Abstract
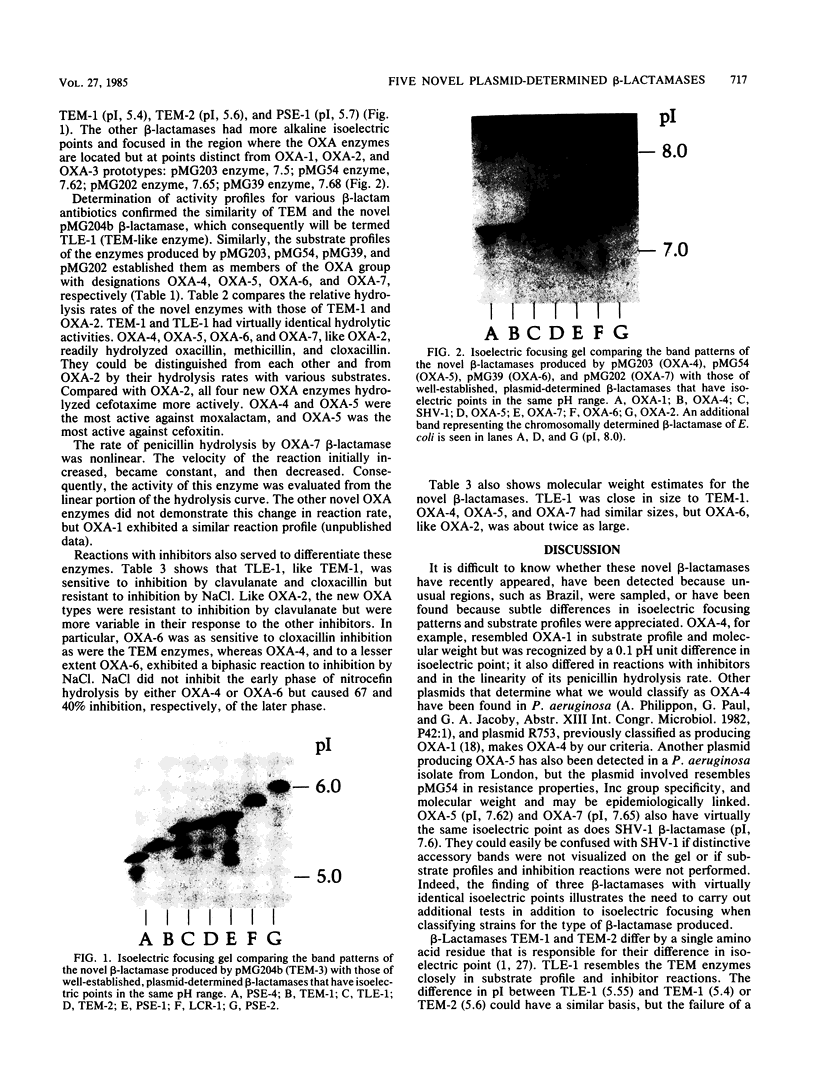
Five novel plasmid-determined beta-lactamases named TLE-1, OXA-4, OXA-5, OXA-6, and OXA-7 were detected in ampicillin-resistant isolates of Escherichia coli and carbenicillin-resistant strains of Pseudomonas aeruginosa. TLE-1 resembled TEM-1 in substrate profile and reactions with inhibitors but differed in isoelectric point (5.55) and enzyme banding pattern on flat-bed electrofocusing.OXA-4, OXA-5, OXA-6, and OXA-7 hydrolyzed oxacillin, methicillin, and cloxacillin readily but differed from OXA-1, OXA-2, and OXA-3 in substrate profiles, inhibitor reactions, and isoelectric points (7.5 to 7.8).OXA-4 and OXA-6 were unusual for members of the OXA group in their sensitivity to inhibition by cloxacillin. OXA-5 and OXA-7 had isoelectric points close to that of SHV-1, emphasizing the need in beta-lactamase classification for studies in addition to isoelectric focusing. These five new enzymes bring the number of plasmid-determined beta-lactamases known in gram-negative organisms to more than 20. The evolution of such enzymatic diversity remains to be explored.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Scott G. K. Partial amino acid sequence of penicillinase coded by Escherichia coli plasmid R6K. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3732–3736. doi: 10.1073/pnas.75.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Smith J. T. The dimeric nature of an R-factor mediated beta-lactamase. Biochem Biophys Res Commun. 1976 Feb 9;68(3):1000–1005. doi: 10.1016/0006-291x(76)91245-6. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacoby G. A. Compatibility and molecular properties of plasmid Rms 149 in Pseudomonas aeruginosa and Escherichia coli. Plasmid. 1980 Jan;3(1):1–6. doi: 10.1016/s0147-619x(80)90029-3. [DOI] [PubMed] [Google Scholar]
- Kratz J., Schmidt F., Wiedemann B. Transposition of a gene encoding OXA-2 beta-lactamase. J Gen Microbiol. 1983 Sep;129(9):2951–2957. doi: 10.1099/00221287-129-9-2951. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Labia R., Andrillon J., Le Goffic F. Computerized microacidimetric determination of beta lactamase Michaelis-Menten constants. FEBS Lett. 1973 Jun 15;33(1):42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- Labia R., Guionie M., Barthélémy M. Properties of three carbenicillin-hydrolysing beta-lactamases (CARB) from Pseudomonas aeruginosa: identification of a new enzyme. J Antimicrob Chemother. 1981 Jan;7(1):49–56. doi: 10.1093/jac/7.1.49. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque R., Roy P. H., Letarte R., Pechère J. C. A plasmid-mediated cephalosporinase from Achromobacter species. J Infect Dis. 1982 May;145(5):753–761. doi: 10.1093/infdis/145.2.753. [DOI] [PubMed] [Google Scholar]
- Levesque R., Roy P. H. Mapping of the plasmid (pLQ3) from Achromobacter and cloning of its cephalosporinase gene in Escherichia coli. Gene. 1982 Apr;18(1):69–75. doi: 10.1016/0378-1119(82)90057-9. [DOI] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Matthew M., Hedges R. W. Analytical isoelectric focusing of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1976 Feb;125(2):713–718. doi: 10.1128/jb.125.2.713-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M., Hedges R. W., Smith J. T. Types of beta-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979 Jun;138(3):657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A., Hedges R. W., Jacoby G. A. Spread of a "Pseudomonas-specific" beta-lactamase to plasmids of enterobacteria. J Bacteriol. 1982 Feb;149(2):700–707. doi: 10.1128/jb.149.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A. M., Paul G. C., Jacoby G. A. Properties of PSE-2 beta-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1983 Sep;24(3):362–369. doi: 10.1128/aac.24.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin L. G., Medeiros A. A., Yolken R. H., Moxon E. R. Ampicillin treatment failure of apparently beta-lactamase-negative Haemophilus influenzae type b meningitis due to novel beta-lactamase. Lancet. 1981 Nov 7;2(8254):1008–1010. doi: 10.1016/s0140-6736(81)91214-9. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I., Tsukamoto K., Harada M., Sawai T. Carbenicillin-hydrolyzing penicillinases of Proteus mirabilis and the PSE-type penicillinases of Pseudomonas aeruginosa. Microbiol Immunol. 1983;27(12):995–1004. doi: 10.1111/j.1348-0421.1983.tb02934.x. [DOI] [PubMed] [Google Scholar]
- Vecoli C., Prevost F. E., Ververis J. J., Medeiros A. A., O'Leary G. P., Jr Comparison of polyacrylamide and agarose gel thin-layer isoelectric focusing for the characterization of beta-lactamases. Antimicrob Agents Chemother. 1983 Aug;24(2):186–189. doi: 10.1128/aac.24.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]





