Abstract
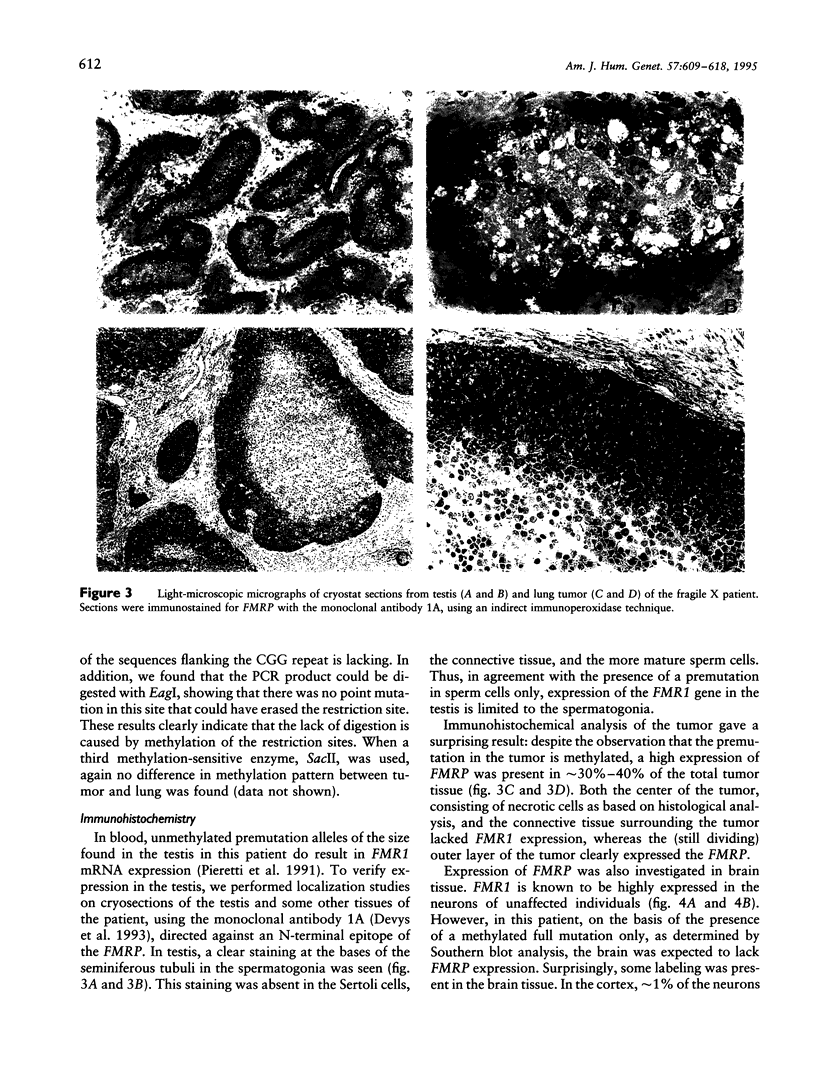
The molecular mechanism of the fragile X syndrome is based on the expansion of an CGG repeat in the 5' UTR of the FMR1 gene in the majority of fragile X patients. This repeat displays instability both between individuals and within an individual. We studied the instability of the CGG repeat and the expression of the FMR1 protein (FMRP) in several different tissues derived from a male fragile X patient. Using Southern blot analysis, only a full mutation is detected in 9 of the 11 tissues tested. The lung tumor contains a methylated premutation of 160 repeats, whereas in the testis, besides the full mutation, a premutation of 60 CGG repeats is detected. Immunohistochemistry of the testis revealed expression of FMR1 in the spermatogonia only, confirming the previous finding that, in the sperm cells of fragile X patients with a full mutation in their blood cells, only a premutation is present. Immunohistochemistry of brain and lung tissue revealed that 1% of the cells are expressing the FMRP. PCR analysis demonstrated the presence of a premutation of 160 repeats in these FMR1-expressing cells. This indicates that the tumor was derived from a lung cell containing a premutation. Remarkably, despite the methylation of the EagI and BssHII sites, FMRP expression is detected in the tumor. Methylation of both restriction sites has thus far resulted in a 100% correlation with the lack of FMR1 expression, but the results found in the tumor suggest that the CpGs in these restriction sites are not essential for regulation of FMR1 expression. This indicates a need for a more accurate study of the exact promoter of FMR1.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abitbol M., Menini C., Delezoide A. L., Rhyner T., Vekemans M., Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993 Jun;4(2):147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Jr, Wilkinson K. D., Reines D., Warren S. T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993 Oct 22;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Bronner C. E., Baker S. M., Morrison P. T., Warren G., Smith L. G., Lescoe M. K., Kane M., Earabino C., Lipford J., Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994 Mar 17;368(6468):258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Brown W. T., Houck G. E., Jr, Jeziorowska A., Levinson F. N., Ding X., Dobkin C., Zhong N., Henderson J., Brooks S. S., Jenkins E. C. Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA. 1993 Oct 6;270(13):1569–1575. [PubMed] [Google Scholar]
- Brown W. T., Jenkins E. C. The fragile X syndrome. Mol Genet Med. 1992;2:39–66. doi: 10.1016/b978-0-12-462002-5.50007-8. [DOI] [PubMed] [Google Scholar]
- Bächner D., Manca A., Steinbach P., Wöhrle D., Just W., Vogel W., Hameister H., Poustka A. Enhanced expression of the murine FMR1 gene during germ cell proliferation suggests a special function in both the male and the female gonad. Hum Mol Genet. 1993 Dec;2(12):2043–2050. doi: 10.1093/hmg/2.12.2043. [DOI] [PubMed] [Google Scholar]
- Chong S. S., Eichler E. E., Nelson D. L., Hughes M. R. Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet. 1994 Jul 15;51(4):522–526. doi: 10.1002/ajmg.1320510447. [DOI] [PubMed] [Google Scholar]
- Devys D., Biancalana V., Rousseau F., Boué J., Mandel J. L., Oberlé I. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):208–216. doi: 10.1002/ajmg.1320430134. [DOI] [PubMed] [Google Scholar]
- Devys D., Lutz Y., Rouyer N., Bellocq J. P., Mandel J. L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993 Aug;4(4):335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hagerman R. J., Hull C. E., Safanda J. F., Carpenter I., Staley L. W., O'Connor R. A., Seydel C., Mazzocco M. M., Snow K., Thibodeau S. N. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994 Jul 15;51(4):298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Canfield T. K., Lamb M. M., Gartler S. M., Laird C. D. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993 Jul 2;73(7):1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M., Scott C. R., Chen S. H., Laird C. D. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992 Nov;1(8):571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- Hinds H. L., Ashley C. T., Sutcliffe J. S., Nelson D. L., Warren S. T., Housman D. E., Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993 Jan;3(1):36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Grewal P. K., Davies K. E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994 Sep;3(9):1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Hornstra I. K., Nelson D. L., Warren S. T., Yang T. P. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet. 1993 Oct;2(10):1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- Hwu W. L., Lee Y. M., Lee S. C., Wang T. R. In vitro DNA methylation inhibits FMR-1 promoter. Biochem Biophys Res Commun. 1993 May 28;193(1):324–329. doi: 10.1006/bbrc.1993.1627. [DOI] [PubMed] [Google Scholar]
- Knight S. J., Flannery A. V., Hirst M. C., Campbell L., Christodoulou Z., Phelps S. R., Pointon J., Middleton-Price H. R., Barnicoat A., Pembrey M. E. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993 Jul 16;74(1):127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- Kunst C. B., Warren S. T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994 Jun 17;77(6):853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Laird P. W., Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3(Spec No):1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- Leach F. S., Nicolaides N. C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomäki P., Sistonen P., Aaltonen L. A., Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993 Dec 17;75(6):1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Li C. Y., Ziesmer S. C., Lazcano-Villareal O. Use of azide and hydrogen peroxide as an inhibitor for endogenous peroxidase in the immunoperoxidase method. J Histochem Cytochem. 1987 Dec;35(12):1457–1460. doi: 10.1177/35.12.2824601. [DOI] [PubMed] [Google Scholar]
- Meijer H., de Graaff E., Merckx D. M., Jongbloed R. J., de Die-Smulders C. E., Engelen J. J., Fryns J. P., Curfs P. M., Oostra B. A. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994 Apr;3(4):615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Nicolaides N. C., Papadopoulos N., Liu B., Wei Y. F., Carter K. C., Ruben S. M., Rosen C. A., Haseltine W. A., Fleischmann R. D., Fraser C. M. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994 Sep 1;371(6492):75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Nolin S. L., Glicksman A., Houck G. E., Jr, Brown W. T., Dobkin C. S. Mosaicism in fragile X affected males. Am J Med Genet. 1994 Jul 15;51(4):509–512. doi: 10.1002/ajmg.1320510444. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Oostra B. A., Jacky P. B., Brown W. T., Rousseau F. Guidelines for the diagnosis of fragile X syndrome. National Fragile X Foundation. J Med Genet. 1993 May;30(5):410–413. doi: 10.1136/jmg.30.5.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Reiss A. L., Kazazian H. H., Jr, Krebs C. M., McAughan A., Boehm C. D., Abrams M. T., Nelson D. L. Frequency and stability of the fragile X premutation. Hum Mol Genet. 1994 Mar;3(3):393–398. doi: 10.1093/hmg/3.3.393. [DOI] [PubMed] [Google Scholar]
- Reyniers E., Vits L., De Boulle K., Van Roy B., Van Velzen D., de Graaff E., Verkerk A. J., Jorens H. Z., Darby J. K., Oostra B. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat Genet. 1993 Jun;4(2):143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Kozman H., Kremer E., Lynch M., Pritchard M., Yu S., Mulley J., Sutherland G. R. Fragile X syndrome: genetic localisation by linkage mapping of two microsatellite repeats FRAXAC1 and FRAXAC2 which immediately flank the fragile site. J Med Genet. 1991 Dec;28(12):818–823. doi: 10.1136/jmg.28.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Biancalana V., Blumenfeld S., Kretz C., Boué J., Tommerup N., Van Der Hagen C., DeLozier-Blanchet C., Croquette M. F. Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med. 1991 Dec 12;325(24):1673–1681. doi: 10.1056/NEJM199112123252401. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Oberlé I., Mandel J. L. Selection in blood cells from female carriers of the fragile X syndrome: inverse correlation between age and proportion of active X chromosomes carrying the full mutation. J Med Genet. 1991 Dec;28(12):830–836. doi: 10.1136/jmg.28.12.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Tarleton J., MacPherson J., Malmgren H., Dahl N., Barnicoat A., Mathew C., Mornet E., Tejada I. A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am J Hum Genet. 1994 Aug;55(2):225–237. [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994 Apr 8;77(1):33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C., Nussbaum R. L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993 Jul 30;74(2):291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Snow K., Doud L. K., Hagerman R., Pergolizzi R. G., Erster S. H., Thibodeau S. N. Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet. 1993 Dec;53(6):1217–1228. [PMC free article] [PubMed] [Google Scholar]
- Snow K., Tester D. J., Kruckeberg K. E., Schaid D. J., Thibodeau S. N. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994 Sep;3(9):1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Verheij C., Bakker C. E., de Graaff E., Keulemans J., Willemsen R., Verkerk A. J., Galjaard H., Reuser A. J., Hoogeveen A. T., Oostra B. A. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993 Jun 24;363(6431):722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Hennig I., Vogel W., Steinbach P. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet. 1993 Jun;4(2):140–142. doi: 10.1038/ng0693-140. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Hirst M. C., Kennerknecht I., Davies K. E., Steinbach P. Genotype mosaicism in fragile X fetal tissues. Hum Genet. 1992 Apr;89(1):114–116. doi: 10.1007/BF00207057. [DOI] [PubMed] [Google Scholar]
- Wölfl S., Schräder M., Wittig B. Lack of correlation between DNA methylation and transcriptional inactivation: the chicken lysozyme gene. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):271–275. doi: 10.1073/pnas.88.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J. C., Warren S. T., Schlessinger D. Fragile X genotype characterized by an unstable region of DNA. Science. 1991 May 24;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- Zavodny P. J., Petro M. E., Kumar C. C., Dailey S. H., Lonial H. K., Narula S. K., Leibowitz P. J. The nucleotide sequence of chicken smooth muscle myosin light chain two. Nucleic Acids Res. 1988 Feb 11;16(3):1214–1214. doi: 10.1093/nar/16.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaff E., Rouillard P., Willems P. J., Smits A. P., Rousseau F., Oostra B. A. Hotspot for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum Mol Genet. 1995 Jan;4(1):45–49. doi: 10.1093/hmg/4.1.45. [DOI] [PubMed] [Google Scholar]
- de Vries B. B., Fryns J. P., Butler M. G., Canziani F., Wesby-van Swaay E., van Hemel J. O., Oostra B. A., Halley D. J., Niermeijer M. F. Clinical and molecular studies in fragile X patients with a Prader-Willi-like phenotype. J Med Genet. 1993 Sep;30(9):761–766. doi: 10.1136/jmg.30.9.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ouweland A. M., de Vries B. B., Bakker P. L., Deelen W. H., de Graaff E., van Hemel J. O., Oostra B. A., Niermeijer M. F., Halley D. J. DNA diagnosis of the fragile X syndrome in a series of 236 mentally retarded subjects and evidence for a reversal of mutation in the FMR-1 gene. Am J Med Genet. 1994 Jul 15;51(4):482–485. doi: 10.1002/ajmg.1320510437. [DOI] [PubMed] [Google Scholar]






