Abstract
Mitotic arrest deficiency 2 (Mad2) is a component of mitotic spindle checkpoint proteins and is essential for accurate chromosome segregation. We investigated a role for Mad2 in hematopoiesis using Mad2-haploinsufficient (Mad2+/−) mice. Mad2+/− bone marrow (BM) and spleen manifested decreased absolute numbers and cycling status of immature, but not mature, hematopoietic progenitor cells. Mad2+/− BM granulocyte-macrophage colony-forming units (CFU-GMs) did not manifest synergistic proliferation in response to stem cell factor (SCF) plus GM-CSF. The percentage of annexin V+ cells was higher in Mad2+/− than Mad2+/+c-Kit+lin− BM after culture with SCF and GM-CSF. However, no significant difference in phosphorylation of extracellular signal–related kinase (Erk1/2) at Thr202/Tyr204 and Akt at Ser473 between Mad2+/− and Mad2+/+BM c-Kit+lin− cells was observed. Immunoprecipitation assays performed in human MO7e cells demonstrated physical association of c-Kit with Mad2. Moreover, stimulation with SCF plus GM-CSF led to dissociation of Mad2 from c-Kit. Confocal microscopy demonstrated that Mad2 colocalized with c-Kit in the cytoplasm of MO7e cells. These results suggest that Mad2 is involved in synergistic growth of immature hematopoietic progenitor cells in response to SCF plus GM-CSF, effects that may be mediated via physical association of Mad2 with c-Kit.
Introduction
c-Kit is a receptor tyrosine kinase (RTK) belonging to subclass III, which includes platelet-derived growth factor receptor (PDGFR), Fms-like tyrosine kinase 3 (Flt3), and macrophage colony-stimulating factor/colony-stimulating factor-1 (M-CSF/CSF-1) receptor (CSF-1R).1 c-Kit is expressed on hematopoietic progenitor cells, mature mast cells, germ cells, melanocytes, and interstitial cells of Cajal, and plays an essential role in regulating proliferation, survival, and migration of primitive hematopoietic cells.1–4 Binding of stem cell factor (SCF) to c-Kit induces dimerization and subsequent activation through autophosphorylation of the receptor, followed by activation of several signaling pathways, including Src kinases, mitogen-activated protein (MAP) kinases, c-Jun N-terminal kinases (JNKs), signal transducers and activators of transcription (STATs), and phosphatidylinositol 3 (PI3)–kinase/Akt.4 Importantly, SCF acts in synergy with other growth factors, including granulocyte-macrophage CSF (GM-CSF), interleukin-3 (IL-3), erythropoietin (Epo), and granulocyte CSF (G-CSF) to stimulate growth of hematopoietic stem/progenitor cells.5–10 This synergism is important for biologic and clinical implications, such as in vitro expansion of primitive hematopoietic stem and progenitor cells, as well as for other therapeutic uses. Recent information indicates that c-Kit interacts with the common β chain of the GM-CSF receptor, and kinase-dependent and -independent components contribute to the synergistic growth by SCF combined with GM-CSF.11 Proliferation and synergistic activation of early signal transduction pathways, including extracellular signal–related kinase (Erk) proteins in response to SCF plus GM-CSF, has been reported.12–15 We reported that SCF and GM-CSF synergistically activate Erk, but not stress-activated protein kinase JNK and p38MAP kinase pathways in MO7e cells.16 We further showed that concurrent stimulation with SCF and GM-CSF synergistically induces phosphorylation of the Rb protein as well as increasing cyclin-dependent kinase inhibitor p21Cip-1Waf1 and decreasing p27Kip-1 expression in MO7e cells.17 Thus, multiple intracellular mechanisms contribute to synergistic proliferation induced by SCF in combination with GM-CSF, and additional mechanisms mediating this synergy remain to be defined.
The mitotic checkpoint, also known as a spindle checkpoint, ensures that cells do not exit from mitosis until all chromosomes align and attach to the spindle. To date, genetic analyses have identified 7 genes (Mad1-Mad3, Bub1-Bub3, and Mps1) that are required for mitotic checkpoint control in Saccharomyces cerevisiae.18–20 The mitotic checkpoint proteins are likely to be highly conserved in mammals.21,22 Of these proteins, Mad2 is crucial for generating the “wait” signal to prevent onset of anaphase in the presence of microtubule disruption.23–25 Mad2-null mice are embryonically lethal by day 7.5 after coitus, and the embryos undergo widespread chromosome missegregation and apoptosis.26 Deletion of 1 allele of the Mad2 gene results in premature separation of sister chromatids and a high frequency of chromosome missegregation in the presence of spindle inhibitor, nocodazole, in both primary murine embryonic fibroblasts and human cancer cells.27 Thus, Mad2 plays a role in accurate chromosome segregation in mitotic cells. In this context, Mad2 regulates metaphase-anaphase transition by associating with checkpoint proteins, including anaphase-promoting complex/cyclosome (APC/C), cdc20, and Mad1.28–31 During interphase, Mad2 binds to Mad1 and is preferentially found on the nuclear periphery.32 Upon onset of mitosis, Mad2 translocates into the nucleus and is guided to unattached kinetochores by Mad1.32 On the other hand, Mad2 has been shown to bind to checkpoint-unrelated proteins such as insulin receptor, estrogen receptor β, and tumor necrosis factor α convertase.33–35 However, the significance of these interactions remains unclear. Recently, Mad2 also has been shown to physically associate with the common β chain of the GM-CSF receptor in a cell-cycle–dependent manner.36 Therefore, it is possible that Mad2 may be involved in cytokine signaling and regulation of mitosis in primary hematopoietic progenitor cells. Here we show that Mad2 is necessary for optimal progenitor cell numbers and cycling and for the synergistic proliferative effect of progenitor cells to SCF combined with GM-CSF, and that Mad2 associates with c-Kit receptor in the human growth factor–dependent cell line MO7e.
Materials and methods
Cytokines, antibodies, and other agents
Purified recombinant preparations of human (rhu) and murine (rmu) GM-CSF were kind gifts from Immunex/Amgen (Seattle, WA), and purified rhuEpo was purchased from Amgen (Thousand Oaks, CA). Pokeweed mitogen mouse spleen cell-conditioned medium (PWMSCM) was prepared as described.37 For flow cytometric analysis, anti–c-Kit, anti–Sca-1, and antibodies (Abs) to CD3E, B220, Gr-1, Mac-1, and isotype controls were purchased from BD Biosciences (San Diego, CA). For protein assay, rabbit polyclonal anti–c-Kit, anti-Akt, anti–phospho-Akt (Ser473), and anti-Erk1/2, horseradish peroxidase (HRP)–linked anti–rabbit IgG, and mouse monoclonal anti–phospho-Erk1/2 (Thr202/Tyr204) Abs were purchased from Cell Signaling Technology (Beverly, MA). Agarose-conjugated goat anti–c-Kit and isotype control, and HRP-linked anti–goat and anti–mouse IgG Abs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-Mad2 Ab was purchased from Bethyl Laboratories (Montgomery, TX). Polyclonal rabbit isotype Ab was obtained from Upstate (Charlottesville, VA).
Mice
Mad2+/+ wild-type and Mad2+/− mutant mice were generated by interbreeding the wild-type and mutant mice (kindly provided by Dr Robert Benezra, Memorial Sloan-Kettering Cancer Center, New York, NY). All of the mice were used at 6 to 10 weeks of age. All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine.
Isolation of c-Kit+lineage marker− cells from BM
After harvesting bone marrow (BM) cells from femurs and tibias of mice, cells were washed once with phosphate-buffered saline (PBS) and resuspended in PBS supplemented with 0.5% bovine serum albumin (BSA; Sigma, St Louis, MO) and 2 mM EDTA (Gibco, Grand Island, NY). Lineage cell depletion was then performed by magnetic separation according to the manufacturer's instruction (Miltenyi Biotech, Auburn, CA). Briefly, cells were stained with biotin-conjugated Ab cocktail (anti-CD5, -B220, -CD11b, –Gr-1, –7-4, and –Ter-119 Abs) and were incubated for 10 minutes at 4°C. Cells were then incubated with antibiotin microbeads for 15 minutes at 4°C. After washing, cell suspension was subjected to magnetic separation. Purity was 90% by FACScan flow cytometer (Becton Dickinson, San Jose, CA). For cytokine signaling and apoptosis assay, lineage depleted cells were stained with allophycocyanin (APC)–conjugated antibiotin Ab (Miltenyi Biotech) and fluorescein isothiocyanate (FITC)–conjugated anti–mouse c-Kit Ab for 20 minutes on ice in the dark. After washing once, c-Kit+lineage marker− (KL) cells were sorted by FACSvantage flow cytometer (Becton Dickinson).
The measurement of KSL cell numbers and expression levels of c-Kit
After harvesting BM from femurs, cells were washed twice with PBS and were resuspended in PBS supplemented with 0.5% BSA and 2 mM EDTA. Then cells were stained with FITC-conjugated anti–Sca-1 Ab, phycoerythrin (PE)–conjugated anti–c-Kit Ab, and APC-conjugated Abs to CD3E, B220, Gr-1, and Mac-1, or FITC-, PE-, or APC-conjugated isotype-matched control Abs for 20 minutes at 4°C in the dark. After washing twice with PBS, cells were fixed with BD cytofix buffer (BD Pharmingen, San Diego, CA). Cells (106) were analyzed on a FACSCalibur flow cytometer using CellQuest software (Becton Dickinson). For measurement of c-Kit+Sca-1+lin− (KSL) cell numbers, the percentage of c-Kit+Sca-1+ cells were analyzed on electronically gated lin− cells. Absolute numbers of KSL cells were calculated from the nucleated cellularity per femur. c-Kit expression levels were analyzed on gated Sca-1+lin− cells and the mean fluorescence intensity (MFI) of c-Kit+ subpopulations was measured.
Clonogenic progenitor cell assay
BM and spleen cells from mice were assessed for granulocyte-macrophage colony-forming unit (CFU-GM), erythroid burst-forming unit (BFU-E), and multipotential granulocyte, erythroid, monocyte, megakaryocyte CFU (CFU-GEMM) progenitor cells, as described elsewhere.38 In short, for methylcellulose assay, 5 × 104 cells were plated and cultured in 0.9% methylcellulose culture medium with the combination of 30% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT), 5% vol/vol PWMSCM, 50 ng/mL rmuSCF, 1 U/mL rhuEpo, 2 mM glutamine (Cambrex Bio Science, Walkersville, MD), 10−4 M 2-mercaptoethanol, and 0.1 mM hemin (Sigma). For agar assay, 5 × 104 cells were plated and cultured in 0.3% agar culture medium in the presence of 50 ng/mL rmuSCF, 10 ng/mL rmuGM-CSF, or a combination of the two. Colonies were scored after 7 days of incubation at 37°C, in 5% O2 and 5% CO2. Absolute numbers of progenitors were calculated from the nucleated cellularity per femur or spleen and the number of colonies formed per number of cells plated. The percentage of progenitors in the S phase of the cell cycle was estimated by use of the tritiated thymidine kill technique, as described previously.39
Immunoprecipitation and immunoblotting
KL BM cells were stimulated with rmuSCF (50 ng/mL), rmuGM-CSF (10 ng/mL), or a combination of the two at the indicated time after serum starvation for 4 hours. Then cell lysates were prepared by ice-cold lysis buffer (0.5% NP-40, 10 mM Tris base, 200 mM NaCl, 10% glycerol, 5 mM NaF, and 0.5 mM sodium orthovanadate [pH 7.4]) with 1% protease inhibitor cocktail (EMD Biosciences, San Diego, CA). Human growth factor–dependent cell line MO7e cells were obtained and maintained as previously described.13 MO7e cells were starved for 18 hours in RPMI 1640 medium supplemented with 0.5% BSA. After washing, cells were placed in RPMI 1640 medium supplemented with 0.5% BSA and rhuSCF (50 ng/mL), rhuGM-CSF (2 ng/mL), or a combination of the two. After washing with ice-cold PBS, total-cell lysates were prepared in ice-cold lysis buffer (0.5% TritonX-100, 20 mM Tris base, 50 mM NaCl, 1mM EDTA, 10 mM NaF, 25 mM β-glycerophosphate, 10 mM sodium orthovanadate, and 30 mM tetrasodium pyrophosphate [pH 7.4]) with 1% protease inhibitor cocktail, and then incubated for 30 minutes at 4°C and centrifuged at 13 148g, for 30 minutes; supernatants were retained. For Mad2 immunoprecipitation, cell lysates were incubated with anti-Mad2 Ab for 4 hours at 4°C and then were incubated with protein A agarose beads (EMD Biosciences) at 4°C for 1 hour with rotation. For c-Kit immunoprecipitation, cell lysates were incubated with agarose-conjugated anti–c-Kit Ab for 4 hours at 4°C. Beads were washed with lysis buffer 4 times. Immunoprecipitates and cell lysates were eluted by incubation with SDS sample buffer (Invitrogen, Carlsbad, CA) for 5 minutes at 95°C. Samples were loaded in 10% or 12% tris-glycine gel (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membrane (Invitrogen). Immunoblotting was then performed according to the antibody manufacturer's instructions. Blots were visualized using HRP-conjugated secondary Abs and an enhanced chemiluminescence system (ECL; Amersham, Piscataway, NJ). To reprobe with other Abs, membranes were incubated in stripping buffer (Pierce, Rockford, IL) according to the manufacturer's instructions.
Immunofluorescence staining
Fixation, permeabilization, and staining procedures were performed as previously described.40 Briefly, MO7e cells were fixed in 3.7% paraformaldehyde, permeabilized with 0.5% Triton-X100, blocked in blocking buffer (PBS and 3% BSA), and stained with mouse monoclonal anti-Mad2 Ab (BD Biosciences) and rabbit polyclonal anti–c-Kit Ab (Santa Cruz Biotechnology). Secondary staining was performed using AlexaFluor 488–conjugated anti–rabbit IgG and AlexaFluor 647–conjugated chicken anti–mouse IgG (Molecular Probes/Invitrogen, Carlsbad, CA). DNA was stained with 4,6-diaminido-2-phenylindole (DAPI). Images were visualized and collected using a Zeiss-LSM510 confocal microscope (Zeiss, Oberkochen, Germany) with an oil immersion lens (100×/1.4 numerical aperture oil objective). Images were subsequently processed using Photoshop 7.0 (Adobe Systems, San Jose, CA).
Apoptosis assay
KL BM cells (2 × 105) from mice were incubated in Iscove modified Dulbecco media (IMDM; Cambrex Bio Science) containing 10% FBS, 2 mM glutamine, 50 ng/mL rmuSCF, and 10 ng/mL rmuGM-CSF for 6 days at 37°C in 5% CO2. Cells were then stained with PE-conjugated annexin V and 7-AAD (BD Biosciences) according to the manufacturer's instructions. The percentage of annexin V+ cells was analyzed on whole cells using the FACSCalibur flow cytometer and measured using the Cell Quest software (Becton Dickinson).
Statistical analysis
Data were analyzed statistically using the Student t test. A P value less than .05 was considered statistically significant.
Results
Influence of Mad2 haploinsufficiency on hematopoiesis in vivo
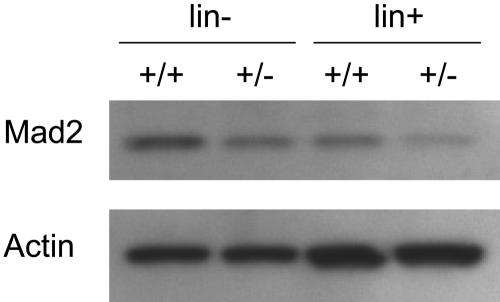
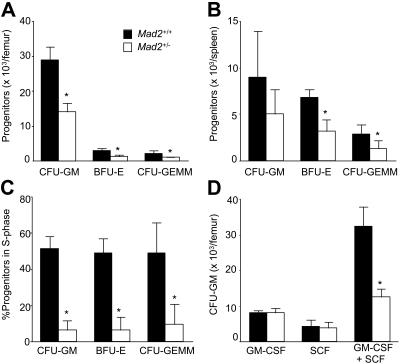
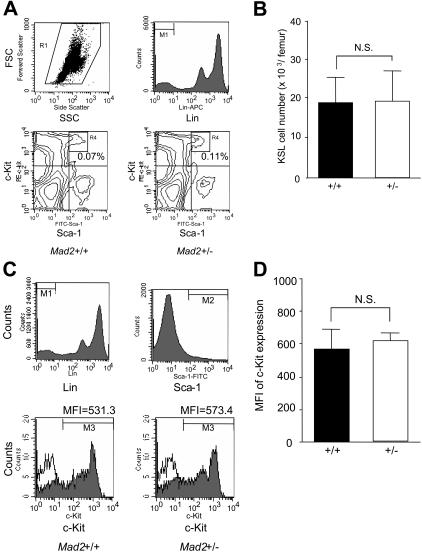
Mad2 protein levels of BM cells were compared between Mad2+/+ and Mad2+/− mice. As shown in Figure 1, Mad2 protein levels of lineage-depleted and lineage+ BM cells from Mad2+/− mice were about half the levels of those from Mad2+/+ mice, respectively. In contrast, the protein levels of β-actin were equal between Mad2+/+ and Mad2+/− mice. These results indicate that Mad2 protein of Mad2+/− BM cells is expressed at half the level of wild-type BM cells. The numbers of immature hematopoietic progenitor cells, those which are responsive in vitro to the proliferative effects of 2 or more synergistically acting cytokines, were evaluated in the BM and spleen of Mad2+/+ and Mad2+/− mice. As shown in Figure 2A, significantly decreased numbers of immature progenitors were observed in Mad2+/− BM compared with that of wild-type BM. In spleens of Mad2+/− mice, immature progenitors were also significantly decreased with the exception of CFU-GM compared with that of Mad2+/+ mice (Figure 2B). To determine whether the decreases in progenitor cell numbers involved the proliferative status of the cells, we analyzed the cell-cycling status of progenitors using the high-specific–activity tritiated thymidine kill assay. As shown in Figure 2C, the percentage of immature progenitor cells in the S phase of the cell cycle was significantly decreased in Mad2+/− BM. The spleen progenitors from Mad2+/+ and Mad2+/− were in a slow or noncycling state, with no significant differences between the Mad2 groups (data not shown). We next evaluated the mature CFU-GMs (those responsive to the proliferative stimulating effects of single cytokines) in both mice. There was no significant difference in total numbers of GM-CSF– or of SCF-responsive BM CFU-GM Mad2+/− and Mad2+/+ mice (Figure 2D). In contrast, total numbers of CFU-GMs responsive to both SCF and GM-CSF were significantly decreased in Mad2+/− BM (Figure 2D). These results suggest that total numbers of immature, but not mature, progenitors are decreased in Mad2+/− BM compared with that in wild-type BM. We also found that Mad2+/− progenitors did not proliferate synergistically in response to SCF plus GM-CSF, while wild-type progenitor cells did ([colonies responsive to SCF + GM-CSF] / [colonies responsive to SCF] + [colonies responsive to GM-CSF]; 1.04 vs 2.58 when CFU-GM per 5 × 104 BM cells was calculated), demonstrating a lack of response of Mad2+/− BM progenitors to synergistic stimulation. Changes in numbers of immature subsets of progenitors did not translate into changes in blood counts or in cellularity of BM and spleen between both sets of mice (Table 1). Because the Mad2+/− progenitor cell number was decreased, we evaluated the absolute numbers of KSL cells and the expression levels of c-Kit on Sca-1+lin− cells in BM from both mice using flow cytometry. There was no significant difference in KSL cell numbers (Figure 3A-B) and in c-Kit expression levels (Figure 3C-D) between both mice. This suggests that Mad2+/− BM is not deficient in KSL cells, and that lack of c-Kit expression is not responsible for the decreased number of Mad2+/− progenitor cells.
Figure 1.
Mad2 protein expression in bone marrow cells of Mad2+/− and Mad2+/+ mice. Lineage-depleted (lin−) and -positive (lin+) cells from Mad2+/+ and Mad2+/− mice BM were obtained by magnetic separation. Total-cell lysates were dissolved in SDS-PAGE. Membrane was immunoblotted with anti-Mad2 and antiactin antibodies. +/+ indicates Mad2+/+; +/−, Mad2+/−.
Figure 2.
Comparative analysis of hematopoietic progenitor cells in bone marrow and spleen of Mad2+/− and Mad2+/+ mice. Absolute numbers of immature hematopoietic progenitor cells (responsive to stimulation by multiple cytokines (5% [vol/vol] PWMSCM, 50 ng/mL rmuSCF, and 1 U/mL rhuEpo in vitro) in bone marrow (A) and spleen (B), and the percentage of BM progenitor cells in S phase (C). Absolute numbers of immature (responsive to 50 ng/mL rmuSCF and 10 ng/mL rmuGM-CSF) and mature (responsive to stimulation by 50 ng/mL rmuSCF or 10 ng/mL rmuGM-CSF) CFU-GM in BM (D). ■ and □ indicate Mad2+/+ and Mad2+/−, respectively. Data represent the average of a total of 5 mice of each phenotype from 2 independent experiments. Error bars represent 1 SD. *P < .05 (+/− vs +/+).
Table 1.
Cellularity of peripheral blood, BM, and spleen of Mad2+/− compared with age-matched control (Mad2+/+)
| Mad2+/− | Mad2+/+ | P | |
|---|---|---|---|
| Peripheral blood | |||
| WBCs, × 109/L (8) | 10.03 ± 5.06 | 11.36 ± 3.84 | .56 |
| RBCs, × 1012/L (8) | 10.1 ± 0.7 | 10.6 ± 1.1 | .21 |
| Hct, proportion of 1 (8) | .532 ± .037 | .570 ± .074 | .42 |
| PLTs, × 109/L (8) | 1280 ± 233 | 1184 ± 223 | .80 |
| BM | |||
| Cells, × 106/femur (10) | 24.9 ± 4.9 | 25.5 ± 5.8 | .80 |
| Spleen | |||
| Cells, × 106 (10) | 126.9 ± 22.3 | 131.8 ± 30.1 | .70 |
Numbers in parentheses indicate the number of mice used for the data presented. Data indicate mean ± SD of 8 to 10 mice. WBCs indicates white blood cells; RBCs, red blood cells; Hct, hematocrit; and PLTs, platelets.
Figure 3.
Absolute numbers of KSL cells and expression of c-Kit on Sca1+lin− cells. Absolute numbers of KSL cells in BM. (A) Representative cytograms of forward scatter (FSC) versus side scatter (SSC), lineage marker (top panels), and Sca-1 versus c-Kit (bottom panels) for measurements of percentages of KSL cells in Mad2+/+ and Mad2+/− BM cells are shown. The percentage of c-Kit+Sca-1+ cells (R4) was analyzed on gated lin− cells (M1 + R1). Numbers indicate the percentage of KSL cells. (B) Data show KSL cell numbers per femur. Error bars indicate mean ± SD of measurements of 6 mice each. (C) Representative histograms of lineage marker and Sca-1 (top panels) for gating the Sca-1+lin− cell population, and c-Kit expression (bottom panels) are shown. c-Kit expression was analyzed on gated lin− cells (M1) and Sca-1+ cells (M2). The shaded curves show staining with anti–c-Kit antibody; the open curves show the isotype control (bottom panels). Numbers indicate the mean fluorescence intensity (MFI) of c-Kit. (D) Expression level of c-Kit on Sca-1+lin− cells in BM. Error bars indicate mean ± SD of measurements of 4 mice each. NS indicates not significant.
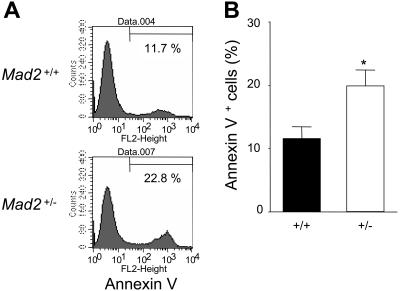
Percentage of cell death of progenitor cells of Mad2+/− mice is higher than that of Mad2+/+ mice
To better understand decreased numbers of immature progenitor cells and decreased responsiveness of Mad2+/− progenitor cells to stimulation by combinations of cytokines, we next compared the percentages of cell death in hematopoietic progenitor cells between Mad2+/+ and Mad2+/− BM. KL cells from BM (enriched for progenitors) were cultured in the presence of SCF and GM-CSF for 6 days. The percentage of annexin V+ cells in whole-cultured cells at day 6 was significantly higher in Mad2+/− mice compared with that of Mad2+/+ mice (Figure 4A-B).
Figure 4.
Cell death 6 days after culture with SCF plus GM-CSF. (A) c-Kit+lin− bone marrow cells (2 × 105) from wild-type and Mad2+/− mice were cultured in a 12-well tissue-culture plate in the presence of SCF (50 ng/mL) plus GM-CSF (10 ng/mL) for 6 days. Representative histograms of Mad2+/+ and Mad2+/− BM cells are shown. Numbers in histograms correspond to the percentage of annexin V+ cells. (B) Data represent the average of triplicate samples, and error bars represent 1 SD from the mean for triplicate wells. Result shown is representative of 2 independent experiments. *P < .01 (+/− vs +/+).
No difference in activation of Erk1/2 and Akt in c-Kit+lin− between Mad2+/+ and Mad2+/− BM cells
Because Mad2+/− BM progenitors showed fewer proliferative effects and increased cell death in response to SCF in combination with GM-CSF compared with wild-type progenitors, we examined the effect of SCF and GM-CSF on activation of Erk1/2 and Akt in KL BM cells, which have been shown to be involved in synergistic proliferation and cell survival of hematopoietic cells.16,41,42 Stimulation with SCF plus GM-CSF enhanced phosphorylation of Erk1/2 at Thr202/Tyr204, but not Akt at Ser473, beyond that seen by SCF alone in both Mad2+/− and Mad2+/+ KL BM cells. However, we did not detect significant differences in phosphorylation of Erk1/2 at Thr202/Tyr204 and Akt at Ser473 after stimulation with SCF plus GM-CSF between Mad2+/− and Mad2+/+ BM cells as well as after stimulation with SCF or GM-CSF alone (data not shown). These results indicate that the lowered proliferative effect and increased cell death in response to SCF plus GM-CSF in Mad2+/− progenitor cells were not due to the reduced activation of these signaling molecules.
Mad2 physically associates with c-Kit in MO7e cells
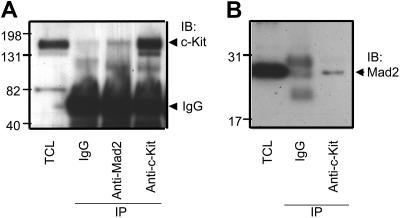
Because Mad2 has been shown to interact with the common β chain of the GM-CSF receptor,36 we hypothesized that Mad2 may mediate the synergistic proliferative effect induced by SCF and GM-CSF through physical interaction between Mad2 and c-Kit. To determine if Mad2 associates with c-Kit, we used the human growth factor–responsive cell line MO7e, which expresses c-Kit and GM-CSF receptor and responds to the synergistic proliferative effects of GM-CSF plus SCF.13,14 We first examined the physical interaction between Mad2 and c-Kit using asynchronous MO7e cells that were cultured with GM-CSF. Cell lysates were immunoprecipitated with anti-Mad2 Ab and then immunoblotted with anti–c-Kit Ab. As shown in Figure 5A, we observed coimmunoprecipitation of c-Kit with the anti-Mad2 Ab, but not with control IgG. To confirm this relationship, we tested the reciprocal interaction. As shown in Figure 5B, we also observed coimmunoprecipitation of Mad2 with the anti–c-Kit Ab. These findings suggest a physical association between Mad2 and c-Kit in MO7e cells.
Figure 5.
Mad2 physically associates with c-Kit in MO7e cells. Total-cell lysates (TCL) were immunoprecipitated with either control IgG, anti-Mad2, or anti–c-Kit Abs, and then immunoblotted with anti–c-Kit Abs (A) or anti-Mad2 Abs (B). The results of 1 representative of 3 experiments are shown. IB indicates immunoblot; IP, immunoprecipitation.
Mad2 dissociates from c-Kit after stimulation with SCF plus GM-CSF in MO7e cells
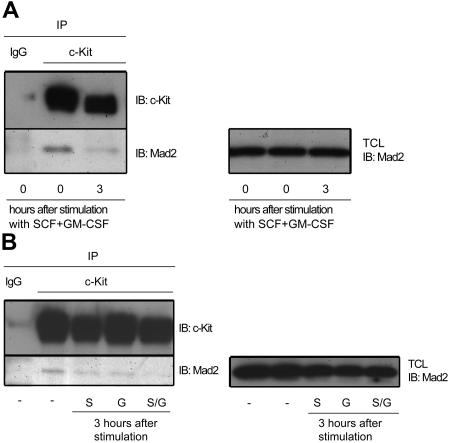
To determine whether the association between Mad2 and c-Kit is cytokine dependent or not, we next examined the interaction of Mad2 with c-Kit before or after stimulation with SCF plus GM-CSF. MO7e cells were cultured in factor- and serum-free medium for 18 hours before cytokine stimulation. As shown in Figure 6A, we observed coimmunoprecipitation of Mad2 with anti–c-Kit Ab before cytokine stimulation. In contrast, the interaction was decreased 3 hours after stimulation with SCF plus GM-CSF. Addition of this cytokine combination did not change total Mad2 protein levels. These results indicate that Mad2 may dissociate from c-Kit after stimulation with SCF plus GM-CSF. Because immunoprecipitated c-Kit was decreased after stimulation, it is also possible that reduced level of immunoprecipitated Mad2 with c-Kit after cytokine stimulation may be due to decreased level of c-Kit itself. To examine this possibility, we next compared immunoprecipitated c-Kit and Mad2 levels after stimulation with SCF, GM-CSF, or a combination of the two. As shown in Figure 6B, immunoprecipitated Mad2 was scarcely detected 3 hours after stimulation with SCF plus GM-CSF. In contrast, a more intense band of immunoprecipitated Mad2 was observed when cells were stimulated with SCF alone. c-Kit levels were equally decreased after stimulation with either SCF alone or SCF combined with GM-CSF. In addition, the Mad2 levels in total-cell lysates were similar between stimulation with SCF alone and SCF plus GM-CSF. These results indicate that reduced levels of coimmunoprecipitated Mad2 after stimulation with SCF plus GM-CSF is due to the dissociation of Mad2 from c-Kit rather than decreased levels of c-Kit receptor. On the other hand, immunoprecipitated Mad2 levels after stimulation with GM-CSF alone was equal to that with SCF alone. Together, the data suggest that Mad2 dissociates from c-Kit 3 hours after stimulation of MO7e cells with SCF plus GM-CSF, but not in the presence of either SCF or GM-CSF alone.
Figure 6.
Mad2 dissociates from c-Kit after stimulation with SCF plus GM-CSF in MO7e cells. (A) After serum starvation, cells were stimulated with 50 ng/mL SCF plus 2 ng/mL GM-CSF for 3 hours. (B) Cells were unstimulated or stimulated with 50 ng/mL SCF, 2 ng/mL GM-CSF, or SCF plus GM-CSF for 3 hours. Total-cell lysates (TCL) were separated into 2 tubes for immunoblotting and immunoprecipitation, respectively. Cell lysates were immunoprecipitated with control IgG or anti–c-Kit Abs, and then immunoblotted with anti–c-Kit Abs (left panel, top blot) or anti-Mad2 Abs (left panel, bottom blot). To compare the Mad2 proteins, separated cell lysates also were immunoblotted with anti-Mad2 Abs (right panel). The results of 1 representative of 2 experiments are shown. IB indicates immunoblot; IP, immunoprecipitation; −, no cytokine stimulation; S, SCF; G, GM-CSF; and S/G, SCF plus GM-CSF.
Mad2 colocalizes with c-Kit in cytoplasm before stimulation with cytokines in MO7e cells
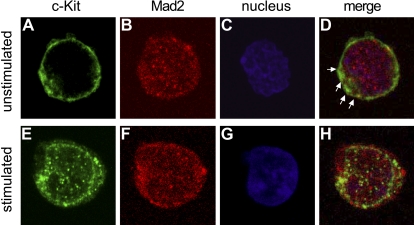
In order to determine the localization of Mad2 and c-Kit, we performed confocal microscopic examination using MO7e cells (Figure 7). Confocal microscopy showed that Mad2 is distributed diffusely in the cytoplasm with some Mad2 in the nucleus (Figure 7B), and partially colocalizes with c-Kit before cytokine stimulation (Figure 7D). After stimulation with SCF plus GM-CSF, c-Kit translocated toward the nuclear membrane (Figure 7H), indicating receptor internalization. Localization of Mad2 3 hours after cytokine stimulation was similar to that before stimulation (Figure 7F). These results suggest that Mad2 colocalizes with c-Kit in cytoplasm of factor- and serum-starved MO7e cells.
Figure 7.
Mad2 colocalizes with c-Kit in cytoplasm in MO7e cells. Top (A-D) and bottom (E-H) panels show unstimulated and cytokine-stimulated MO7e cells, respectively. Images were acquired by confocal microscopy with an oil immersion lens (magnification, × 100). c-Kit is shown in green (A,E), Mad2 in red (B,F), and nucleus in blue (C,G). Merged images are shown in panels D and H. Arrows indicate representative colocalization of Mad2 with c-Kit.
Discussion
Our data demonstrated that Mad2 plays a role in the in vivo and in vitro proliferation of hematopoietic progenitor cells. Mad2+/− mice showed decreased total numbers of immature hematopoietic progenitor cells in both the BM and spleen compared with wild-type control mice as well as decreased cell cycling of BM progenitors. In contrast, there was no significant difference in more mature progenitors between Mad2+/− and wild-type mice. Consistent with these findings, Mad2+/− BM CFU-GMs did not respond to the synergistic proliferative effects of the combination of SCF plus GM-CSF. These results suggest that Mad2 plays a role in the proliferation of immature, but not more mature, hematopoietic progenitors. Decreases in numbers and cycling of Mad2+/− hematopoietic progenitors did not manifest in decreased nucleated cellularity in BM, blood, or spleen. This is not necessarily surprising since this has been seen with a number of different mouse models in which deficiencies in the expression of intracellular or surface proteins that manifest in decreased progenitor cell numbers or cell cycling do not result in nucleated cell changes. These characteristics of Mad2+/− mice are also observed in the CD34−/−, c-mpl−/−, and dual CD34/c-mpl−/− mice among a number of different gene “knockout” mice, where normal nucleated cellularity persists in the face of decreased progenitor cell numbers.38,43–46 This may relate to the massive reserve of hematopoietic progenitor cells that exist in the BM of normal mice.38 We also showed no significant difference in the absolute numbers of KSL cells in BM between Mad2+/− mice and wild-type control mice, indicating that the defect of 1 allele of the Mad2 gene does not affect the maintenance of this phenotypically defined population of hematopoietic stem cells in steady-state conditions. Whether or not Mad2 plays a regulatory role in hematopoietic stem cell function remains to be determined. The strain background that the Mad2+/− mice are on currently precludes analysis of stem cell transplantation studies using congenic mice.
Recently, the function of BubR1 in hematopoiesis, which is also a mitotic checkpoint protein, was investigated using BubR1-haploinsufficient (BubR1+/−) mice.47 The authors showed that the spleens of adult BubR1+/− mice were enlarged compared with that of wild-type mice, and BubR1+/− was associated with increased splenic megakaryocytes as well as megakaryocytic progenitors in BM. In contrast, we did not see any differences in spleen size and megakaryocyte morphology in BM between Mad2+/− and wild-type mice (data not shown). The differences in phenotype between Mad2+/− and BubR1+/− mice may be due to different functions or cell-lineage specificities of these intracellular proteins.
SCF is known to act in synergy with GM-CSF to stimulate proliferation of hematopoietic progenitor cells.6,7,11–15 However, the underlying mechanism of this synergy has not been fully defined. Interestingly, we observed no synergistic proliferative effect induced by SCF plus GM-CSF on Mad2+/− progenitors in in vitro colony assay. This observation led us to investigate the underlying mechanism of lack of synergy. We examined Erk1/2 and Akt activation in response to SCF in combination with GM-CSF using primary BM cells from Mad2+/– mice. However, we did not observe significant differences in phosphorylation of Erk1/2 at Thr202/Tyr204 and Akt at Ser473 in response to SCF plus GM-CSF between Mad2+/+ and Mad2+/− KL BM cells. It is possible that other cytokine-signaling molecules may relate to lack of synergy with SCF plus GM-CSF in Mad2+/− BM progenitor cells.
Interestingly, we found a higher percentage of cell death in KL BM cells of Mad2+/− mice compared with that of wild-type mice when these cells were cultured in the presence of SCF and GM-CSF. This indicates that Mad2 may play a role in not only synergistic proliferation but also antiapoptotic effects of SCF plus GM-CSF. It is possible that inactivation of 1 Mad2 allele directly leads to cell death, since Mad2-null embryos and Mad2-defective somatic cells are involved in cell death by extensive chromosome missegregation.26,48 However, this possibility is not absolutely clear because there was no significant difference between Mad2+/− and wild-type mice in the percentage of cell death of BM cells before cell culture (data not shown). Thus, the greater cell death of Mad2+/− progenitor cells may occur in stress conditions but not in steady-state conditions. Recently, it has been shown that down-regulation of Mad2 inhibits anticancer drug–induced apoptosis by up-regulating Bcl-2 and interfering with the mitochondrion pathway.49 It is therefore possible that Mad2 may regulate apoptosis-related proteins in hematopoietic progenitor cells.
Our observations raise the question of how Mad2 regulates the proliferative response to cytokines. Mad2 is known to interact with the cytoplasmic domain of the insulin receptor and tumor necrosis factor α convertase, and with estrogen receptor β.33–35 However, the significance of these interactions is unknown. Recently, Mad2 has been shown to physically interact with the common β chain of the GM-CSF receptor in a cell-cycle–dependent manner.36 Because Mad2 was associated with box1/2 within the β chain, which is important for the binding of the Janus kinase Jak2, we examined whether Jak2 and Mad2 compete with each other for binding to the β chain. However, no clear biochemical result was obtained. In addition, Mad2 overexpression neither enhanced nor inhibited the proliferation of cells in response to GM-CSF. In our study, Mad2 haploinsufficiency did not affect the proliferation of hematopoietic progenitors in response to GM-CSF alone. These findings indicate that Mad2 may not be involved in GM-CSF receptor signaling in the proliferation of hematopoietic progenitors. Moreover, there was no significant difference in the proliferation of hematopoietic progenitors in response to SCF alone between both mice, suggesting that Mad2 may not play a role in c-Kit signaling in proliferation of hematopoietic progenitors to SCF alone. In the present study, we showed that Mad2 is physically associated with c-Kit in the human growth factor dependent cell line MO7e. Interestingly, Mad2 dissociated from c-Kit after stimulation with SCF plus GM-CSF. In contrast, Mad2 protein did not dissociate from c-Kit when cells were stimulated with SCF or GM-CSF alone for at least 3 hours, suggesting that this dissociation requires concurrent stimulation with SCF and GM-CSF. Subcellular localization of Mad2 during interphase is not well known. Li and Benezra demonstrated that Mad2 was distributed throughout the cell, with prominent perinuclear localization during interphase in HeLa cells.23 In our study, confocal microscopy demonstrated that Mad2 localizes diffusely in the cytoplasm and colocalizes with c-Kit near the cell membrane of MO7e cells before stimulation with SCF plus GM-CSF. These results suggest that Mad2 localizes in the cytoplasm of MO7e cells during interphase and may be involved in the synergistic proliferation in response to SCF in combination with GM-CSF via the interaction of Mad2 with c-Kit. Mad2 appears in the kinetochore in the G2/M phase but disappears after cytokinesis.23 In this scenario, after Mad2 participates in cytokine signaling in the cytoplasm during the G1/S phase, it may translocate to the nucleus to regulate the mitotic checkpoint.
In summary, our paper describes for the first time that the mitotic spindle checkpoint protein Mad2 physically associates with c-Kit. Mad2 plays a role in the synergistic proliferation of hematopoietic progenitor cells in response to SCF in combination with GM-CSF. Mad2 may have a role in synergistic cytokine signaling through the interaction with c-Kit that is distinct from its function as a mitotic spindle checkpoint protein.
Acknowledgments
The authors thank Denessa Luckett for cell sorting.
This work was supported by US Public Health Service grants HL56416, HL67384, and DK53674 from the National Institutes of Health (H.E.B.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.I. designed and performed the study, analyzed the data, and drafted the paper. C.R.M. participated in the design of the study and preparation of mice, and assisted in drafting the manuscript. M.-K.H. designed the study and analyzed data. S.B. designed and performed the study, and analyzed the data. S.F. participated in design of the study. S.C. performed the study. H.E.B. participated in design, coordination, and performance of the study; assisted in drafting the manuscript; and funded the study. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hal E. Broxmeyer, Department of Microbiology & Immunology and the Walther Oncology Center, Indiana University School of Medicine, R2 Rm 302, 950 West Walnut St, Indianapolis, IN 46202; e-mail: hbroxmey@iupui.edu.
References
- 1.Ashman LK. The biology of stem cell factor and its receptor C-kit. Int J Biochem Cell Biol. 1999;31:1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- 2.Boissan M, Feger F, Guillosson JJ, Arock M. c-Kit and c-kit mutations in mastocytosis and other hematological diseases. J Leukoc Biol. 2000;67:135–148. doi: 10.1002/jlb.67.2.135. [DOI] [PubMed] [Google Scholar]
- 3.Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- 4.Linnekin D. Early signaling pathway activated by c-Kit in hematopoietic cells. Int J Biochem Cell Biol. 1999;31:1053–1074. doi: 10.1016/s1357-2725(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 5.Duarte R, Franf D. The synergy between stem cell factor (SCF) and granulocyte colony-stimulating factor (G-CSF): molecular basis and clinical relevance. Leuk Lymphoma. 2002;43:1179–1187. doi: 10.1080/10428190290026231. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Broxmeyer HE, Mantel C, Kwon HJ, et al. Synergistic activation of p70S6 kinase associated with stem cell factor in MO7e cells. Exp Mol Med. 2003;35:222–226. doi: 10.1038/emm.2003.30. [DOI] [PubMed] [Google Scholar]
- 7.Mantel C, Hendrie P, Broxmeyer HE. Steel factor regulates cell cycle asymmetry. Stem Cells. 2001;19:483–491. doi: 10.1634/stemcells.19-6-483. [DOI] [PubMed] [Google Scholar]
- 8.Pearson MA, O'Farrell AM, Dexter TM, Whetton AD, Owen-Lynch PJ, Heyworth CM. Investigation of the molecular mechanisms underlying growth factor synergy: the role of ERK2 activation in synergy. Growth Factors. 1998;15:293–306. doi: 10.3109/08977199809017484. [DOI] [PubMed] [Google Scholar]
- 9.Broudy VC, Morgan DA, Lin N, Zsebo KM, Jacobsen FW, Papayannopoulou T. Stem cell factor influences the proliferation and erythroid differenciation of the MB-02 human erythroleukemia cell line by binding to a high-affinity c-kit receptor. Blood. 1993;82:436–444. [PubMed] [Google Scholar]
- 10.Rafael FD, David AF. SCF and G-CSF lead to the synergistic induction of proliferation and gene expression through complementary signaling pathways. Blood. 2000;96:3422–3430. [PubMed] [Google Scholar]
- 11.Lennartsson J, Shivakrupa R, Linnekin D. Synergistic growth of stem cell factor and granulocyte macrophage colony-stimulating factor involves kinase-dependent and -independent contributions from c-Kit. J Biol Chem. 2004;279:44544–44553. doi: 10.1074/jbc.M404085200. [DOI] [PubMed] [Google Scholar]
- 12.Kamijo T, Koike K, Takeuchi K, et al. Analysis of synergism between stem cell factor and granulocyte-macrophage colony-stimulating factor on human megakaryoblastic cells: an increase in tyrosine phosphorylation of 145kDa subunit of c-kit in two-factor combination. Leuk Res. 1997;21:1097–1106. doi: 10.1016/s0145-2126(97)00086-6. [DOI] [PubMed] [Google Scholar]
- 13.Hendrie PC, Miyazawa K, Yang YC, et al. Mast cell growth factor (c-kit ligand) enhances cytokine stimulation of proliferation of the human factor-dependent cell line, MO7e. Exp Hematol. 1991;19:1031–1398. [PubMed] [Google Scholar]
- 14.Hallek M, Drucker B, Lepisto EM, et al. Granulocyte-macrophage colony-stimulating factor and steel factor induce phosphorylation of both unuque and overlapping signal transduction intermediates in a human factor-dependent hematopoietic cell line. J Cell Physiol. 1992;153:176–186. doi: 10.1002/jcp.1041530122. [DOI] [PubMed] [Google Scholar]
- 15.Miyazawa K, Hendrie PC, Mantel C, et al. Comparative analysis of signaling pathway between mast cell growth factor (c-kit ligand) and granulocyte-macrophage colony-stimulating factor in a human factor-dependent myeloid cell line involves phosphorylation of Raf-1, GTPase-activating protein and mitogen-activated protein kinase. Exp Hematol. 1991;19:1110–1123. [PubMed] [Google Scholar]
- 16.Lee Y, Mantel C, Anzai N, et al. Transcriptional and ERK1/2-dependent synergistic upregulation of p21 (cip1/waf1) associated with steel factor synergy in MO7e. Biochem Biophys Res Commun. 2001;280:675–683. doi: 10.1006/bbrc.2000.4215. [DOI] [PubMed] [Google Scholar]
- 17.Mantel C, Luo Z, Canfield J, Braun S, Deng C, Broxmeyer HE. Involvement of p21cip-1 and p27kip-1 in the molecular mechanisms of steel factor-induced proliferative synergy in vitro and of p21cip-1 in the maintenance of stem/progenitor cells in vivo. Blood. 1996;88:3710–3719. [PubMed] [Google Scholar]
- 18.Hoyt MA, Totis L, Roberts BT. S cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 19.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 20.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle checkpoint disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 21.Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signaling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 22.Page AM, Hieter P. The anaphase-promoting complex: new subunits and regulators. Annu Rev Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 24.Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 25.Wassmann K, Benezra R. Mitotic checkpoints: from yeast to cancer. Curr Opin Genet Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 26.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:636–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 27.Michel LS, Liberal V, Chatterjee A, et al. Mad2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 28.Jin DY, Spencer F, Jeang KT. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein Mad1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. Mad2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci U S A. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang G, Yu H, Kirschner MW. The checkpoint protein Mad2 and the mitotic regulator cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang LH, Lau LF, Smith DL, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:999–1000. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 32.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill TJ, Zhu Y, Gustafson TA. Interaction of Mad2 with the carboxyl terminus of the insulin receptor but not with the IGFIR. J Biol Chem. 1997;272:10035–10040. doi: 10.1074/jbc.272.15.10035. [DOI] [PubMed] [Google Scholar]
- 34.Poelzl G, Kasai Y, Mochizuki N, Shaul PW, Brown M, Mendelsohn ME. Specific association of estrogen receptor beta with the cell cycle spindle assembly checkpoint protein, MAD2. Proc Natl Acad Sci U S A. 2000;97:2836–2839. doi: 10.1073/pnas.050580997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson KK, Schlöndorff J, Blobel CP. Evidence for an interaction of the metalloprotease-disintegrin tumor necrosis factor α convertase (TACE) with mitotic arrest deficient 2 (Mad2), and of the metalloprotease-disintegrin MDC9 with a novel Mad2-related protein, Mad2beta. Biochem J. 1999;343:673–680. [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda M, Dohmae N, Takio K, Arai K, Watanabe S. Cell cycle-dependent interaction of Mad2 with conserved box1/2 region of human granulocyte-macrophage colony-stimulating factor receptor common βc. J Biol Chem. 2001;276:41803–41809. doi: 10.1074/jbc.M101488200. [DOI] [PubMed] [Google Scholar]
- 37.Cooper S, Broxmeyer HE. Clonogenic methods in vitro for the enumeration of granulocyte-macrophage progenitor cells (CFU-GM) in human bone marrow and mouse bone marrow and spleen. J Tissue Cult Methods. 1991;13:77–82. [Google Scholar]
- 38.Broxmeyer HE, Cooper S, Lasky LA, de Sauvage F. Identification of a massive reserve of hematopoietic progenitors in mice. Stem Cells Dev. 2005;14:105–110. doi: 10.1089/scd.2005.14.105. [DOI] [PubMed] [Google Scholar]
- 39.Cooper S, Mantel C, Broxmeyer HE. Myelosuppressive effects in vivo with very low dosages of monomeric recombinant murine macrophage inflammatory protein-1α. Exp Hematol. 1994;22:186–193. [PubMed] [Google Scholar]
- 40.Pines J. Localization of cell cycle regulations by immunofluorescence. Methods Enzymol. 1997;283:99–113. doi: 10.1016/s0076-6879(97)83010-8. [DOI] [PubMed] [Google Scholar]
- 41.Huang HM, Huang CJ, Yen JJ. Mcl-1 is a common target of stem cell factor and interleukin-5 for apoptosis prevention activity via MEK/MAPK and PI-3K/Akt pathways. Blood. 2000;96:1764–1771. [PubMed] [Google Scholar]
- 42.Blume-Jensen P, Janknecht R, Hunter T. The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- 43.Cheng J, Baumhueter S, Thibodeaux H, et al. Hematopoietic defects in mice lacking the sialomucin CD34. Blood. 1996;87:479–490. [PubMed] [Google Scholar]
- 44.Gurney AL, Carver-Moore K, de Sauvage FJ, Moore MW. Thrombocytopenia in c-mpl-deficient mice. Science. 1994;265:1445–1447. doi: 10.1126/science.8073287. [DOI] [PubMed] [Google Scholar]
- 45.Carver-Moore K, Broxmeyer HE, Luoh SM, et al. Low levels of erythroid and myeloid progenitors in TPO and c-mpl deficient mice. Blood. 1996;88:803–808. [PubMed] [Google Scholar]
- 46.Broxmeyer HE. Regulation of myelopoiesis as assessed by gene deletion and gene transduction. In: Zon LI, editor. Hematopoiesis: A Developmental Approach. New York, NY: Oxford University Press; 2001. pp. 247–257. [Google Scholar]
- 47.Wang Q, Liu T, Fang Y, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 48.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VVVS, Benezra R. Complete loss of the tumor suppressor Mad2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Y, Yin F, Liu C, et al. Depression of Mad2 inhibits apoptosis of gastric cancer cells by upregulating Bcl-2 and interfering mitochondrion pathway. Biochem Biophys Res Commun. 2006;345:1092–1098. doi: 10.1016/j.bbrc.2006.04.172. [DOI] [PubMed] [Google Scholar]