Abstract
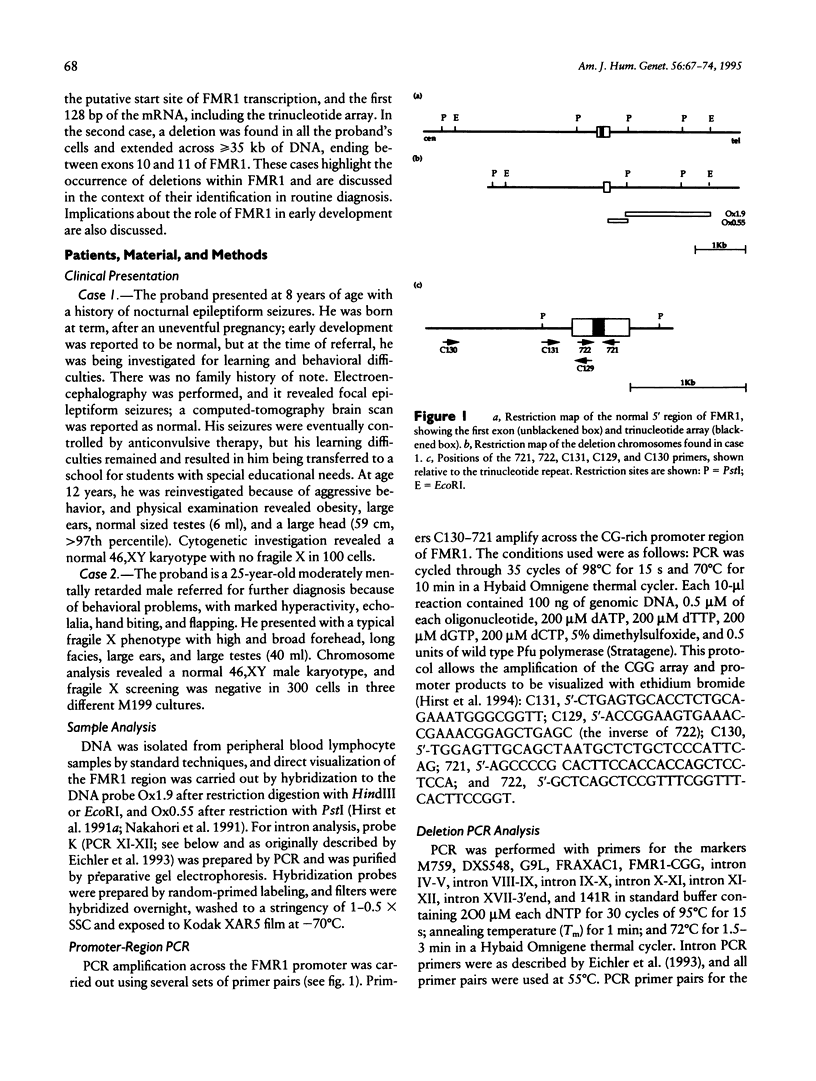
Screening of families clinically ascertained for the fragile X syndrome phenotype revealed two mentally impaired males who were cytogenetically negative for the fragile X chromosome. In both cases, screening for the FMR1 trinucleotide expansion mutation revealed a rearrangement within the FMR1 gene. In the first case, a 660-bp deletion is present in 40% of peripheral lymphocytes. PCR and sequence analysis revealed it to include the CpG island and the CGG trinucleotide repeat, thus removing the FMR1 promoter region and putative mRNA start site. In the second case, PCR analysis demonstrated that a deletion extended from a point proximal to FMR1 to 25 kb into the gene, removing all the region 5' to exon 11. The distal breakpoint was confirmed by Southern blot analysis and localized to a 600-bp region, and FMR1-mRNA analysis in a cell line established from this individual confirmed the lack of a transcript. These deletion patients provide further confirmatory evidence that loss of FMR1 gene expression is indeed responsible for mental retardation. Additionally, these cases highlight the need for the careful examination of the FMR1 gene, even in the absence of cytogenetic expression, particularly when several fragile X-like clinical features are present.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abitbol M., Menini C., Delezoide A. L., Rhyner T., Vekemans M., Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993 Jun;4(2):147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Sutcliffe J. S., Kunst C. B., Leiner H. A., Eichler E. E., Nelson D. L., Warren S. T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat Genet. 1993 Jul;4(3):244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- Bell M. V., Hirst M. C., Nakahori Y., MacKinnon R. N., Roche A., Flint T. J., Jacobs P. A., Tommerup N., Tranebjaerg L., Froster-Iskenius U. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991 Feb 22;64(4):861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- De Boulle K., Verkerk A. J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van den Bos F., de Graaff E., Oostra B. A., Willems P. J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993 Jan;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Devys D., Lutz Y., Rouyer N., Bellocq J. P., Mandel J. L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993 Aug;4(4):335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Richards S., Gibbs R. A., Nelson D. L. Fine structure of the human FMR1 gene. Hum Mol Genet. 1993 Aug;2(8):1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- Gedeon A. K., Baker E., Robinson H., Partington M. W., Gross B., Manca A., Korn B., Poustka A., Yu S., Sutherland G. R. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992 Aug;1(5):341–344. doi: 10.1038/ng0892-341. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Rice P. M., Thompson J. D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem Sci. 1993 Sep;18(9):331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M., Scott C. R., Chen S. H., Laird C. D. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992 Nov;1(8):571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- Hinds H. L., Ashley C. T., Sutcliffe J. S., Nelson D. L., Warren S. T., Housman D. E., Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993 Jan;3(1):36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Grewal P. K., Davies K. E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994 Sep;3(9):1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Knight S. J., Christodoulou Z., Grewal P. K., Fryns J. P., Davies K. E. Origins of the fragile X syndrome mutation. J Med Genet. 1993 Aug;30(8):647–650. doi: 10.1136/jmg.30.8.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M. C., Nakahori Y., Knight S. J., Schwartz C., Thibodeau S. N., Roche A., Flint T. J., Connor J. M., Fryns J. P., Davies K. E. Genotype prediction in the fragile X syndrome. J Med Genet. 1991 Dec;28(12):824–829. doi: 10.1136/jmg.28.12.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M. C., Rack K., Nakahori Y., Roche A., Bell M. V., Flynn G., Christadoulou Z., MacKinnon R. N., Francis M., Littler A. J. A YAC contig across the fragile X site defines the region of fragility. Nucleic Acids Res. 1991 Jun 25;19(12):3283–3288. doi: 10.1093/nar/19.12.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu W. L., Lee Y. M., Lee S. C., Wang T. R. In vitro DNA methylation inhibits FMR-1 promoter. Biochem Biophys Res Commun. 1993 May 28;193(1):324–329. doi: 10.1006/bbrc.1993.1627. [DOI] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Meijer H., de Graaff E., Merckx D. M., Jongbloed R. J., de Die-Smulders C. E., Engelen J. J., Fryns J. P., Curfs P. M., Oostra B. A. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994 Apr;3(4):615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Mornet E., Bogyo A., Deluchat C., Simon-Bouy B., Mathieu M., Thépot F., Grisard M. C., Leguern E., Boué J., Boué A. Molecular analysis of a ring chromosome X in a family with fragile X syndrome. Hum Genet. 1993 Oct;92(4):373–378. doi: 10.1007/BF01247338. [DOI] [PubMed] [Google Scholar]
- Nakahori Y., Knight S. J., Holland J., Schwartz C., Roche A., Tarleton J., Wong S., Flint T. J., Froster-Iskenius U., Bentley D. Molecular heterogeneity of the fragile X syndrome. Nucleic Acids Res. 1991 Aug 25;19(16):4355–4359. doi: 10.1093/nar/19.16.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola N. A., Metcalf D. Subunit promiscuity among hemopoietic growth factor receptors. Cell. 1991 Oct 4;67(1):1–4. doi: 10.1016/0092-8674(91)90564-f. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Kozman H., Kremer E., Lynch M., Pritchard M., Yu S., Mulley J., Sutherland G. R. Fragile X syndrome: genetic localisation by linkage mapping of two microsatellite repeats FRAXAC1 and FRAXAC2 which immediately flank the fragile site. J Med Genet. 1991 Dec;28(12):818–823. doi: 10.1136/jmg.28.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994 Apr 8;77(1):33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C., Nussbaum R. L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993 Jul 30;74(2):291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Vincent A., Heitz D., Petit C., Kretz C., Oberlé I., Mandel J. L. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991 Feb 14;349(6310):624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]
- Weber C., Oudet C., Johnson S., Pilia G., Schlessinger D., Hanauer A. Dinucleotide repeat polymorphism close to IDS gene in Xq27.3-q28 (DXS1113). Hum Mol Genet. 1993 May;2(5):612–612. doi: 10.1093/hmg/2.5.612-a. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Kotzot D., Hirst M. C., Manca A., Korn B., Schmidt A., Barbi G., Rott H. D., Poustka A., Davies K. E. A microdeletion of less than 250 kb, including the proximal part of the FMR-I gene and the fragile-X site, in a male with the clinical phenotype of fragile-X syndrome. Am J Hum Genet. 1992 Aug;51(2):299–306. [PMC free article] [PubMed] [Google Scholar]
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J. C., Warren S. T., Schlessinger D. Fragile X genotype characterized by an unstable region of DNA. Science. 1991 May 24;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]




