the X chromosome with an average inter-marker distance of 7.5 cM. The total map length corresponds to 1480.2 cM. From comparison of sex-averaged meiotic linkage maps of the fox and dog genomes, suppression of recombination in pericentromeric regions of the metacentric fox chromosomes was apparent, relative to the corresponding segments of acrocentric dog chromosomes. Alignment of the fox meiotic map against the 7.6x canine genome sequence revealed high conservation of marker order between homologous regions of the two species. The fox meiotic map provides a critical tool for genetic studies in foxes and identification of genetic loci and genes implicated in fox domestication.
The silver fox is a coat color variant of the red fox (Vulpes vulpes) (Nes et al. 1988). In nature, the red fox has a wider geographical distribution than any other Carnivora species, inhabiting ecological zones ranging from tundra to desert, and demonstrates remarkable variation in size and morphology (Nowak 1991; Sheldon 1992; MacDonald and Reynolds 2004). Multiple genetically determined coat color phenotypes have been maintained in farm-raised foxes since the early twentieth century (Nes et al. 1988; Vage et al. 1997). Although bred in captivity, farm-bred foxes normally exhibit a pattern of aggressive, fear-aggressive, and avoidance behavior toward humans. At the Institute of Cytology and Genetics of the Russian Academy of Sciences (ICG) the silver fox was chosen as a model for experimental domestication. Starting with a large commercial population, foxes were selected for tame behavior for over 45 generations to produce a strain with behavioral responses to humans like those of domestic dogs (Belyaev 1969, 1979; Trut 1980, 1999, 2001; Trut et al. 2004). In parallel, selection for aggressive behavior produced extremely aggressive and difficult-to-handle animals (see http://cbsu.tc.cornell.edu/ccgr/behaviour/index.html). Domesticated foxes are eager to establish human contact from a very early age (Trut 1999). Like dogs, they develop a close attachment to their owners when raised in human homes. Although selection focused solely on behavioral criteria, other characteristics of domestication including a curled tail, coat spotting, and other morphological, developmental, and neurohormonal changes emerged in these fox populations (Belyaev et al. 1984; Trut and Oskina 1985; Plyusnina et al. 1991; Oskina and Tinnikov 1992; Oskina 1996; Popova et al. 1997; Trut 2001). Phenotypic variation has also emerged in physical parameters of size, shape, and appearance (Trut et al. 2006) that draws parallels to those morphological phenotypes mapped in Portuguese water dogs (Chase et al. 2002), and provides an opportunity for a comparative approach to studying the genetic architecture of the mammalian skeleton (Lark et al. 2006).
Unlike modern dogs, the strain of domesticated foxes was created rapidly and recently by selection focused only on specific behavioral traits, suggesting that the imposed selection may be acting on relatively few loci influencing behavior. To map the genetic loci underlying these behavioral traits, a meiotic linkage map of the silver fox genome is required.
The red fox and the dog (Canis familiaris) are both members of the family Canidae, whose karyotypes present one of the most shuffled sets within mammalian families (Yang et al. 1999). The dog and fox diverged from a common ancestor about 10 million years ago (Wayne et al. 1997), and it is believed that the fox karyotype evolved from that of an ancestral dog-like animal (Yang et al. 1999; Wienberg 2004). The correspondence of chromosomal arms between the dog and fox has been established (Yang et al. 1999, 2000; Graphodatsky et al. 2000, 2001), and a comparative cytogenetic map of the dog and red fox genomes was developed by reciprocal chromosomal painting (Yang et al. 1999). The dog karyotype comprises 37 pairs of acrocentric autosomes and a pair of sex chromosomes while that of the red fox is 16 pairs of metacentric autosomes, a pair of sex chromosomes, and 0–8 B chromosomes (Wipf and Scackeford 1942; Belyaev et al. 1974). Although the dog lacks B chromosomes, in mammals these supernumerary chromosomes are generally considered to lack major genes, and FISH with fox B-chromosomal probes did not reveal any specific signals in the dog (Yang et al. 1999).
The evolutionary closeness of the dog and fox expedites investigation of the fox genome. The dog has well developed meiotic and radiation hybrid linkage maps (Mellersh et al. 1997; Neff et al. 1999, 2006; Werner et al. 1999; Breen et al. 2004), and the canine genome sequence (Kirkness et al. 2003; Hitte et al. 2005; Lindblad-Toh et al. 2005) anchors these maps. In a preliminary study, over 60% of tested canine-derived microsatellite markers robustly amplified fox DNA were polymorphic in foxes and were thus applicable for genotyping fox pedigrees (Kukekova et al. 2004). This confirmed previous demonstrations that microsatellites identified in well studied genomes can be used for genotyping closely related species (de Gortari et al. 1998; Rogers et al. 2000, 2006; Reed et al. 2005; Rexroad et al. 2005; Varshney et al. 2005).
To construct a meiotic linkage map of the fox genome we used markers that were either identified from the Marshfield canine microsatellite set, adapted from the canine integrated map, and/or identified directly from the canine genome sequence. In total, 320 markers were placed on the fox map, of which 318 could be uniquely identified in the 7.6x sequence of the dog. This fox linkage map is thus directly anchored to the dog genome sequence, enabling detailed comparisons to be made between corresponding chromosomal fragments of the two species and indirect comparisons between fox and human chromosomes. This map provides opportunities to evaluate how structural differences in genome arrangements affect the species–specific recombination landscape, facilitates exploitation of the dog genome for genetic studies in foxes, and presents an essential tool for mapping segregating traits of interest in foxes.
Results
Fox meiotic linkage map construction
The LOD 2.0 fox map
The 320 microsatellite markers placed on the fox meiotic linkage map included 162 canine markers identified previously (Kukekova et al. 2004) plus 158 markers newly adapted from the canine genome (Supplemental Table 1). On average, 195 (SD ± 62) informative meioses per marker were observed. Allele numbers varied among loci with a mean of 5.4 (SD ± 2.4) alleles per marker. Average marker PIC value was 0.5 (SD ± 0.16) (Supplemental Fig. 1).
The fox meiotic linkage map was constructed in a two-step process. First, using pairwise linkage analysis and LOD ≥4.0 threshold, 315 markers were assembled into 17 linkage groups, each containing six or more markers. A further small linkage group comprised only two markers (CM24.48, FH3287), and another three markers (FH2457, FH2001, and FH2233) were unlinked. The two-marker group and marker FH2457 were unambiguously assigned to single groups when tested at lower threshold (LOD score ≥3.5). Each linkage group was then evaluated, at LOD ≥3.0 to establish marker order; 199 markers were ordered at a confidence level of 1000:1 (LOD ≥3.0), as shown in Figure 1 (markers in plain text). The two initially unlinked markers (FH2001 and FH2233) were predicted from fox and dog comparative cytogenetic data (Yang et al. 1999, 2000; Graphodatsky et al. 2000, 2001) to be on the same chromosome as linkage group 11. Therefore, these two markers were analyzed together with the 19 markers initially placed in linkage group 11 to produce a framework map for this linkage group.
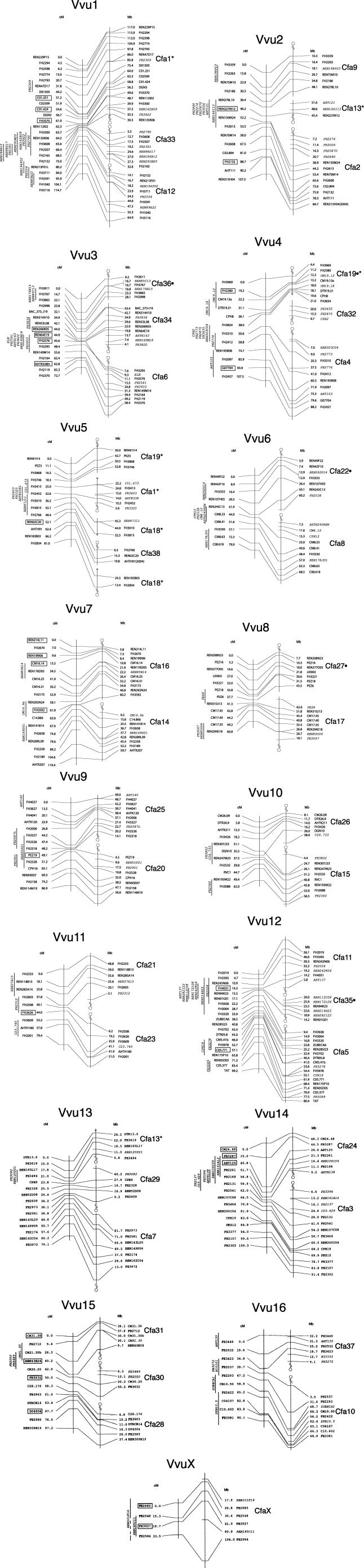
Figure 1.
Integrated meiotic linkage map of the silver fox (Vulpes vulpes). Each linkage group corresponding to a fox chromosome (VVU1 through VVU16 and VVUX) is presented on the left side of each panel and aligned with the corresponding segments of the 7.6x dog genome sequence (CanFam2.0). Lines connect markers mapped onto the fox meiotic linkage map and identified in the assembly of the dog genome. Markers in plain format map to unique locations with confidence ≥1000:1 (LOD ≥3.0). Boxed markers were placed to unique locations with confidence ≥100:1 (LOD ≥2). Markers in italics are drawn in their most likely position, determined at the latter confidence threshold. Genetic distances between markers were calculated using the Kosambi mapping function. In general, most dog chromosomes each map to a single fox chromosome. Dog chromosomes which have their homologs divided among more than one fox chromosome are marked by asterisks (*) (see VVU1, 2, 4, 5, and 13). Dog chromosomal segments for which the orientation on the fox meiotic map was reversed compared to the orientation reported in previous cytogenetic studies (Graphodatsky et al. 2002) are marked by dots (●) (see VVU3, 4, 6, 8, and 12). Centromere positions of dog chromosomes are indicated in accordance with the dog genome sequence, assuming that the centromere is located at the beginning of each chromosome. Where different fragments of a single dog chromosome correspond to different fox chromosomal segments, a tilde (∼) indicates the breakage point along the dog chromosome. Positions of markers in the dog sequence are indicated in accordance with the CanFam2.0 assembly, except for REN315H04 and AHTH91. In the present study, marker REN315H04 mapped to VVU2 in a region corresponding to CFA2, which is in agreement with the Breen et al. (2001) and Guyon et al. (2003) canine maps, and CanFam1.0 assembly of the canine genome (chr2:84,742,789–84,742,951). The CanFam2.0 assembly, however, locates REN315H04 on CFA9 (chr9:21,100,622–21,100,777). Marker AHTH91 was identified only in the CanFam1.0 assembly.
An additional 29 markers were placed uniquely on the fox map at a confidence level of 100:1 (LOD ≥2.0), as shown in Figure 1 (markers in boxed text). Ninety-two markers could not be uniquely placed on the fox LOD 2.0 map; the most likely positions for these at this confidence threshold (LOD ≥2.0) are also shown in Figure 1 (markers in italics). The majority of these 92 markers mapped to pericentromeric regions of fox chromosomes (Fig. 1).
In total, meiotic maps for 17 fox linkage groups were constructed and assigned to the 16 fox autosomes and the X chromosome (Fig. 1). Previously, the relationship between dog and fox chromosomes has been well defined by comparative cytogenetic mapping and reciprocal chromosomal painting (Yang et al. 1999; Graphodatsky et al. 2000, 2001). The unique location in the canine genome sequence of all but two markers was identified in silico (Supplemental Table 1). Markers AHTK338 and FH2087 each amplified two bands in the fox genome and each mapped to two different chromosomes (see legend to Supplemental Table 1). Alignment of fox linkage groups with homologous regions of the integrated canine genome maps and sequence, and comparative cytogenetic maps of the fox and dog genomes enabled assignment of linkage groups to fox chromosomes. Fox chromosomes were identified using nomenclature established for the fox karyotype (Andres 1938; Wipf and Scackeford 1942; Graphodatsky and Radjably 1981; Makinen 1985).
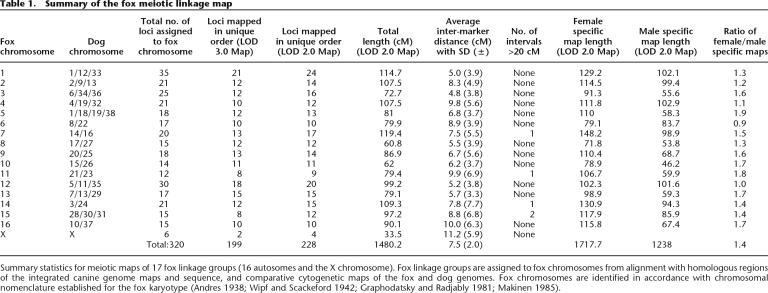
The number of markers mapped to each autosome ranged from 12 to 35 (Table 1). The meiotic lengths of fox autosomes, evaluated as the distance between the outermost markers in the linkage group, ranged from 60.8 (VVU8) to 119.4 (VVU7) centimorgans (cM) (Table 1; Fig. 1). Sex chromosomes were less well covered than the autosomes—six markers were mapped on the X chromosome and none on the Y. The total length of the fox map, calculated as the sum of all chromosome lengths, was 1480.2 cM. The average spacing between markers mapped to unique locations was 7.5 cM (SD ±2.0). There were five intervals >20 cM, with the largest inter-marker distance being 30.8 cM (Table 1). Differences in recombination length between sex specific maps were observed for most fox chromosomes. In total, the female map was 1.4 times longer than the male map (comparative statistics for the two maps are presented in Table 1).
Table 1.
Summary of the fox meiotic linkage map
Summary statistics for meiotic maps of 17 fox linkage groups (16 autosomes and the X chromosome). Fox linkage groups are assigned to fox chromosomes from alignment with homologous regions of the integrated canine genome maps and sequence, and comparative cytogenetic maps of the fox and dog genomes. Fox chromosomes are identified in accordance with chromosomal nomenclature established for the fox karyotype (Andres 1938; Wipf and Scackeford 1942; Graphodatsky and Radjably 1981; Makinen 1985).
The comprehensive fox map
Solely to enable comparison of corresponding meiotic distances between markers on the fox and dog maps, a comprehensive map of the fox genome was constructed at reduced confidence levels. This allowed specific locations to be assigned on the fox map for all markers that were also placed on the dog map. This generated a map for the fox with low statistical support for order among some tightly linked markers, but with unique map positions assigned with adequate precision (because the markers were so tightly linked). This permitted comparison of recombination distances between marker pairs mapped meiotically in both the fox and dog (Supplemental Table 2). Except for three instances (order of markers FH2087U, FH3006, and REN150M24 on VVU2; FH2087L, FH2526, and FH2318 on VVU9; and FH2261, AHTH138, and FH2168 on VVU14) the order best supported for the fox (even at low confidence) was in agreement with that in the dog map (Supplemental Fig. 2) and sequence (Supplemental Table 2).
Comparison of the fox LOD 2.0 meiotic linkage map with the sequence of the dog genome
Forty-three fox blocks were identified, and each corresponded to a unique continuous interval in the 7.6x whole genome assembly of the dog (CanFam2.0) (Fig. 1). Thirty-five fox chromosomal blocks correspond to a single uninterrupted dog chromosome. Each of the remaining eight blocks corresponds to two segments on each of dog chromosomes 1, 13, 18, and 19 (Table 1; Fig. 1). To examine the colinearity between fox and dog chromosomes at a higher resolution we compared the order of markers on the fox meiotic linkage map with their corresponding location in CanFam2.0 (Fig. 1; Supplemental Table1).
Five segments on the fox meiotic linkage map were orientated in reverse direction (centromere to telomere) compared to the orientation of these fragments expected from the fox–dog comparative cytogenetic map (Graphodatsky et al. 2000). These five segments (see Fig. 1) were on VVU3 (segment corresponding to CFA36), VVU4 (CFA19), VVU6 (CFA22), VVU8 (CFA27), and VVU12 (CFA35). As well, the order of markers in fox chromosomal blocks corresponding to CFA32 (VVU4) and a segment of CFA13 (VVU2) could not be determined because of suppressed recombination on the fox chromosome; even on the fox comprehensive map the distances between these markers were too small to unambiguously establish order. Third, a small putative inversion (2.8 cM) was observed on VVU3 between markers FH3295 and FH2576 and a rearrangement between syntenic blocks corresponding to dog chromosomes 15 and 26 was observed on VVU10 (markers DGN10 and REN307J23). In both cases rearrangements were observed between single pairs of markers.
To compare regions of conserved synteny between the fox and human genomes we anchored markers from the fox meiotic map to the dog sequence, identified genes uniquely located near these markers on the dog sequence, located the same genes on the human map, and then compared corresponding regions of the fox, dog, and human genomes. Alignment of fox chromosomes 4 and 13 to the human genome revealed two fox chromosomal regions, each corresponding to a single human chromosome (HSA4 and HSA8, respectively), but to separate dog chromosomes (CFA19 and 32 for VVU4 and CFA13 and 29 for VVU13), demonstrating homology of these fox chromosomes with the longer continuous regions of the human genome than on the corresponding dog chromosomes (Supplemental Fig. 3A,B). Genes selected for comparison were located in the dog genome at an average distance of 3.2 Mb (SE ±0.7) from the closest marker mapped in the fox. A part of VVU4 corresponding to the “north” (i.e., centromeric) end of CFA32 was not covered by markers at high resolution, and the distance between the most distal marker (FH3635) and the outermost distal gene (PDE5A) was 21.2 Mb (Supplemental Fig. 3A).
Comparison of fox and dog meiotic linkage maps
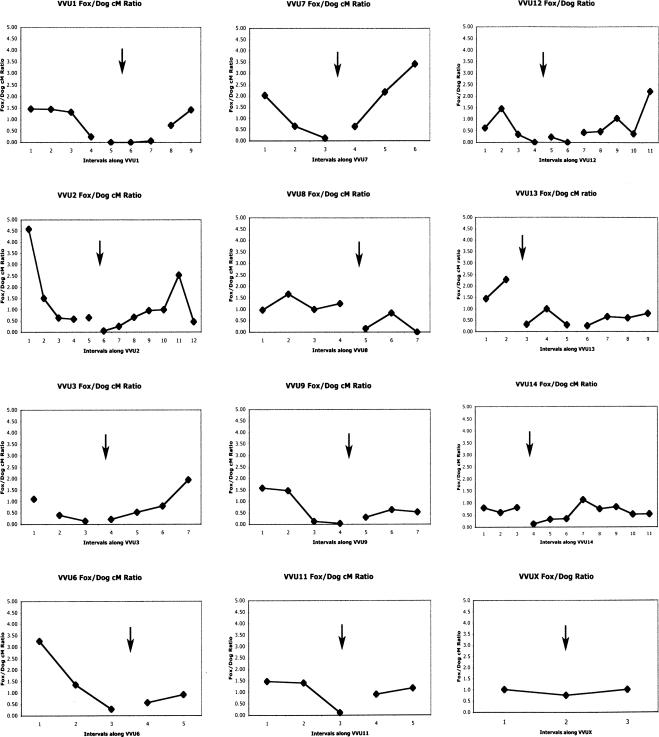
To compare recombination rates on corresponding chromosomal segments of the fox and dog genomes we constructed a meiotic linkage map of the dog genome with markers in common with the fox meiotic map (Supplemental Fig. 2). Of the 181 common markers, 155 markers were uniquely assigned to the dog LOD 1.0 map (Supplemental Fig. 2; see Methods section “Map Construction”). We compared recombination distances between adjacent markers that were mapped in both the dog and fox, for each of 32 corresponding blocks of conserved synteny (Supplemental Table 2). Recombination distances between markers on the fox comprehensive map have been compared to the distances between corresponding markers on homologous dog chromosomes using the dog LOD 1.0 map (Supplemental Table 2; Fig. 2). The largest differences in recombination distance between corresponding markers on the fox and dog maps were observed for marker pairs located near the centromeres of fox chromosomes. The highest ratio of recombination distance for homologous segments of the dog and fox meiotic maps was observed for a region corresponding to part of CFA33 and the pericentromeric region of VVU1 (Supplemental Table 2; Fig. 2). The interval on VVU1 flanked by markers FH2361 and FH2507 at one end, and FH2165 at the opposite end, measures only 2.1 cM on the fox LOD 2.0 map (Fig. 1) and about 1 cM on the fox comprehensive map (Supplemental Table 2). The corresponding interval on CFA33 measures ∼27 cM on the dog meiotic map (see Supplemental Fig. 2) and represents ∼15 Mb of canine genome sequence. Furthermore, neither markers FH2790 nor FH3608, both of which map to this region of suppressed recombination and are identified in the corresponding region of CFA33 on CanFam2.0, have been placed on a canine meiotic linkage map, either in the present study or previous studies. It is likely that, if and when the position of these markers is determined on the dog map, the difference in recombination distance between the homologous regions of VVU1 and CFA33 will prove to be even greater.
Figure 2.
Comparisons of recombination rates between fox and dog chromosomes. Each point on the graph represents the ratio (fox to dog) of the recombination distances between a pair of markers mapped in both the fox and the dog (fox and dog distances between these markers and recombination ratios are presented in Supplemental Table 2). Points connected by a line correspond to a single dog chromosome. Arrows indicate putative centromere positions of fox chromosomes. Relative recombination suppression (ratio <1.0) is evident in the pericentromeric region of several fox chromosomes.
Suppressed recombination was also apparent in other fox pericentromeric regions (VVU3, VVU12, and VVU13), corresponding to dog chromosomes CFA34, 35, and 30 where recombination distance ratios of 1:3.6, 1:12.9, and 1:2.5, respectively, were observed (Supplemental Table 2; Fig. 2). In these intervals, also, only those portions of dog chromosomes that contained markers common to both the dog and fox meiotic maps could be directly compared to the fox homologous region.
Discussion
This first meiotic linkage map of the silver fox genome uses 320 microsatellite markers adapted from the dog genome (Fig. 1). The fox map covers 16 autosomes and the X chromosome with an average inter-marker distance of 7.5 cM. Five inter-marker intervals ≥20 cM remain to be filled. Alignment of the fox meiotic map against the 7.6x sequence of the dog genome revealed high conservation of marker order between homologous regions of the two species. The approximate length of fox telomeric regions not covered by markers on the current map was estimated by comparison with corresponding regions in the dog genome. On average, the distance from the most distal marker placed in a fox linkage group, to the end of the homologous dog chromosome, corresponded to 8.9 Mb (SE ±1.0 Mb); the largest uncovered telomeric region—on the X chromosome—corresponded to 22.9 Mb. Although this version of the fox meiotic map has several gaps, the map provides a sufficient tool for initial mapping of traits of interest in the fox. Conserved synteny between the domestic dog and fox indicates that the 7.6x sequence of the dog genome will be an important resource for fine mapping and cloning fox genes.
Alignment of the fox meiotic linkage map against dog sequence did not reveal any large unpredicted rearrangements between the fox and dog genomes. The largest gap on the fox map (30.8 cM) was observed on VVU14 between markers FH3287 and AHT125. This distance corresponds to 14.6 Mb in the dog genome sequence. The large interval between these markers on the fox map could indicate either that the physical distance of this region of VVU14 is longer than on CFA24, that in the fox this region represents a recombination hot spot, or that it contains an unrecognized insertion representing part of another dog chromosome. Two small inversions in marker order have been observed on VVU3 and VVU10 but additional markers need to be placed on the fox map to confirm that these rearrangements do not simply result from an insufficient number of coinformative meioses.
Comparison of the fox map with the current assembly of the dog genome sequence identified 35 dog chromosomes, each represented by a single block in the fox genome, and four dog chromosomes (1, 13, 18, 19) corresponding to two blocks each in the fox. These results agree with the previous comparative cytogenetic map of the dog and fox (Yang et al. 1999). The orientation of blocks corresponding to dog chromosomes 22, 27, 35, 36, and a fragment of CFA19 on the fox meiotic map was reversed compared to the orientation reported in previous cytogenetic studies (Graphodatsky et al. 2002).
From the analysis of marker order on homologous dog and fox chromosomes, the comparative orientation in terms of telomere to centromere direction, or reverse, could be examined. In several cases the orientation of dog chromosomes and corresponding fox chromosomal arms was concordant (for example, CFA8 and one arm of VVU6, CFA14 and one arm of VVU7; Fig. 1). In other cases this orientation is reversed. For example, the centromeric regions of CFA22 and CFA16 correspond to the telomeric regions of VVU6 and VVU7, respectively. The effect of concordant and reverse orientation of homologous chromosomal segments between the genomes of the two species remains to be investigated.
Anchoring the fox meiotic map to the dog genome map and sequence allows indirect comparison between corresponding regions of the fox, dog, and human genomes. In general, similar breakpoint locations were observed between dog and human chromosomes and between fox and human chromosomes. Using this indirect comparison we identified two fox chromosomes (VVU4 and VVU13) which show homology with human chromosomes over longer continuous regions than do the corresponding dog chromosomes. This was in agreement with comparative cytogenetic maps of human, fox, and dog (Yang et al. 1999). Each of these blocks of HSA4 and HSA8 corresponds to a single block in the genomes of other mammalian species including Arctic fox (Alopex lagopus), cats, bovids, and pigs (Graphodatsky et al. 2000; Yang et al. 2000; Fronicke and Wienberg 2001; Biltueva et al. 2004). Such evolutionary conservation suggests that these chromosomal blocks were formed before the divergence of primates and carnivores (Graphodatsky et al. 2001) and further indicates that the dog genome has undergone rearrangements since the separation of canine and vulpine lines. Alignment of the fox meiotic map with the dog genome sequence provides an opportunity to refine the breakpoints between dog and fox chromosomes with higher resolution than it was previously possible, thus yielding further insight into the evolution of Canidae genomes.
Estimating the total lengths of the sex-averaged and sex-specific fox meiotic maps as the sum of distances between distal markers on each fox chromosome, a difference in the lengths of fox sex-specific maps was observed, as is seen in other mammals. The observed ratio between the fox female and male maps was close to 1.4 (Table 1), which is similar to the difference reported for the dog sex-specific maps (Neff et al. 1999). The total length of the last published dog meiotic map, constructed with 341 markers (Werner et al. 1999) and calculated by the same method, corresponds to 21.3 M, which is ∼30% longer than the length of the current fox meiotic map. Although comparison of the fox and dog sex-averaged maps revealed a significant difference in map lengths, the observed difference may be attributed to the low marker density of the current maps. Preliminary results for a more comprehensive meiotic linkage map of the dog genome have been presented (Neff et al. 2006), which might help address some of these issues in the near future.
To perform more detailed analysis of differences in recombination rate between homologous regions of the fox and dog genomes we constructed a new dog map using the Marshfield marker set which includes 181 markers mapped in foxes. Preliminary results of comparative analysis of the current fox and dog meiotic maps indicate that, overall, the fox meiotic map is shorter than the dog map, and differences in recombination rates between the two species appear to be associated with pericentromeric suppression in foxes. Pericentromeric suppression has been observed in Drosophila, human, and other species (True et al. 1996; Yu et al. 2001; Kong et al. 2002; Jensen-Seaman et al. 2004). Among other factors considered to influence recombination rate are CpG islands, GC content, polyA/polyT content, simple repeats, LINE, SINE elements, and other sequence features (Yu et al. 2001; Kong et al. 2002; Hellman et al. 2003; Jensen-Seaman et al. 2004). When sequence of the fox genome becomes available, a comparative analysis of the fox and dog genomes would allow for a better understanding of the factors that influence recombination in the two species.
The meiotic linkage map of the fox genome provides a critical tool for identification of loci responsible for phenotypic variation observed in natural and farm bred fox populations. In particular, the fox map can now be exploited to map loci and genes underlying the unique heritable behavioral traits in the fox strains selectively bred at the ICG in Russia. This can be predicted to yield insights into the genetics of social behavior and its underlying molecular mechanisms, not only in foxes, but in other mammals, including humans, and also contribute to our understanding of the animal domestication process.
Methods
Pedigrees and DNA preparation
Fox
Thirty-seven three-generation silver fox pedigrees, produced at the Institute of Cytology and Genetics (ICG) of the Russian Academy of Sciences, Novosibirsk, Russia, were selected for genotyping and map construction. Thirty-four fox pedigrees were developed by breeding foxes from the tame and aggressive strains and then backcrossing the F1 progeny to the tame strain; one pedigree was produced by crossing parents from tame and aggressive strains; and two pedigrees were selected from the tame strain. Samples from grandparents were available for all but 10 pedigrees. Blood samples from 286 available individuals (180 animals in the third generation) in these 37 pedigrees were collected at ICG.
Dog
Fourteen mixed-breed three-generation dog pedigrees comprising 148 individuals (126 animals in the third generation) were studied. Dog pedigrees were produced at the Retinal Disease Studies Facility in Kennett Square, PA. Tissue samples (blood, spleen) collected from these 148 dogs included all grandparents but two in one pedigree. DNA from fox and dog samples was extracted using Qiagen Maxi Blood kits (Qiagen) or phenol-chloroform extraction methods (Gilbert and Vance 1994).
Markers for genotyping fox pedigrees
Canine microsatellites were adapted for genotyping fox pedigrees. Two marker sets were used: (1) the Marshfield canine screening set and (2) a marker set developed at Cornell University specifically for mapping fox and dog pedigrees. In all, 181 markers from the Marshfield 249 marker set and 139 markers from the Cornell set were used. The Cornell set included 78 markers from the canine MSS-2 set (Clark et al. 2004), 38 markers from the integrated canine genome maps (Breen et al. 2001; Guyon et al. 2003), and 23 new microsatellite markers developed from dog genome sequence data. Markers selected at Cornell were tested for amplification and polymorphism on DNA samples of seven dogs and 48 foxes using unlabeled primers. PCRs were performed under standard conditions: an initial 2 min denaturation at 96°C; then 30 cycles of 96°C (20 sec), 58°C (20 sec), 72°C (20 sec); and a final extension step at 72°C for 5 min. PCR products were resolved on 10% native polyacrylamide gel and visualized by ethidium bromide staining. Polymorphic markers that amplified robustly were selected for genotyping with fluorescently labeled primers.
Genotyping
Dog
Dog pedigrees comprising 148 individuals were genotyped at the Mammalian Genotyping Service of Marshfield Laboratories with the Marshfield set of 249 canine microsatellite markers, which are distributed relatively evenly across the canine genome. Dog pedigrees used in the current study have not been previously used for construction of the dog meiotic map.
Fox
Fox pedigrees were genotyped using two complementary strategies: (1) 263 fox DNA samples comprising 35 pedigrees were genotyped at Marshfield with the same canine markers used for dog genotyping; (2) 286 individuals, including the same 263 samples genotyped at Marshfield plus two additional pedigrees, were genotyped with the Supplemental 139-marker Cornell set (Supplemental Table 1). Microsatellites were amplified on fox DNA with fluorescent primers under the same conditions used for amplification with unlabeled primers, but a final extension step at 72°C was extended for 1 h. PCRs were performed in 15 μL containing 1× Invitrogen Taq Polymerase buffer, 1.5 mM MgCl2, 0.2 mM of dNTP, 0.3 pmol of each primer, 1.5 ng/μL of fox DNA, and 0.5 units of Invitrogen Taq Polymerase. From four to seven microsatellites were combined, post PCR, in multiplex sets and resolved on an ABI3730 Genetic Analyzer (PE Biosystems). PCR products were sized relative to an internal size standard using ABI Genemapper 3.5 software package (PE Biosystems).
Selection of marker genotypes for map construction
Genotypes for both fox and dog that were generated by Marshfield with a confidence value ≥0.99 were retained for analysis. Genotyping data for all genotyped markers (249) were retained for dog, and for 181 markers (72% of all genotyped markers) for fox. Quality parameters for genotypes produced at Cornell University were established using ABI Genemapper 3.5 (PE Biosystems) and included data clarity, number of alleles, peak height, and percentage of failed samples, and these genotypes were double-scored by independent investigators. Fox genotypes for 320 nonoverlapping markers from both sets (181 markers from the Marshfield set and 139 markers from Cornell) that passed initial evaluation criterion were selected for linkage analysis.
All genotypes obtained for the dog and fox pedigrees at Marshfield and Cornell were checked for Mendelian segregation using the prepare option of MultiMap (Green et al. 1990; Matise et al. 1994) and manually corrected for errors.
To evaluate each marker, heterozygosity, PIC (Polymorphism Information Content) value, and the number of informative meioses were estimated using MultiMap. Markers with PIC >0.1 and more than 40 informative meioses were retained for map construction.
Map construction
In the current study, three meiotic linkage maps were generated and used for different comparative analyses: (1) a fox meiotic linkage map constructed at a confidence level of 100:1 (LOD ≥2); (2) a fox comprehensive meiotic linkage map with markers ordered without strong statistical support; and (3) a dog meiotic linkage map generated at a confidence level of 10:1 (LOD ≥1).
The fox LOD 2.0 map and subsequent maps were each generated using MultiMap (Matise et al. 1994), in a two-step process. First, fox markers were assigned to linkage groups (find-all-linkage-groups function) if linked to at least one other marker in the group with a recombination fraction ≤0.25 and a LOD score ≥4. A sex-averaged, framework map was then constructed for each fox linkage group beginning with the pair of markers in each group with the highest joint PIC value and for which a recombination fraction of 0.1–0.4 was supported with a LOD score ≥3.0. Further markers were added to each linkage group in decreasing order of informativeness until no further markers could be localized to a unique interval at a confidence level of 1000:1 (LOD ≥3). When each framework map was considered complete, the FLIPS function of MultiMap was used to ensure that the odds in favor of the final order of each set of three to six markers were at least 1000:1 over alternative orders. Subsequent markers were added to the framework map at a confidence of 100:1 (LOD ≥2.0) to generate the fox LOD 2.0 map. Genetic distances were calculated using the Kosambi mapping function. Both sex-averaged and sex-specific maps were computed in similar manner.
Some markers could not be assigned to unique positions on the fox LOD 2.0 map because of the low recombination rate between neighboring marker pairs. For the fox comprehensive map, order for these tightly linked markers was assigned without strong statistical support, as position rather than order was the needed datum.
To enable direct comparisons between the fox and dog meiotic linkage maps, using the same set of markers, we constructed a meiotic linkage map of the dog genome using data generated at Marshfield Laboratories (Kukekova et al. 2006) with markers used for fox mapping. This permitted comparative studies of recombination distances between corresponding pairs of markers in the dog and fox genomes. Construction of the dog genetic map was performed as for the LOD 2.0 fox meiotic map, except that initial assignment of markers to dog chromosomes was based on the canine RH map and dog sequence information. A dog framework map was first computed with a criterion LOD score ≥3.0. Additional markers were then added to the map at LOD ≥1.0 to generate the dog LOD 1.0 map.
In silico PCR and map integration
The genetic maps constructed in this study for the dog and fox genomes were integrated with the physical map of the canine genome. Positions of 318 genetically mapped microsatellite loci were mapped in the dog genome (CanFam2.0, May 2005 assembly) in silico using In-Silico PCR and BLAT at the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/).
Acknowledgments
We thank Irina V. Pivovarova, Tatyana I. Semenova, Anna Stepika, Vasiliy V. Ivaykin, Vera I. Vladimirova, Tatyana V. Konovalova, Vera L. Haustova, and all the animal keepers at the ICG experimental farm for research assistance. We thank K. Gordon Lark and Kevin Chase for help, advice, and important discussions. We express our profound gratitude to Marshfield Laboratories Mammalian Genotyping Service for genotyping fox pedigrees. We thank Elaine Ostrander for critical reading of the manuscript. Research was supported by NIH grants MH069688, EY06855, EY13729, and NIH FIRCA grant no. RO3 TW007056 awarded to the University of Utah, grant nos. 05-04-4837 and 06-04-48142 of the Russian Fund for Basic Research, Program of the Russian Academy of Sciences: “Biodiversity and Genome Dynamics”, MCB, and Cornell VERGE Initiative.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are online at http://www.genome.org/cgi/doi/10.1101/gr.5893307
References
- Andres A.H. On the chromosome complex in several Canidae. Cytologia (Tokyo) 1938;9:35–37. [Google Scholar]
- Belyaev D.K. Domestication of animals. Science. 1969;5:47–52. [Google Scholar]
- Belyaev D.K. The Wilhelmine E. Key 1978 invitational lecture. Destabilizing selection as a factor in domestication. J. Hered. 1979;70:301–308. doi: 10.1093/oxfordjournals.jhered.a109263. [DOI] [PubMed] [Google Scholar]
- Belyaev D.K., Volobuev V.T., Radzhabli S.I., Trut L.N., Volobuev V.T., Radzhabli S.I., Trut L.N., Radzhabli S.I., Trut L.N., Trut L.N. Supernumary chromosome polymorphism and mosaicism in silver foxes. Genetika. 1974;10:58–67. [PubMed] [Google Scholar]
- Belyaev D.K., Plyusnina I.Z., Trut L.N., Plyusnina I.Z., Trut L.N., Trut L.N. Domestication in the silver fox (Vulpes fulvusdesm.): Changes in physiological boundaries of the sensitive period of primary socialization. Appl. Anim. Behav. Sci. 1984;13:359–370. [Google Scholar]
- Biltueva L.S., Yang F., Vorobieva N.V., Graphodatsky A.S., Yang F., Vorobieva N.V., Graphodatsky A.S., Vorobieva N.V., Graphodatsky A.S., Graphodatsky A.S. Comparative map between the domestic pig and dog. Mamm. Genome. 2004;15:809–818. doi: 10.1007/s00335-004-2391-3. [DOI] [PubMed] [Google Scholar]
- Breen M., Jouquand S., Renier C., Mellersh C.S., Hitte C., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Jouquand S., Renier C., Mellersh C.S., Hitte C., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Renier C., Mellersh C.S., Hitte C., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Mellersh C.S., Hitte C., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Hitte C., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Holmes N.G., Cheron A., Suter N., Vignaux F., Bristow A.E., Cheron A., Suter N., Vignaux F., Bristow A.E., Suter N., Vignaux F., Bristow A.E., Vignaux F., Bristow A.E., Bristow A.E., et al. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M., Hitte C., Lorentzen T.D., Thomas R., Cadieu E., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Hitte C., Lorentzen T.D., Thomas R., Cadieu E., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Lorentzen T.D., Thomas R., Cadieu E., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Thomas R., Cadieu E., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Cadieu E., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Sabacan L., Scott A., Evanno G., Parker H.G., Kirkness E.F., Scott A., Evanno G., Parker H.G., Kirkness E.F., Evanno G., Parker H.G., Kirkness E.F., Parker H.G., Kirkness E.F., Kirkness E.F., et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K., Carrier D.R., Adler F.R., Jarvik T., Ostrander E.A., Lorentzen T.D., Lark K.G., Carrier D.R., Adler F.R., Jarvik T., Ostrander E.A., Lorentzen T.D., Lark K.G., Adler F.R., Jarvik T., Ostrander E.A., Lorentzen T.D., Lark K.G., Jarvik T., Ostrander E.A., Lorentzen T.D., Lark K.G., Ostrander E.A., Lorentzen T.D., Lark K.G., Lorentzen T.D., Lark K.G., Lark K.G. Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proc. Natl. Acad. Sci. 2002;99:9930–9935. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.A., Tsai K.L., Steiner J.M., Williams D.A., Guerra T., Ostrander E.A., Galibert F., Murphy K.E., Tsai K.L., Steiner J.M., Williams D.A., Guerra T., Ostrander E.A., Galibert F., Murphy K.E., Steiner J.M., Williams D.A., Guerra T., Ostrander E.A., Galibert F., Murphy K.E., Williams D.A., Guerra T., Ostrander E.A., Galibert F., Murphy K.E., Guerra T., Ostrander E.A., Galibert F., Murphy K.E., Ostrander E.A., Galibert F., Murphy K.E., Galibert F., Murphy K.E., Murphy K.E. Chromosome-specific microsatellite multiplex sets for linkage studies in the domestic dog. Genomics. 2004;84:550–554. doi: 10.1016/j.ygeno.2004.06.006. [DOI] [PubMed] [Google Scholar]
- de Gortari M.J., Freking B.A., Cuthbertson R.P., Kappes S.M., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Freking B.A., Cuthbertson R.P., Kappes S.M., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Cuthbertson R.P., Kappes S.M., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Kappes S.M., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Leymaster K.A., Dodds K.G., Crawford A.M., Beattie C.W., Dodds K.G., Crawford A.M., Beattie C.W., Crawford A.M., Beattie C.W., Beattie C.W. A second-generation linkage map of the sheep genome. Mamm. Genome. 1998;9:204–209. doi: 10.1007/s003359900726. [DOI] [PubMed] [Google Scholar]
- Fronicke L., Wienberg J., Wienberg J. Comparative chromosome painting defines the high rate of karyotype changes between pigs and bovids. Mamm. Genome. 2001;12:442–449. doi: 10.1007/s003350010288. [DOI] [PubMed] [Google Scholar]
- Gilbert J.R., Vance J.M., Vance J.M. Isolation of genomic DNA from mammalian cells. In: Dracopoli N.C., et al., editors. Current protocols in human genetics. John Wiley and Sons; New York: 1994. pp. 1–6. Appendix A.3B. [DOI] [PubMed] [Google Scholar]
- Graphodatsky A.S., Radjably S.I., Radjably S.I. Comparative cytogenetics of three canids species (Carnivora, Canidae). II. Distribution of C-heterochromatin. Genetika. 1981;17:1500–1504. [Google Scholar]
- Graphodatsky A.S., Yang F., O'Brien P.C., Serdukova N., Milne B.S., Trifonov V., Ferguson-Smith M.A., Yang F., O'Brien P.C., Serdukova N., Milne B.S., Trifonov V., Ferguson-Smith M.A., O'Brien P.C., Serdukova N., Milne B.S., Trifonov V., Ferguson-Smith M.A., Serdukova N., Milne B.S., Trifonov V., Ferguson-Smith M.A., Milne B.S., Trifonov V., Ferguson-Smith M.A., Trifonov V., Ferguson-Smith M.A., Ferguson-Smith M.A. A comparative chromosome map of the Arctic fox, red fox and dog defined by chromosome painting and high resolution G-banding. Chromosome Res. 2000;8:253–263. doi: 10.1023/a:1009217400140. [DOI] [PubMed] [Google Scholar]
- Graphodatsky A.S., Yang F., O'Brien P.C., Perelman P., Milne B.S., Serdukova N., Kawada S.I., Ferguson-Smith M.A., Yang F., O'Brien P.C., Perelman P., Milne B.S., Serdukova N., Kawada S.I., Ferguson-Smith M.A., O'Brien P.C., Perelman P., Milne B.S., Serdukova N., Kawada S.I., Ferguson-Smith M.A., Perelman P., Milne B.S., Serdukova N., Kawada S.I., Ferguson-Smith M.A., Milne B.S., Serdukova N., Kawada S.I., Ferguson-Smith M.A., Serdukova N., Kawada S.I., Ferguson-Smith M.A., Kawada S.I., Ferguson-Smith M.A., Ferguson-Smith M.A. Phylogenetic implications of the 38 putative ancestral chromosome segments for four canid species. Cytogenet. Cell Genet. 2001;92:243–247. doi: 10.1159/000056911. [DOI] [PubMed] [Google Scholar]
- Graphodatsky A.S., Yang F., Perelman P.L., O'Brien P.C.M., Serdukova N.A., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Yang F., Perelman P.L., O'Brien P.C.M., Serdukova N.A., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Perelman P.L., O'Brien P.C.M., Serdukova N.A., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., O'Brien P.C.M., Serdukova N.A., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Serdukova N.A., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Milne B.S., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Biltueva L.S., Fu B., Vorobieva N.V., Kawada S.-I., Fu B., Vorobieva N.V., Kawada S.-I., Vorobieva N.V., Kawada S.-I., Kawada S.-I., et al. Comparative molecular cytogenetic studies in the order Carnivora: Mapping chromosomal rearrangements onto the phylogenetic tree. Cytogenet. Genome Res. 2002;96:137–145. doi: 10.1159/000063032. [DOI] [PubMed] [Google Scholar]
- Green P., Fall K., Crooks S., Fall K., Crooks S., Crooks S. Documentation for CRI-MAP, version 2.4. Washington University School of Medicine; St. Louis, MO: 1990. [Google Scholar]
- Guyon R., Lorentzen T.D., Hitte C., Kim L., Cadieu E., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Lorentzen T.D., Hitte C., Kim L., Cadieu E., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Hitte C., Kim L., Cadieu E., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Kim L., Cadieu E., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Cadieu E., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Parker H.G., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Quignon P., Lowe J.K., Renier C., Gelfenbeyn B., Lowe J.K., Renier C., Gelfenbeyn B., Renier C., Gelfenbeyn B., Gelfenbeyn B., et al. A 1-Mb resolution radiation hybrid map of the canine genome. Proc. Natl. Acad. Sci. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S.E., Paabo S., Przeworski M., Ebersberger I., Ptak S.E., Paabo S., Przeworski M., Ptak S.E., Paabo S., Przeworski M., Paabo S., Przeworski M., Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitte C., Madeoy J., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Madeoy J., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Thomas D., Derrien T., Ramirez C., Scott C., Derrien T., Ramirez C., Scott C., Ramirez C., Scott C., Scott C., et al. Facilitating genome navigation: Survey sequencing and dense radiation-hybrid gene mapping. Nat. Rev. Genet. 2005;6:643–648. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman M.I., Furey T.S., Payseur B.A., Lu Y., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Furey T.S., Payseur B.A., Lu Y., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Payseur B.A., Lu Y., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Lu Y., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Chen C.F., Thomas M.A., Haussler D., Jacob H.J., Thomas M.A., Haussler D., Jacob H.J., Haussler D., Jacob H.J., Jacob H.J. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness E.F., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Delcher A.L., Pop M., Wang W., Fraser C.M., Pop M., Wang W., Fraser C.M., Wang W., Fraser C.M., Fraser C.M., et al. The dog genome: Survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Barnard J., Hallbeck B., Masson G., Hallbeck B., Masson G., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kukekova A.V., Trut L.N., Oskina I.N., Kharlamova A.V., Shikhevich S.G., Kirkness E.F., Aguirre G.D., Acland G.M., Trut L.N., Oskina I.N., Kharlamova A.V., Shikhevich S.G., Kirkness E.F., Aguirre G.D., Acland G.M., Oskina I.N., Kharlamova A.V., Shikhevich S.G., Kirkness E.F., Aguirre G.D., Acland G.M., Kharlamova A.V., Shikhevich S.G., Kirkness E.F., Aguirre G.D., Acland G.M., Shikhevich S.G., Kirkness E.F., Aguirre G.D., Acland G.M., Kirkness E.F., Aguirre G.D., Acland G.M., Aguirre G.D., Acland G.M., Acland G.M. A marker set for construction of a genetic map of the silver fox (Vulpes vulpes) J. Hered. 2004;95:185–194. doi: 10.1093/jhered/esh033. [DOI] [PubMed] [Google Scholar]
- Kukekova A.V., Nelson J., Kuchtey R.W., Lowe J.K., Johnson J.L., Ostrander E.A., Aguirre G.D., Acland G.M., Nelson J., Kuchtey R.W., Lowe J.K., Johnson J.L., Ostrander E.A., Aguirre G.D., Acland G.M., Kuchtey R.W., Lowe J.K., Johnson J.L., Ostrander E.A., Aguirre G.D., Acland G.M., Lowe J.K., Johnson J.L., Ostrander E.A., Aguirre G.D., Acland G.M., Johnson J.L., Ostrander E.A., Aguirre G.D., Acland G.M., Ostrander E.A., Aguirre G.D., Acland G.M., Aguirre G.D., Acland G.M., Acland G.M. Linkage mapping of canine rod cone dysplasia type 2 (rcd2) to CFA7, the canine orthologue of human 1q32. Invest. Ophthalmol. Vis. Sci. 2006;47:1210–1215. doi: 10.1167/iovs.05-0861. [DOI] [PubMed] [Google Scholar]
- Lark K.G., Chase K., Sutter N.B., Chase K., Sutter N.B., Sutter N.B. Genetic architecture of the dog: Sexual size dimorphism and functional morphology. Trends Genet. 2006;22:537–544. doi: 10.1016/j.tig.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Chang J.L., Kulbokas E.J., III, Zody M.C., Kulbokas E.J., III, Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- MacDonald D.W., Reynolds J.C., Reynolds J.C. Red fox (Vulpes vulpes) In: Sillero-Zubiri C., et al., editors. Canids: Foxes, wolves, jackals and dogs. Status survey and conservation action plan. IUCN; Gland, Switzerland: Cambridge, UK: 2004. pp. 129–136. [Google Scholar]
- Makinen A. The standard karyotype of the silver fox (Vulpes fulvus Desm.). Committee for the standard karyotype of Vulpes fulvus Desm. Hereditas. 1985;103:289–297. doi: 10.1111/j.1601-5223.1985.tb00498.x. [DOI] [PubMed] [Google Scholar]
- Matise T.C., Perlin M., Chakravarti A., Perlin M., Chakravarti A., Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): A human genome linkage map. Nat. Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Mellersh C.S., Langston A.A., Acland G.M., Fleming M.A., Ray K., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Langston A.A., Acland G.M., Fleming M.A., Ray K., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Acland G.M., Fleming M.A., Ray K., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Fleming M.A., Ray K., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Ray K., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Wiegand N.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Francisco L.V., Gibbs M., Aguirre G.D., Ostrander E.A., Gibbs M., Aguirre G.D., Ostrander E.A., Aguirre G.D., Ostrander E.A., Ostrander E.A. A linkage map of the canine genome. . Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Neff M.W., Broman K.W., Mellersh C.S., Ray K., Acland G.M., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Broman K.W., Mellersh C.S., Ray K., Acland G.M., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Mellersh C.S., Ray K., Acland G.M., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Ray K., Acland G.M., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Acland G.M., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Aguirre G.D., Ziegle J.S., Ostrander E.A., Rine J., Ziegle J.S., Ostrander E.A., Rine J., Ostrander E.A., Rine J., Rine J. A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics. 1999;151:803–820. doi: 10.1093/genetics/151.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.W., Wong A., Ruhe A., Bruce S., Robertson K., Ziegle J., Broman K.W., Wong A., Ruhe A., Bruce S., Robertson K., Ziegle J., Broman K.W., Ruhe A., Bruce S., Robertson K., Ziegle J., Broman K.W., Bruce S., Robertson K., Ziegle J., Broman K.W., Robertson K., Ziegle J., Broman K.W., Ziegle J., Broman K.W., Broman K.W. A comprehensive linkage map of the dog genome. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes N.N., Einarsson E.J., Lohi O., Jorgensen G., Einarsson E.J., Lohi O., Jorgensen G., Lohi O., Jorgensen G., Jorgensen G. Beautiful fur animals—And their colour genetics. SCIENTIFUR; Denmark: 1988. [Google Scholar]
- Nowak R.M. Walker’s mammals of the world. John Hopkins University Press; Baltimore, MD: 1991. [Google Scholar]
- Oskina I.N. Analysis of the functional state of the pituitary-adrenal axis during postnatal development of domesticated foxes (Vulpes Vulpes) Scientifur. 1996;20:159–167. [Google Scholar]
- Oskina I.N., Tinnikov A.A., Tinnikov A.A. Interaction between cortisol and cortisol-binding protein in silver foxes (Vulpes fulvus) Comp. Biochem. Physiol. Comp. Physiol. 1992;101:665–668. doi: 10.1016/0300-9629(92)90341-m. [DOI] [PubMed] [Google Scholar]
- Plyusnina I.Z., Oskina I.N., Trut L.N., Oskina I.N., Trut L.N., Trut L.N. An analysis of fear and aggression during early development of behavior in silver foxes (Vulpes vulpes) Appl. Anim. Behav. Sci. 1991;32:253–268. [Google Scholar]
- Popova N.K., Kulikov A.V., Avgustinovich D.F., Voitenko N.N., Trut L.N., Kulikov A.V., Avgustinovich D.F., Voitenko N.N., Trut L.N., Avgustinovich D.F., Voitenko N.N., Trut L.N., Voitenko N.N., Trut L.N., Trut L.N. Effect of domestication of the silver fox on the main enzymes of serotonin metabolism and serotonin receptors. Genetika. 1997;33:370–374. [PubMed] [Google Scholar]
- Reed K.M., Chaves L.D., Hall M.K., Knutson T.P., Harry D.E., Chaves L.D., Hall M.K., Knutson T.P., Harry D.E., Hall M.K., Knutson T.P., Harry D.E., Knutson T.P., Harry D.E., Harry D.E. A comparative genetic map of the turkey genome. Cytogenet. Genome Res. 2005;111:118–127. doi: 10.1159/000086380. [DOI] [PubMed] [Google Scholar]
- Rexroad C.E., III, Rodriguez M.F., Coulibaly I., Gharbi K., Danzmann R.G., Dekoning J., Phillips R., Palti Y., Rodriguez M.F., Coulibaly I., Gharbi K., Danzmann R.G., Dekoning J., Phillips R., Palti Y., Coulibaly I., Gharbi K., Danzmann R.G., Dekoning J., Phillips R., Palti Y., Gharbi K., Danzmann R.G., Dekoning J., Phillips R., Palti Y., Danzmann R.G., Dekoning J., Phillips R., Palti Y., Dekoning J., Phillips R., Palti Y., Phillips R., Palti Y., Palti Y. Comparative mapping of expressed sequence tags containing microsatellites in rainbow trout (Oncorhynchus mykiss) BMC Genomics. 2005;6:54. doi: 10.1186/1471-2164-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Mahaney M.C., Witte S.M., Nair S., Newman D., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Mahaney M.C., Witte S.M., Nair S., Newman D., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Witte S.M., Nair S., Newman D., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Nair S., Newman D., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Newman D., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Wedel S., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Rodriguez L.A., Rice K.S., Slifer S.H., Perelygin A., Rice K.S., Slifer S.H., Perelygin A., Slifer S.H., Perelygin A., Perelygin A., et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- Rogers J., Garcia R., Shelledy W., Kaplan J., Arya A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Garcia R., Shelledy W., Kaplan J., Arya A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Shelledy W., Kaplan J., Arya A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Kaplan J., Arya A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Arya A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Johnson Z., Bergstrom M., Novakowski L., Nair P., Vinson A., Bergstrom M., Novakowski L., Nair P., Vinson A., Novakowski L., Nair P., Vinson A., Nair P., Vinson A., Vinson A., et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Sheldon J.W. Wild dogs: The natural history of the nondomestic Canidae. Academic Press; San Diego, CA: 1992. [Google Scholar]
- True J.R., Mercer J.M., Laurie C.C., Mercer J.M., Laurie C.C., Laurie C.C. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics. 1996;142:507–523. doi: 10.1093/genetics/142.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trut L.N. Proceeding of the XIV International Congress of Genetics. Vol. 2. MIR Publishers; Moscow, Russia: 1980. The genetics and phenogenetics of domestic behaviour. Problems in general genetics; pp. 123–136. Book 2. [Google Scholar]
- Trut L.N. Early canid domestication: The farm fox experiment. Am. Sci. 1999;87:160–169. [Google Scholar]
- Trut L.N. Experimental studies of early canid domestication. In: Ruvinsky A., Sampson J., Sampson J., editors. The Genetics of the Dog. CABI; Wallingford, UK: 2001. pp. 15–43. [Google Scholar]
- Trut L.N., Oskina I.N., Oskina I.N. Developmental changes of corticosteroid level in foxes with different behavior. Reports of Russian Academy of Sciences. 1985;281:1010–1014. [Google Scholar]
- Trut L.N., Pliusnina I.Z., Oskina I.N., Pliusnina I.Z., Oskina I.N., Oskina I.N. An experiment on fox domestication and debatable issues of evolution of the dog. Genetika. 2004;40:794–807. [PubMed] [Google Scholar]
- Trut L.N., Kharlamova A.V., Carrier D.R., Chase K., Kukekova A.V., Acland G.M., Lark K.G., Kharlamova A.V., Carrier D.R., Chase K., Kukekova A.V., Acland G.M., Lark K.G., Carrier D.R., Chase K., Kukekova A.V., Acland G.M., Lark K.G., Chase K., Kukekova A.V., Acland G.M., Lark K.G., Kukekova A.V., Acland G.M., Lark K.G., Acland G.M., Lark K.G., Lark K.G. The Genome of the Domestic Dog (Cold Spring Harbor Monograph Series 44) Cold Spring Harbor Press; Cold Spring Harbor, NY: 2006. Morphology and behavior: Are they coupled at the genome level? pp. 81–93. [Google Scholar]
- Vage D.I., Lu D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Lu D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Klungland H., Lien S., Adalsteinsson S., Cone R.D., Lien S., Adalsteinsson S., Cone R.D., Adalsteinsson S., Cone R.D., Cone R.D. A non-epistatic interaction of agouti and extension in the fox, Vulpes vulpes. Nat. Genet. 1997;15:311–315. doi: 10.1038/ng0397-311. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Sigmund R., Borner A., Korzun V., Stein N., Sorrels M.E., Langridge P., Graner A., Sigmund R., Borner A., Korzun V., Stein N., Sorrels M.E., Langridge P., Graner A., Borner A., Korzun V., Stein N., Sorrels M.E., Langridge P., Graner A., Korzun V., Stein N., Sorrels M.E., Langridge P., Graner A., Stein N., Sorrels M.E., Langridge P., Graner A., Sorrels M.E., Langridge P., Graner A., Langridge P., Graner A., Graner A. Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci. 2005;168:195–202. [Google Scholar]
- Wayne R.K., Geffen E., Girman D.J., Koepfli K.P., Lau L.M., Marshall C.R., Geffen E., Girman D.J., Koepfli K.P., Lau L.M., Marshall C.R., Girman D.J., Koepfli K.P., Lau L.M., Marshall C.R., Koepfli K.P., Lau L.M., Marshall C.R., Lau L.M., Marshall C.R., Marshall C.R. Molecular systematics of the Canidae. . Syst. Biol. 1997;46:622–653. doi: 10.1093/sysbio/46.4.622. [DOI] [PubMed] [Google Scholar]
- Werner P., Mellersh C.S., Raducha M.G., DeRose S., Acland G.M., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Mellersh C.S., Raducha M.G., DeRose S., Acland G.M., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Raducha M.G., DeRose S., Acland G.M., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., DeRose S., Acland G.M., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Acland G.M., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Prociuk U., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Wiegand N., Aguirre G.D., Henthorn P.S., Patterson D.F., Aguirre G.D., Henthorn P.S., Patterson D.F., Henthorn P.S., Patterson D.F., Patterson D.F., et al. Anchoring of canine linkage groups with chromosome-specific markers. Mamm. Genome. 1999;10:814–823. doi: 10.1007/s003359901096. [DOI] [PubMed] [Google Scholar]
- Wienberg J. The evolution of eutherian chromosomes. Curr. Opin. Genet. Dev. 2004;14:657–666. doi: 10.1016/j.gde.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Wipf L., Scackeford R.M., Scackeford R.M. Chromosomes of red fox. Proc. Natl. Acad. Sci. 1942;28:265–268. doi: 10.1073/pnas.28.7.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., O'Brien P.C., Milne B.S., Graphodatsky A.S., Solanky N., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., O'Brien P.C., Milne B.S., Graphodatsky A.S., Solanky N., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., Milne B.S., Graphodatsky A.S., Solanky N., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., Graphodatsky A.S., Solanky N., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., Solanky N., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., Trifonov V., Rens W., Sargan D., Ferguson-Smith M.A., Rens W., Sargan D., Ferguson-Smith M.A., Sargan D., Ferguson-Smith M.A., Ferguson-Smith M.A. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- Yang F., Milne B.S., Schelling C., Dolf G., Schlapfer J., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Milne B.S., Schelling C., Dolf G., Schlapfer J., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Schelling C., Dolf G., Schlapfer J., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Dolf G., Schlapfer J., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Schlapfer J., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Switonski M., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Ladon D., Pienkowska A., Bosma A.A., Sargan D.R., Pienkowska A., Bosma A.A., Sargan D.R., Bosma A.A., Sargan D.R., Sargan D.R., et al. Chromosome identification and assignment of DNA clones in the dog using a red fox and dog comparative map. Chromosome Res. 2000;8:93–100. doi: 10.1023/a:1009251917072. [DOI] [PubMed] [Google Scholar]
- Yu A., Zhao C., Fan Y., Jang W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Zhao C., Fan Y., Jang W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Fan Y., Jang W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Jang W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Doggett N.A., Ghebranious N., Broman K.W., Ghebranious N., Broman K.W., Broman K.W., et al. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]