Abstract
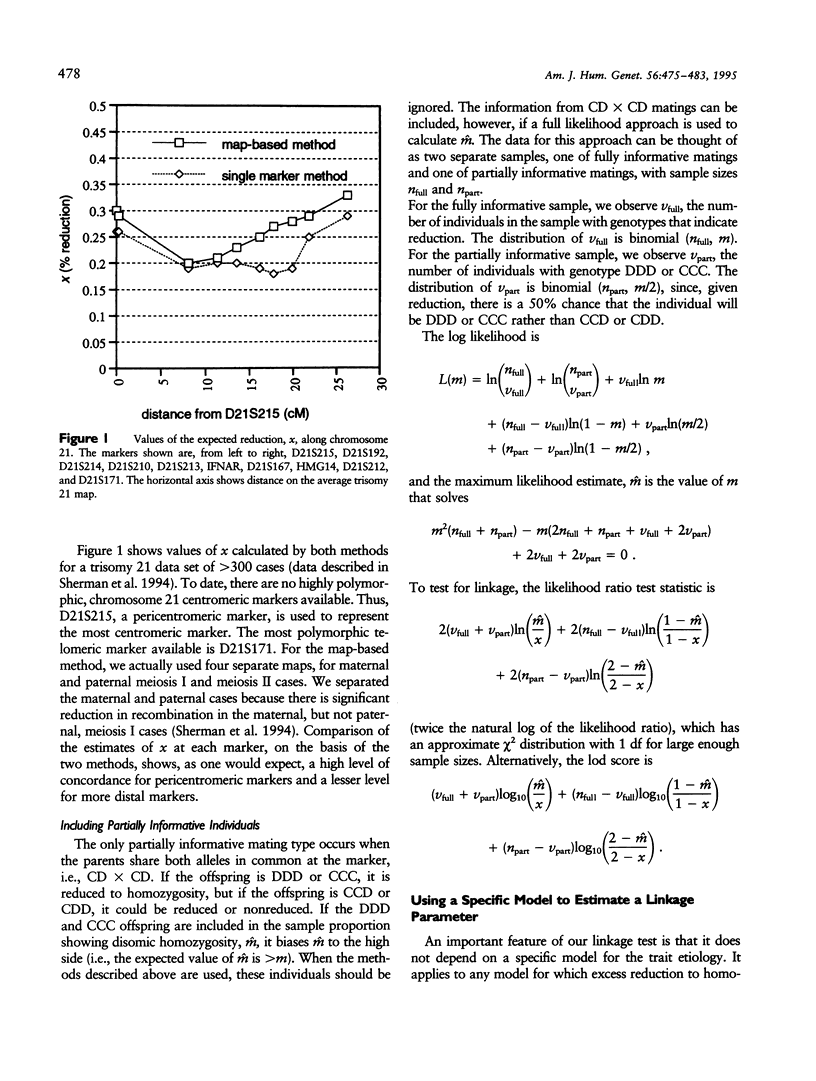
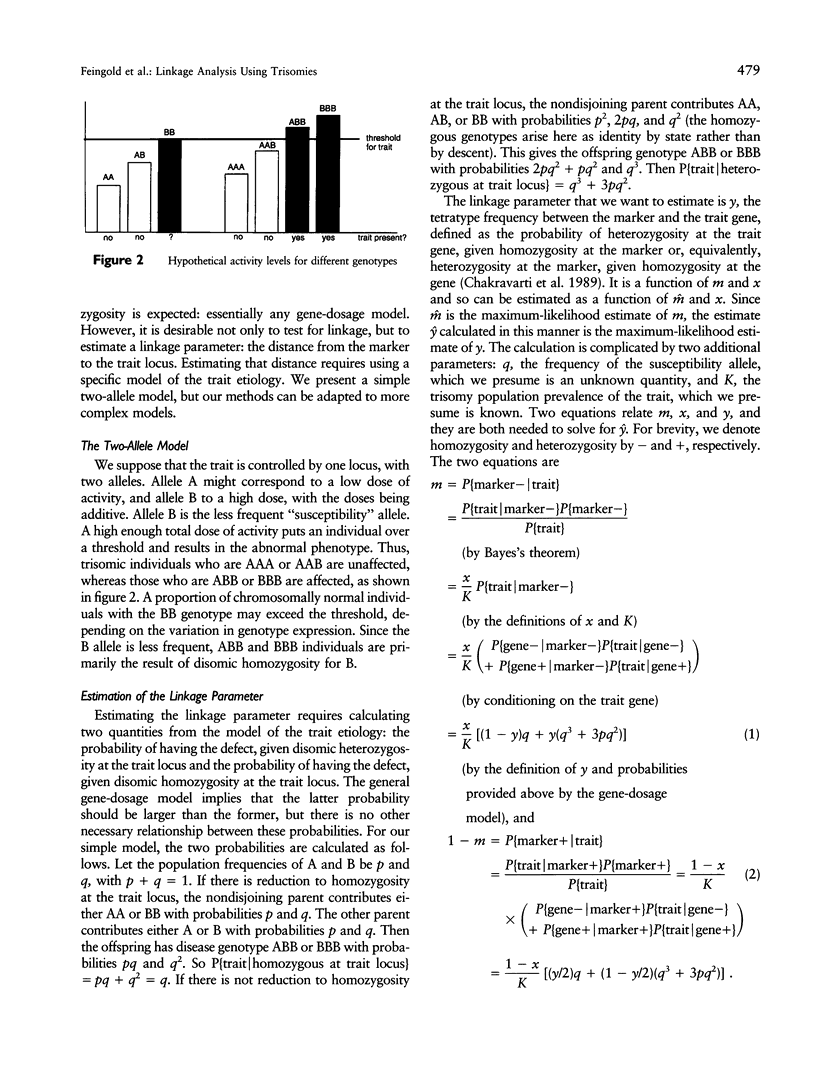
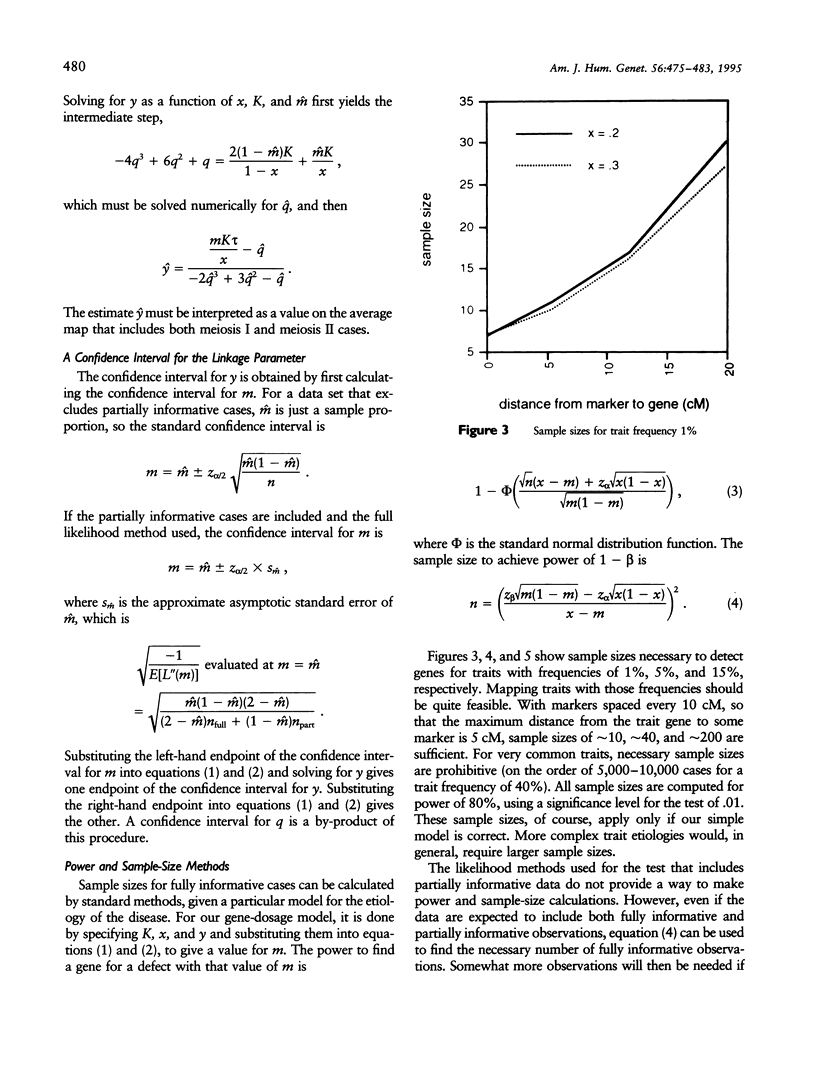
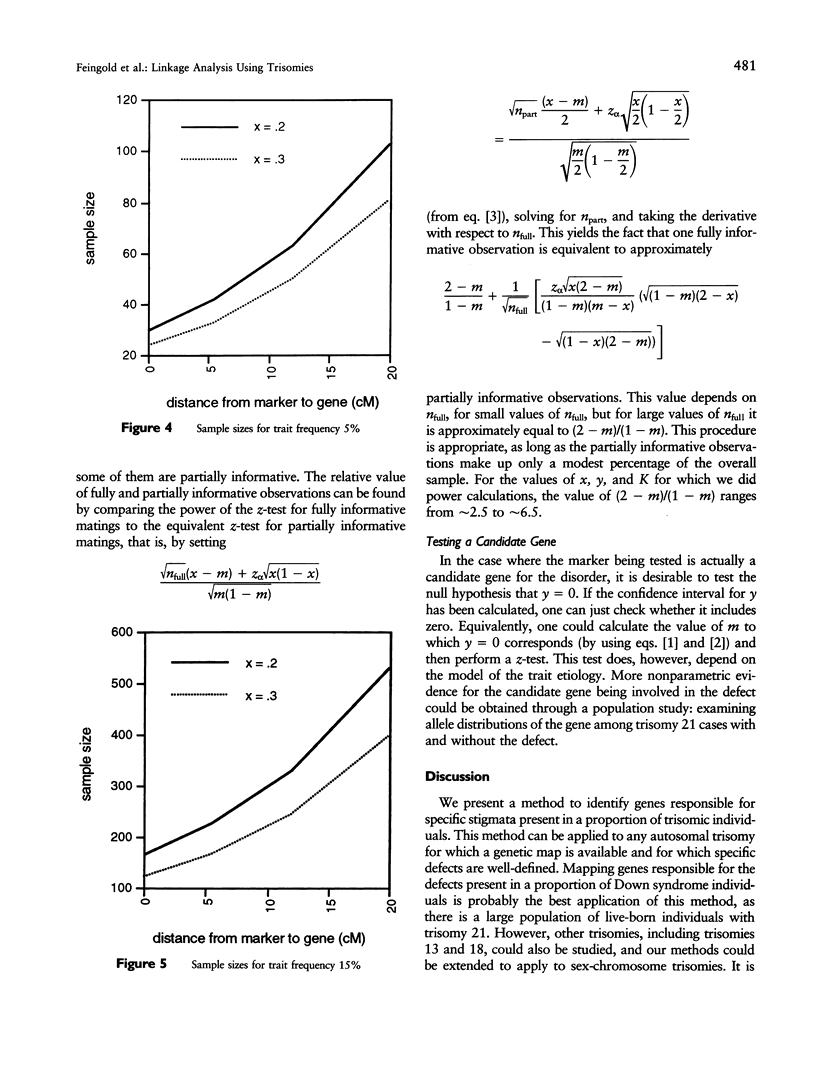
Certain genetic disorders are rare in the general population, but more common in individuals with specific trisomies. Examples of this include leukemia and duodenal atresia in trisomy 21. This paper presents a linkage analysis method for using trisomic individuals to map genes for such traits. It is based on a very general gene-specific dosage model that posits that the trait is caused by specific effects of different alleles at one or a few loci and that duplicate copies of "susceptibility" alleles inherited from the nondisjoining parent give increased likelihood of having the trait. Our mapping method is similar to identity-by-descent-based mapping methods using affected relative pairs and also to methods for mapping recessive traits using inbred individuals by looking for markers with greater than expected homozygosity by descent. In the trisomy case, one would take trisomic individuals and look for markers with greater than expected homozygosity in the chromosomes inherited from the nondisjoining parent. We present statistical methods for performing such a linkage analysis, including a test for linkage to a marker, a method for estimating the distance from the marker to the trait gene, a confidence interval for that distance, and methods for computing power and sample sizes. We also resolve some practical issues involved in implementing the methods, including how to use partially informative markers and how to test candidate genes.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chakravarti A., Majumder P. P., Slaugenhaupt S. A., Deka R., Warren A. C., Surti U., Ferrell R. E., Antonarakis S. E. Gene-centromere mapping and the study of non-disjunction in autosomal trisomies and ovarian teratomas. Prog Clin Biol Res. 1989;311:45–79. [PubMed] [Google Scholar]
- Chakravarti A., Slaugenhaupt S. A. Methods for studying recombination on chromosomes that undergo nondisjunction. Genomics. 1987 Sep;1(1):35–42. doi: 10.1016/0888-7543(87)90102-9. [DOI] [PubMed] [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6(2):137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- Epstein C. J., Korenberg J. R., Annerén G., Antonarakis S. E., Aymé S., Courchesne E., Epstein L. B., Fowler A., Groner Y., Huret J. L. Protocols to establish genotype-phenotype correlations in Down syndrome. Am J Hum Genet. 1991 Jul;49(1):207–235. [PMC free article] [PubMed] [Google Scholar]
- Feingold E., Brown P. O., Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993 Jul;53(1):234–251. [PMC free article] [PubMed] [Google Scholar]
- Gene dosage effects in trisomy: comment on a recent article by B. L. Shapiro. Am J Med Genet. 1983 Dec;16(4):635–639. doi: 10.1002/ajmg.1320160421. [DOI] [PubMed] [Google Scholar]
- Korenberg J. R., Bradley C., Disteche C. M. Down syndrome: molecular mapping of the congenital heart disease and duodenal stenosis. Am J Hum Genet. 1992 Feb;50(2):294–302. [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R. Toward a molecular understanding of Down syndrome. Prog Clin Biol Res. 1993;384:87–115. [PubMed] [Google Scholar]
- Lander E. S., Botstein D. Mapping complex genetic traits in humans: new methods using a complete RFLP linkage map. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):49–62. doi: 10.1101/sqb.1986.051.01.007. [DOI] [PubMed] [Google Scholar]
- Morton N. E., Keats B. J., Jacobs P. A., Hassold T., Pettay D., Harvey J., Andrews V. A centromere map of the X chromosome from trisomies of maternal origin. Ann Hum Genet. 1990 Jan;54(Pt 1):39–47. doi: 10.1111/j.1469-1809.1990.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Morton N. E., MacLean C. J. Multilocus recombination frequencies. Genet Res. 1984 Aug;44(1):99–107. doi: 10.1017/s0016672300026276. [DOI] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet. 1990 Feb;46(2):229–241. [PMC free article] [PubMed] [Google Scholar]
- Shahar S., Morton N. E. Origin of teratomas and twins. Hum Genet. 1986 Nov;74(3):215–218. doi: 10.1007/BF00282536. [DOI] [PubMed] [Google Scholar]
- Sherman S. L., Petersen M. B., Freeman S. B., Hersey J., Pettay D., Taft L., Frantzen M., Mikkelsen M., Hassold T. J. Non-disjunction of chromosome 21 in maternal meiosis I: evidence for a maternal age-dependent mechanism involving reduced recombination. Hum Mol Genet. 1994 Sep;3(9):1529–1535. doi: 10.1093/hmg/3.9.1529. [DOI] [PubMed] [Google Scholar]
- Sherman S. L., Takaesu N., Freeman S. B., Grantham M., Phillips C., Blackston R. D., Jacobs P. A., Cockwell A. E., Freeman V., Uchida I. Trisomy 21: association between reduced recombination and nondisjunction. Am J Hum Genet. 1991 Sep;49(3):608–620. [PMC free article] [PubMed] [Google Scholar]
- Suarez B. K., Rice J., Reich T. The generalized sib pair IBD distribution: its use in the detection of linkage. Ann Hum Genet. 1978 Jul;42(1):87–94. doi: 10.1111/j.1469-1809.1978.tb00933.x. [DOI] [PubMed] [Google Scholar]
- Warren A. C., Chakravarti A., Wong C., Slaugenhaupt S. A., Halloran S. L., Watkins P. C., Metaxotou C., Antonarakis S. E. Evidence for reduced recombination on the nondisjoined chromosomes 21 in Down syndrome. Science. 1987 Aug 7;237(4815):652–654. doi: 10.1126/science.2955519. [DOI] [PubMed] [Google Scholar]


