Abstract
Amyloid deposits are associated with several neurodegenerative diseases, including Alzheimer’s disease and the prion diseases. The amyloid fibrils isolated from these different diseases share similar structural features. However, the protein sequences that assemble into these fibrils differ substantially from one disease to another. To probe the relationship between amino acid sequence and the propensity to form amyloid, we studied a combinatorial library of sequences designed de novo. All sequences in the library were designed to share an identical pattern of alternating polar and nonpolar residues, but the precise identities of these side chains were not constrained and were varied combinatorially. The resulting proteins self-assemble into large oligomers visible by electron microscopy as amyloid-like fibrils. Like natural amyloid, the de novo fibrils are composed of β-sheet secondary structure and bind the diagnostic dye, Congo red. Thus, binary patterning of polar and nonpolar residues arranged in alternating periodicity can direct protein sequences to form fibrils resembling amyloid. The model amyloid fibrils assemble and disassemble reversibly, providing a tractable system for both basic studies into the mechanisms of fibril assembly and the development of molecular therapies that interfere with this assembly.
The deposition of insoluble amyloid plaque is associated with several neurodegenerative diseases, including Alzheimer’s disease, senile systemic amyloidosis, and spongiform encephalopathies (e.g., “mad cow” disease) (1–8). The primary component of amyloid differs from one disease to another: in Alzheimer’s, it is the 42 residue Aβ peptide; in senile systemic amyloidosis, it is the transthyretin protein; and in spongiform encephalopathy, it is the prion protein (2–5). Despite substantial differences in both sequence and length (40–250 residues), these diverse proteins assemble into amyloid structures that are remarkably similar to one another. They all form fibrils composed of β-strands running perpendicular to the fibril axis.
What molecular determinants predispose a protein to form amyloid? The extreme dissimilarity between the various amyloidogenic sequences, coupled with the unavailability of simple model systems, has limited our understanding of the sequence determinants of amyloidogenesis. However, the similarity among the structures of the different amyloids suggests they may share unifying structural determinants. Here we describe an assessment of these determinants through the study of a combinatorial library of protein sequences designed de novo. Because of their combinatorial origins, the sequences in the library differ significantly from one another. Yet, because the combinatorial diversity was constrained by elements of rational design, all sequences in the library share essential global features. Structural and biophysical characterization of the purified de novo proteins suggests that these global features can suffice to predispose amino acid sequences to form amyloid fibrils.
MATERIALS AND METHODS
Construction of a Combinatorial Library of Synthetic Genes.
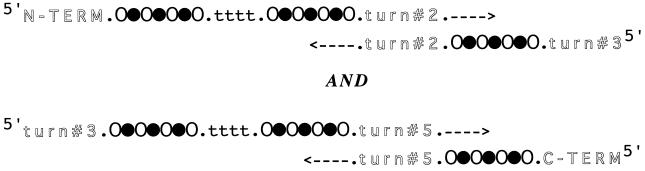
The library of genes was constructed from synthetic oligonucleotides containing partially degenerate sequences (see Fig. 2). Oligonucleotides were designed as follows:
Figure 2.
Construction of a combinatorial library of genes. Each single-stranded DNA oligonucleotide encodes degenerate sequences in the β-strands and in turns 1 and 4. Degenerate codons encoding polar residues are shown as (○), those encoding nonpolar residues are shown as (●), and those encoding turns are shown as (t). Fixed sequences occur in turns 2, 3, and 5 and also in the gene termini. These are shown in shadow font. The fixed sequences at the 3′ ends of each oligonucleotide allow annealing and priming for synthesis of complementary DNA. Elongation of complementary DNA by polymerase is indicated by arrows. The entire procedure was carried out twice: once to construct the first three β-strands (and associated turns) and again to construct the last three β-strands (and associated turns). The two libraries of double-stranded pieces were then used as PCR templates to construct a library of full length genes. The nondegenerate sequence of turn no. 3 served as an internal annealing sequence, and oligonucleotides complementary to the N- and C- terminal sequences served as external primers.
1–8 is sense strand DNA encoding from the N terminus of the proteins through turn no. 2; 2–6 is antisense strand DNA encoding from turn no. 2 through turn no. 3; 3–8 is sense strand DNA encoding from turn no. 3 through turn no. 5; and 4–6 is antisense strand DNA encoding from turn no. 5 through the C terminus.
Oligonucleotides 1–8 and 2–6 were annealed via complementary 3′ ends, and double-stranded DNA was synthesized by using Klenow enzyme. The double-stranded product encodes from the N terminus through turn no. 3. A similar set of reactions with oligonucleotides 3–8 and 4–6 yielded double-stranded DNA encoding from turn no. 3 through the C terminus. These two double-stranded products possess fixed DNA sequences encoding the region surrounding turn no. 3. To produce a library of full-length genes, these two half genes were mixed with two biotinylated external primers and amplified by PCR. The resulting full-length product was cut with NcoI and NheI, and the biotinylated ends were removed with streptavidin agarose (Pierce). The restricted DNA was then ligated into plasmid pET-25bM (9), a derivative of pET-25b (Novagen) in which the herpes simplex virus (HSV) epitope tag is out of frame. The ligation mixture was then used to transform Escherichia coli strain BL21/DE3 (10). Transformants were screened for correct expression of the HSV epitope tag by using the Colony Finder immunoscreening kit (Novagen). Sequences of the oligonucleotides are shown below, with the following abbreviations: B = G, C, or T; W = T or A; H = C, A, or T; K = G or T; V = C, G, or A; M = A or C; D = G, A, or T; R = A or G; S = G or C; Y = C or T.
1–8, 5′-GCG ACG CTC TCC ATG GAT TAT VAS NTC VAS NTC VAS RRT RRT RRT RRT VAS NTC VAS NTC VAS NTC VAC GAT TCT GGT GGT GA-3′; 2–6, 5′-TGG ACA CGG CCA CCC GGG CCG CGG ATC TBA ANW TBA ANW TCA CCA CCA GAA TCGT-3′; 3–8, 5′-AGA TCC GCG GCC CGG GTG GCC GTG TCC ASN TCV ASN TCV ASR RTR RTR RTR RTV ASN TCV ASN TCV ASN TCV ACA ACG ACG GCG GCG A-3′; 4–6, 5′-GCG CAG ACC TCG AGC ATW TBA ANW TBA ANW TCG CCG CCG TCG TTG T-3′; primer P-1, 5′-GCG ACG CTC TCC ATG GAT TAT-3′; primer-NHE, 5′-CCC AAG CTT GCT AGC GGC GCC GAC CTC GAG CAT-3′.
Expression, Purification, and Size-Exclusion Chromatography.
Genes encoding the de novo proteins were subcloned into plasmid pET-3d for high-level expression without signal sequences or C-terminal tags (9). Plasmids were transformed into E. coli strain BL2/DE3 (10). Cultures were grown to an OD600 of 0.6≈0.7 and induced with isopropyl-d-thiogalactoside (final concentration, 0.5 mM). Cells were harvested by centrifugation and resuspended in 50 mM Tris⋅HCl, pH 8.0/25% sucrose/1 mM EDTA. Lysozyme was added to a final concentration of 2 mg/ml, and cells were lysed by incubation for 30 min. MgCl2 and MnCl2 were added to 10 mM and 1 mM, respectively, and DNaseI was added to a final concentration of 20 μg/ml. Inclusion bodies were prepared according to the protocol of Nagi and Thogersen (11). The washed inclusion bodies were solubilized in 10 M urea at pH 8.0, and proteins were purified on a Poros 20HQ anion-exchange column (PerSeptive Biosystems, Framingham, MA) run under denaturing conditions (buffer A, 10 mM sodium phosphate/6 M urea, pH 8.0; buffer B, buffer A plus 1 M NaCl.) After purification, proteins were folded by dialysis into native buffer—typically 10 mM sodium phosphate/100 mM NaCl, pH 7.8. Size-exclusion chromatography was performed by using an Ultraspherogel SEC3000 column (Beckman Coulter) in 10 mM sodium phosphate/100 mM NaCl, pH 7.8.
Electron Microscopy.
Transmission electron microscopy was performed on a JEOL 100C TEM operating at 100 kV. Protein concentration was 20 μM in 25 mM Tris⋅HCl (pH 7.8). Formvar/carbon-coated grids were floated on a drop of the protein sample for 5 min. The grids were blotted by using filter paper and then stained for 30 sec with freshly made 1% uranyl acetate.
CD and Thermal Disassembly.
CD spectra were measured in 10 mM sodium phosphate/100 mM NaCl (pH 7.8) at 4°C in a 1-mm pathlength cuvette by using an Aviv 62 DS spectropolarimeter (Aviv Associates, Lakewood, NJ). Protein concentrations were determined by quantitative amino acid analysis (no. 4, 75 μM; no. 17, 52 μM; no. 23, 86 μM). Thermally induced disassembly was followed by monitoring the loss of β-structure as measured by ellipticity at 217 nm. Data points were collected every 2°C after equilibration for 2 min at each temperature. More than 90% of the initial signal is restored on cooling back to room temperature.
Congo Red Binding.
The method of Klunk et al. (12) was used with slight modifications. Briefly, a 40 μM stock solution of CR (Aldrich) was prepared in 5 mM sodium phosphate/150 mM NaCl (pH 7.4). Twenty to forty microliters of concentrated protein sample (250–500 μM) was added to a Congo red solution to bring the final volume to 400 μl. Final concentration of protein was approximately 25 μM. The suspension was mixed and then incubated at room temperature for 30 min before measurements. [Controls: bovine insulin fibrils (13). The fibrillar form of the Alzheimer’s peptide, Aβ (1–40), yielded a spectrum similar to the insulin control. Aβ (1–40) was a gift from J. D. Harper and P. T. Lansbury, Jr. (Harvard University Medical School) (14).]
RESULTS
Design of Sequences.
Polar/nonpolar patterning of β-strands. Natural amyloid fibrils are composed of β-strands running perpendicular to the long axis of the fibril (“cross-β” structure) (7, 8, 15). Therefore, for model proteins to accurately mimic amyloid, they must be designed to form multimeric β-strands. Previous research demonstrated that for peptides destined to assemble into multimers, secondary structure is dictated by the periodicity of polar and nonpolar residues in the linear sequence (16). When the periodicity of the linear sequence matches the repeat pattern of a particular secondary structure, the peptide will form that structure—irrespective of the intrinsic propensities of the individual amino acids (16). For example, a sequence of polar (○) and nonpolar (●) amino acids with the pattern ●●○○●●○○●○○●●○○● has a nonpolar residue every three or four positions, which matches the α-helical repeat of 3.6 residues/turn. Hence, peptides with this sequence pattern form amphiphilic α-helices that self-assemble into bundles and bury the nonpolar faces of the individual helices in the core of the bundle (16). Conversely, sequences with the alternating pattern ●○●○●○●○● have a periodicity of 2. This matches the structural repeat of β-strands with successive side chains pointing up-down-up-down, etc. Such sequences form amphiphilic β-strands that bury their nonpolar faces by aggregating into large β-sheet structures (16, 17). These earlier findings suggest that proteins designed to contain segments of alternating polar/nonpolar periodicity might be predisposed to oligomerize into cross-β structures.
We designed a library of de novo proteins such that all sequences were constrained to include this β-type pattern of alternating polar and nonpolar residues (9, 18). Combinatorial diversity was incorporated into the library by allowing polar residues to be His, Lys, Asn, Asp, Gln or Glu, and nonpolar residues to be Leu, Ile, Val or Phe. These combinatorial sets of amino acids were encoded by libraries of synthetic genes in which polar residues are encoded by the degenerate DNA codon XAX, and nonpolar residues by the degenerate DNA codon XTX (where X represent a combinatorial mixture of bases).
Length of β-strands.
Natural amyloid fibrils are typically composed of two or more protofilaments, each having a diameter of approximately 30 Å (7, 14). This diameter presumably includes the cross β-strand itself and the turns connecting successive β-strands. Thus the β-strands (not including turns) presumably span 20 Å to 25 Å. Because β-structure has a translation of 3.2 Å per residue for parallel strands and 3.4 Å per residue for antiparallel strands (19), approximately seven residues are required to span this distance.
Relative abundance of polar and nonpolar residues.
β-Strands composed of seven alternating polar and nonpolar residues could contain either an excess of polar or an excess of nonpolar residues. Both options were considered (9). However, earlier work with model peptides showed that alternating sequences with an excess of nonpolar residues form intractable gels (17). Therefore the ○●○●○● ○ pattern was chosen, with four polar (○) and three nonpolar (●) residues per β-strand (9).
Parallel vs. antiparallel β-strands.
Structural studies of amyloid composed of peptides derived from the Alzheimer’s Aβ sequence have found evidence for both parallel and antiparallel β-structure (7, 20). In our designed library, antiparallel β-strands were chosen (9) because they can be linked by relatively short turns or loops. Parallel strands were avoided because long connections between strands are difficult to design.
Topology: Up-down vs. Greek key.
Antiparallel β-strands can be hydrogen bonded either to β-strands adjacent in the primary sequence (up-down topology) or to β-strands distant in the primary sequence (Greek key topology) (21). For any particular amino acid sequence, the choice between these pairings is influenced by the side chains on the respective strands (22). Because different amyloid proteins have different sequences, natural amyloid structures probably include contributions from both topologies. Likewise, because each member of our combinatorial library of de novo proteins has a different sequence, we expect some will favor up-down topology, whereas others will favor Greek key. The combinatorial underpinnings of the binary code strategy allow many different side chain combinations, and therefore both topologies are possible.
Length of turns.
Because the turns between β-strands must accommodate either the up-down or the Greek key topology, they must be long enough to join strands that are not neighbors in the three-dimensional structure. Hence, they must be longer than two residues. Yet, they must also be short enough to favor turns rather than extensive loops or meanders. Turns of three or five residues were ruled out by considerations of “negative design” (23). We wished not only to favor the desired structure—amphiphilic β-strands separated by chain reversals—but also to disfavor undesired structures, such as long uninterrupted amphiphilic β-strands running the length of a fibril. A sequence with an odd number of turn residues (e.g., ○●○●○●○-t-t-t-○●○●○●○) could maintain alternating periodicity, but a sequence with an even number (e.g., ○●○●○●○-t-t-t-t-○●○●○●○) cannot; the turn residues interrupt the periodicity. Therefore, we designed four-residue turns (9).
Sequences of turns.
As the high-resolution structure of amyloid has not been determined, it is not known what types of turns link successive strands in the cross-β structure. In designing our library, we chose combinatorial turn sequences that allow various types of turns (e.g., I, I′, II, II′, etc.). The combinatorial diversity of the designed turn sequences was constrained to residues known to have high turn potentials at all positions irrespective of position or type. The residues having the highest overall turn potential are Gly, Asn, Asp, Pro, and Ser (24). Of these, we excluded Pro because its potential depends on specific positions in particular turn types. Based on these considerations, turn sequences were combinatorially varied among Gly, Asn, Asp, and Ser. This mixture is encoded by the degenerate DNA codon RRT (“R” represents A or G).
Overall length of sequence.
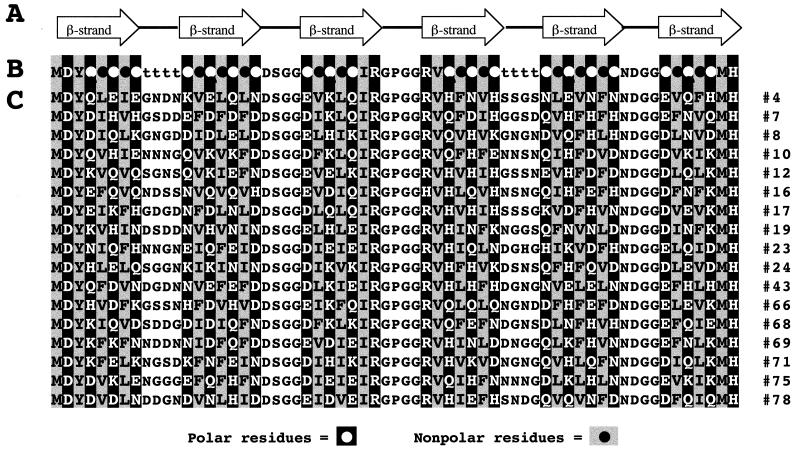
Amyloid structures from different diseases are composed of polypeptides of different lengths (2–8). For example, the dominant sequence in Alzheimer’s plaque is only 42 residues, whereas the sequence of transthyretin in senile systemic amyloidosis is 127 residues. Thus it is clear that sequences of various lengths can assemble into amyloid. We chose to design sequences that would be short enough to facilitate straightforward gene construction but long enough to resemble independent protein domains. The current library was designed to encode sequences encompassing six β-strands (seven residues each) punctuated by five turns (four residues each). A schematic diagram of this pattern is shown in Fig. 1A.
Figure 1.
(A) Schematic illustration of the design of a combinatorial library of de novo proteins containing six β-strands (arrows) punctuated by turns. (B) Designed binary sequence pattern. Alternating pattern in the β-strands is indicated with polar residues (○) as white font in black background and nonpolar residues (●) as black font in gray background. Combinatorial diversity is incorporated at positions marked ○, ●, and t (turn). Fixed residues are incorporated at the termini and in some of the turns. (C) Amino acid sequences (single letter code) of 17 de novo proteins from the combinatorial library.
Construction of a Combinatorial Library of Synthetic Genes.
A diverse library of synthetic genes was constructed such that each member of the library encodes a different amino acid sequence, yet all sequences conform to the designed polar/nonpolar pattern shown in Fig. 1B. The library of genes was constructed by assembling four synthetic oligonucleotides in which most codons were combinatorially degenerate (Fig. 2). These degenerate codons encode amino acid sequences consistent with the alternating polar/nonpolar pattern of the β-strands (○●○●○● ○), as well as combinatorially diverse residues in the first and fourth turns. Within each β-strand, the polar residues (His, Lys, Asn, Asp, Gln, or Glu) were encoded by the degenerate DNA codon VAW, whereas nonpolar residues (Leu, Ile, Val, or Phe) were encoded by NTT (“V” represents G, C, or A; “W” represents A or T; and “N” represents any base). Within the combinatorial turn sequences, the residues Gly, Asn, Asp, or Ser were encoded by the degenerate codon RRT (“R” represents G or A). The sequences of the second, third, and fifth turns were not varied, but were held constant to facilitate annealing and priming for the enzymatic synthesis of complementary strands (Fig. 2).
Synthesis of the full-length genes yielded a highly degenerate library. Yet, because the binary patterning of polar and nonpolar codons is specified explicitly, all DNA sequences are designed to encode the identical polar/nonpolar pattern shown in Fig. 1 A and B).
After assembly of the genes, the library was cloned into a T7 expression vector in which the de novo sequences are preceded by a signal sequence and followed by a 12-residue herpes simplex virus (HSV) epitope tag (9). This construct facilitated rapid screening of the library for genes that assembled in the correct reading frame and thereby expressed the epitope tag. Error sequences causing a shift in frame were thus “weeded out” of the library before further characterization. Initial screening of colonies by using an anti-HSV ELISA demonstrated that ≈40% of the clones expressed the epitope. (The rest presumably contained errors in the synthetic oligonucleotides and were discarded.) Sequences in the correct frame were analyzed by PCR to ensure that the synthetic gene had the expected length, and inserts of the correct length were sequenced. Inferred amino acid sequences of several proteins in the initial collection are shown in Fig. 1C.
Expression and Purification of Proteins.
Genes were subcloned for high-level expression without signal sequences or C-terminal tags. From the sequences shown in Fig. 1C, eight proteins (nos. 4, 17, 19, 23, 66, 68, 69, and 71) were arbitrarily chosen for expression and purification. As expected for sequences designed to form amphiphilic β-strands, all eight expressed as insoluble inclusion bodies. The inclusion bodies were solubilized in 10 M urea, and the de novo proteins were purified by anion exchange chromatography in the presence of urea. Because the proteins are purified in their urea-denatured state, the method is generic and was readily applied to numerous members of the collection—despite their different amino acid sequences. After purification, urea was removed by dialysis into native buffer. The de novo proteins remained in solution. Identities of the purified proteins were confirmed by electrospray mass spectrometry (data not shown).
Biochemical and Structural Characterization of de Novo Amyloid Proteins.
Oligomeric state and hydrodynamic properties. Size and oligomeric state were estimated by size-exclusion chromatography. At moderate protein concentrations (500 μM), the de novo proteins elute in the excluded volume of the sizing column, indicating assembly into an oligomeric state with an apparent molecular mass of >1,000,000 Da (data not shown). For a 63-residue protein, this suggests >140 monomers per oligomer. Sedimentation velocity ultracentrifugation confirmed the presence of high-order oligomers (W.W., R. Fairman, and M.H.H., unpublished results). The large oligomers were dissociated into protomers by lowering the protein concentration 100-fold. For example, when diluted from 500 μM to 5 μM, the oligomers disassemble into a species with an apparent molecular mass of ≈29,000 Da (data not shown). This mass is approximately four times the mass expected from the amino acid sequence (6,925 Da), suggesting that at low concentrations the protein forms tetramers. [The native nonamyloid form of transthyretin is also tetrameric (8)]. These protomers, which form on dilution, are competent to reassemble into the large oligomers and readily do so on reconcentration.
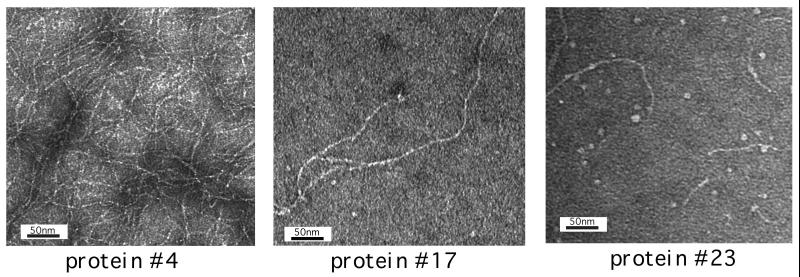
Electron microscopy.
Fig. 3 shows transmission electron microscope images of the large oligomeric form of proteins nos. 4, 17, and 23. These images demonstrate that the de novo sequences self-assemble into fibrils. Like the fibrils observed in the amyloid diseases, these de novo structures are typically straight, not branched. The fibrils have diameters of 25–35 Å, similar to those seen for the protofilaments of natural amyloid (7, 14). Similar images were observed by atomic force microscopy (W.W. and M.H.H., unpublished results).
Figure 3.
Transmission electron microscographs of proteins nos. 4, 17, and 23.
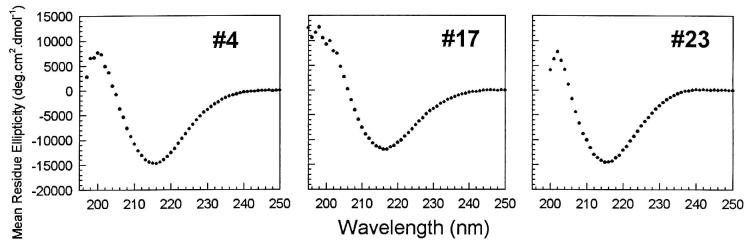
Secondary structure.
Natural amyloid is composed of β-sheet secondary structure. The secondary structures of the designed proteins were determined by CD spectroscopy (Fig. 4). The spectra display a single minimum at ≈217 nm, demonstrating that the de novo fibrils are dominated by β-structure (25). Fitting the experimental spectra to the algorithm of Greenfield and Fasman (25) enables an estimation of 71% β-structure for sequence no. 4, 76% for no. 17, and 78% for no. 23. These estimates are consistent with the design in which 68% of the residues (43 of 63) were designed to be in β-strands (Fig. 1 A and B).
Figure 4.
CD spectra. A minimum at ≈217 nm indicates the presence β-structure (25). The algorithm of Greenfield and Fasman (25) yielded estimates of 71% β-structure for sequence no. 4; 76% for no. 17; and 78% for no. 23.
Binding to Congo red.
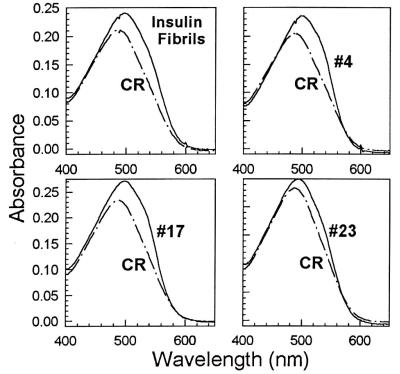
Amyloid structures are routinely identified by their binding to the dye Congo red. Binding appears to be specific for amyloid proteins with a cross-β structure (12). For our designed proteins, Congo red binding was demonstrated spectroscopically as shown in Fig. 5. Visually, this corresponds to a shift from pink to bright red. Binding was observed for the high-order oligomeric form of the de novo proteins, but not for monomeric or tetrameric forms. The results shown in Fig. 5 for the de novo proteins mimic those reported for naturally occurring amyloid fibrils (12).
Figure 5.
Binding to Congo red. Each panel compares the spectrum of Congo red (CR) alone to that of CR in the presence of amyloid protein. [Controls: Bovine insulin fibrils prepared by the method of Burke and Rougvie (13)]. The fibrillar form of the Alzheimer’s peptide, Aβ (1–40) yielded a spectrum similar to the insulin control.
Assembly and disassembly of fibrils.
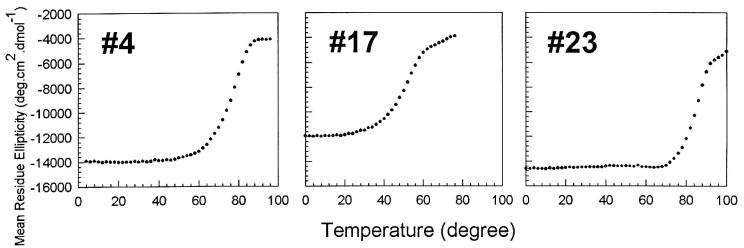
At room temperature (or physiological temperature), the de novo proteins form β-strands that assemble into long fibrils capable of binding Congo red (Figs. 3–5). However, as the temperature is raised, the structure responsible for these properties disassembles (Fig. 6) On cooling back to room temperature, β-structure reforms, and the protein reassembles into large oligomers that elute in the excluded volume of a sizing column. Thus the disassembly and reassembly process is reversible.
Figure 6.
Thermally induced disassembly. Loss of structure was followed by monitoring ellipticity at 217 nm. >90% of the initial signal is restored on cooling back to room temperature.
DISCUSSION
Which features in their design predispose these de novo protein sequences to assemble into amyloid-like fibrils? To address this question, we contrast the sequences described in the current study with those described in our earlier work designing α-helical proteins (26). In both cases, combinatorial libraries were designed by constraining the binary pattern of polar and nonpolar residues. Yet the resulting proteins display dramatically different properties: in the previous work, the sequences formed α-helices that folded intramolecularly into small globular domains (26–28). In contrast, the sequences described in the current work form β-strands and self-assemble intermolecularly into high-order oligomers, which assume fibrillar structures.
What causes these dramatically different structures? The lengths of the sequences are not dramatically different (74 vs. 63), nor are their overall compositions. We propose that the determining difference is the binary patterning itself. In the earlier work, the library was constrained by the pattern ○●○○●●○○●○○●●○, consistent with the periodicity of α-helical structure (26). In contrast, the current library is constrained by the pattern ○●○●○●○, consistent with the periodicity of amphiphilic β-strands.
Amphiphilic β-strands are not stable in isolation: they require pairing to satisfy backbone hydrogen bonding and tertiary structure to bury nonpolar surfaces. Hence, sequences constrained by the alternating β periodicity are inherently predisposed to self-assemble (29). For the designed binary pattern depicted in Fig. 1, one might have expected that pairing of β-strands with the simultaneous burial of hydrophobic surfaces could be accomplished either (i) by intramolecular folding into small globular domains, or (ii) by self assembly into large multimeric structures. Our results demonstrate that for these sequences the second option is favored: we have characterized approximately a dozen proteins from the collection shown in Fig. 1, and at moderate concentrations all of them are capable of assembling into multimeric fibrillar structures. Although it is possible that within our library we will ultimately find some sequences that favor intramolecular folding, it is clear that for sequences incorporating the binary pattern shown in Fig. 1, assembly into amyloid-like fibrils is an option that is easily accessible.
Our findings are consistent with previous observations on several synthetic peptides. In pioneering work, Brack and Orgel demonstrated that poly(Val-Lys), like other polypeptides in which hydrophobic and hydrophilic residues alternate, tends to form β-structures that aggregate into large assemblies (30). Zhang and Rich found that 16-residue peptides in which the sequence alternates between alanine and a charged residue spontaneously assemble into oligomeric structures dominated by β-sheet secondary structure (31). More recently, Lim et al. synthesized a nonnatural 32-residue peptide containing several d-amino acids and found that when their designed peptide was crosslinked by an intermolecular disulfide bond, the resulting dimers assembled into long narrow fibers composed of β-sheet secondary structure (32). Consistent with our model, the sequence of their 32-mer also contained segments of alternating polar/nonpolar patterning.
The amyloidogenic propensity of alternating polar/nonpolar patterns is further supported by a recent survey of the sequences of natural proteins. We analyzed 250,514 sequences comprising 79,708,024 residues in the owl database (33) and asked which binary patterns are favored and which are disfavored. We found that nature disfavors alternating patterns. For example, for seven-residue lengths there are 35 different ways of arranging four polar (○) and three nonpolar (●) residues. Among these, the alternating pattern, ○●○●○●○, ranks 35th. Similar results were found for “windows” shorter or longer than seven (B. Broome and M.H.H., unpublished results). Alternating sequences of polar and nonpolar residues are disfavored. Perhaps evolution has selected against such sequences because they have an inherent propensity to aggregate into deleterious fibrillar plaque.
Finally, we asked whether the sequences of natural amyloid proteins display unusual binary patterns. We found that the polar/nonpolar patterns in such sequences are similar to those of the genomic database as a whole. Although initially surprising, this avoidance of alternating patterns among the natural sequences is exactly what would be expected. The amyloid found in neurodegenerative diseases is a misfolded structure. Amyloidogenic sequences did not evolve to form amyloid plaque. They evolved to fold into globular structures capable of performing functions beneficial to the organism. The transthyretin sequence, for example, was not selected to aggregate as it does in senile systemic amyloidosis; it was selected to fold into soluble tetramers capable of transporting thyroxine hormone. The amyloid structure in diseased tissues is not the correctly folded state for these sequences. It is an off-pathway misfolded structure (6, 8). Hence the alternating patterns that favor fibrilogenesis are not apparent in naturally occurring amyloidogenic sequences.
Herein lies a key advantage of de novo sequences explicitly designed to contain alternating polar/nonpolar patterns. The designed sequences fold into fibrils quite readily. Moreover, the reversible assembly and disassembly of these de novo fibrils suggests their structures are dictated by thermodynamic stability, not kinetic trapping. For these reasons, this library of model amyloid structures may provide important advantages as a tractable system both for basic studies into the mechanisms of fibril assembly and for the development of molecular therapies that interfere with this assembly.
Acknowledgments
We gratefully acknowledge support from the Army Research Office Biological Sciences program, the Culpepper Foundation, the New Jersey Center for Biomaterials, and the National Science Foundation Materials Research Science and Engineering Center (DMR98–09483).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Alzheimer A. Alg Z Psychiatry. 1907;64:146–148. [Google Scholar]
- 2.Lansbury P T., Jr Acc Chem Res. 1996;29:317–321. [Google Scholar]
- 3.Kelly J W. Curr Opin Struct Biol. 1996;6:11–17. doi: 10.1016/s0959-440x(96)80089-3. [DOI] [PubMed] [Google Scholar]
- 4.Kisilevsky R, Fraser P E. Crit Rev Biochem Mol Biol. 1997;32:361–404. doi: 10.3109/10409239709082674. [DOI] [PubMed] [Google Scholar]
- 5.Pruisner S B. Science. 1997;278:245–250. [Google Scholar]
- 6.Wetzel R. Adv Protein Chem. 1997;50:183–242. doi: 10.1016/s0065-3233(08)60322-8. [DOI] [PubMed] [Google Scholar]
- 7.Sunde M, Blake C. Adv Protein Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 8.Kelly J W, Colon W, Lai Z, Lashuel H A, McCulloch J, McCutchen S L, Miroy G J, Peterson S A. Adv Protein Chem. 1997;50:161–181. doi: 10.1016/s0065-3233(08)60321-6. [DOI] [PubMed] [Google Scholar]
- 9.West M W. Ph.D. thesis. Princeton University: Department of Chemistry; 1997. [Google Scholar]
- 10.Studier F W, Rosenberg A H, Dunn J J, Dubendorf J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 11.Nagi K, Thogersen H C. Methods Enzymol. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 12.Klunk W E, Pettegrew J W, Abraham D J. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 13.Burke M J, Rougvie M A. Biochemistry. 1972;11:2435–2439. doi: 10.1021/bi00763a008. [DOI] [PubMed] [Google Scholar]
- 14.Harper J D, Wong S S, Lieber C M, Lansbury P T., Jr Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 15.Lansbury P T, Jr, Costa P R, Griffiths J M, Simon E J, Auger M, Halverson K J, Kocisko D A, Hendsch Z S, Ashburn T T, Spencer R G S, et al. Nat Struct Biol. 1995;2:990–997. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 16.Xiong H, Buckwalter B L, Shieh H M, Hecht M H. Proc Natl Acad Sci USA. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong H. Ph.D. thesis. Princeton University: Department of Chemistry; 1995. [Google Scholar]
- 18.West M W, Hecht M H. Protein Sci. 1995;4:2032–2039. doi: 10.1002/pro.5560041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creighton T E. Proteins: Structures and Molecular Properties. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 20.Benzinger T M, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson J S. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Braun W. Protein Sci. 1999;8:326–342. doi: 10.1110/ps.8.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecht M H, Richardson J S, Richardson D C, Ogden R C. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson E G, Thornton J M. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenfield N, Fasman G D. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 26.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, Ratnaswamy G, Boice J A, Fairman R, McLendon G, Hecht M H. J Am Chem Soc. 1997;119:5302–5306. [Google Scholar]
- 28.Roy S. Ph. D. Dissertation. 1998. Department of Chemistry, Princeton University. [Google Scholar]
- 29.Hecht M H. Proc Natl Acad Sci USA. 1994;91:8729–8730. doi: 10.1073/pnas.91.19.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brack A, Orgel L E. Nature (London) 1975;256:383–387. doi: 10.1038/256383a0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Rich A. Proc Natl Acad Sci USA. 1997;94:23–28. doi: 10.1073/pnas.94.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim A, Saderholm M J, Makhov A M, Kroll M, Yan Y, Perera L, Griffith J D, Erickson B W. Protein Sci. 1998;7:1545–1554. doi: 10.1002/pro.5560070708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleasby A J, Akrigg D, Attwood T K. Nucleic Acids Res. 1994;22:3574–3577. [PMC free article] [PubMed] [Google Scholar]