Abstract
Estrogens induce cell proliferation in target tissues by stimulating progression through the G1 phase of the cell cycle. Induction of cyclin D1 expression is a critical feature of the mitogenic action of estrogen. We have determined a region between −96 and −29 in the cyclin D1 promoter that confers regulation by estrogens in the human mammary carcinoma cells MCF-7. This region encompasses a unique known transcription factor binding site with a sequence of a potential cAMP response element (CRE-D1). The induction is strictly hormone dependent and requires the DNA binding domain as well as both AF-1 and AF-2 domains of the estrogen receptor (ER) α. Destruction of the CRE-D1 motif caused complete loss of estrogen responsiveness. Both c-Jun and ATF-2 transactivated the cyclin D1 promoter in transient transfection experiments, and a clear additional increase was detected when ER was cotransfected with either c-Jun or with c-Jun and ATF-2 but not with ATF-2 alone. Furthermore, the expression of a dominant negative variant of c-Jun, TAM67, completely abolished the induction of the cyclin D1 promoter both in the absence and presence of ER. We show that ATF-2 homodimers and ATF-2/c-Jun heterodimers, but not c-Jun homodimers, were able to bind the CRE of the cyclin D1 promoter. To interpret these results, we propose a mechanism in which ATF-2/c-Jun heterodimers bind to the CRE-D1 element and mediate the activation of cyclin D1 promoter by the ER. This mechanism represents a pathway by which estrogens control the proliferation of target cells.
Estrogens are decisive actors responsible for the proliferation of normal mammary epithelial cells and the development and progression of breast cancer (1). They act in early G1 phase of the cell cycle, and both steroidal and nonsteroidal antiestrogens arrest estrogen-dependent breast cancer cells in the G0/G1 phases (2, 3). Several studies have suggested that cyclin D1 may be involved in mediating the steroid-dependent growth of both normal and malignant mammary epithelial cells. Cyclin D1-deficient mice have a defect in steroid hormone-responsive proliferation of breast epithelium during pregnancy (4). Estrogens reinitiate the cell cycle in simvastatin-arrested MCF-7 breast cancer cells as well as the expression of cyclin D1 and phosphorylation of the retinoblastoma protein even under conditions where mitogen-activated protein kinase activity is inhibited by the continuous presence of simvastatin in the culture medium (5). Moreover, a strong correlation of increased levels of cyclin D1 mRNA with estrogen receptor (ER) overexpression in breast cancer cells has been noted (6). These results suggest a direct regulation of cyclin D1 expression by estrogens even in the absence of peptide growth factors. Thus, cell cycle control by estrogen differs from that by growth factors.
Estrogens act primarily through binding to their receptor, which belongs to the steroid/thyroid nuclear receptor superfamily (7). Like other members of the superfamily, ERα (NR3A1; ref. 32) exhibits a modular structure composed of three functional regions, with the amino-terminal region containing a constitutive transactivation function (AF-1), the DNA binding central domain (DBD), and the carboxyl-terminal region that encompasses the ligand binding domain, a dimerization surface, and a ligand-dependent transactivation function (AF-2) (8). Upon activation by hormone binding, the receptor interacts specifically with a cis-acting DNA sequence called the estrogen response element (ERE), which usually is located upstream from the promoter and displays enhancer properties (7, 8). This interaction rapidly activates the transcription of estrogen-responsive genes. An alternative pathway of ER action has been reported (9) in which the receptor appears to be able to stimulate the transcription from certain promoters containing an AP-1 binding site. The mechanism may involve direct interaction with the AP-1 proteins and synergism between ER activation domains and those of the AP-1 proteins. Such targeting of the ER to c-Fos/c-Jun at their cognate AP-1 site does not require DNA binding of the ER and is independent of the DBD of the receptor.
However, many genes respond to estrogens after a lag of several hours and require RNA and protein synthesis (10). These genes are classified as secondary-response genes and are regulated by a hormone-induced transcriptional cascade. In this model, the primary-response gene(s) code for transcriptional factor(s) that in turn influence the regulation of secondary-response genes.
Estrogen regulation of cyclin D1 expression appears to be at least in part explained by transcriptional activation of its gene (11). First, accumulation of cyclin D1 mRNA, in estrogen-stimulated cells, is blocked by the RNA polymerase II inhibitor actinomycin D. Second, the ability of protein synthesis inhibitors to counter the mRNA induction suggests that synthesis of intermediary proteins is involved in the effect of ER on the cyclin D1 promoter (5). The cyclin D1 promoter contains multiple regulatory elements [TRE, E2F, Oct, Sp1, and cAMP response element (CRE)] and some potential elements that may be involved in the transcriptional regulation of the gene (12). However, no ERE-related sequence has been identified in the proximal cyclin D1 promoter.
To understand the molecular basis of cyclin D1 regulation by estrogens at the transcriptional level, we performed a series of gene transfer studies and examined the mechanisms mediating the effects of estrogens on cyclin D1 promoter. In this study we have mapped a region located between positions −96 and −29, containing a putative CRE (CRE-D1) that contributes to hormonal activation, suggesting a mechanism by which estrogens control cell proliferation.
MATERIALS AND METHODS
Cell Culture and Synchronization.
MCF-7 cells and HeLa cells cells were grown in DMEM supplemented with 10% FCS. For synchronization, exponentially growing MCF-7 cells were incubated for 48 h in phenol red free medium containing 5% charcoal-stripped serum and 5 nM ICI 182,780. Cells were released from the G0/G1 block by placing them in fresh identical medium containing 10 nM 17β-estradiol. Cells fed identical fresh medium without estradiol served as controls.
Expression Vectors and Reporter Constructs.
The cyclin D1 reporter constructs used for luciferase assays termed D1–973, D1–944, D1–848, D1–543, and D1–181 were described in ref. 12. The D1–96 and D1–29 were constructed by using PCR techniques. Site-directed mutagenesis of D1–96 was performed by using a QuickChange Site-Directed Mutagenesis Kit (Stratagene) and a primer containing a three-point mutation in the CRE of the cyclin D1 promoter 5′-TAAgaTCt-3′ to yield the D1–96m reporter construct. ATF-2 cDNA was cloned into pSG5 expression vector (Stratagene).
Transient Transfection Assays.
Cells were transfected either by the calcium phosphate coprecipitation method or by using Lipofectamine following the protocol provided by the supplier. Cells were harvested 48 h after transfection and hormone addition. For kinetics of estradiol-stimulated cyclin D1 luciferase reporter expression, after incubation with DNA for 24 h, cells were washed and allowed to grow for 24 h in medium containing 10% FCS. Thereafter, cells were synchronized and released from the G0/G1 arrest as described above. At different times, cells were harvested for luciferase assay.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay.
Nuclear extracts were prepared according to the method of Andrews and Faller (13). A 40-bp double-stranded oligonucleotide containing the cyclin D1 CRE was end-labeled with [α-32P]dCTP and the Klenow fragment of DNA polymerase I. Binding assays were performed as described (14). The complementary oligonucleotides used for the DNA probe (CRE) were 5′-AGCTTAGGCCTTAACAACAGTAACGTCACACGGACTACAG-3′; the mutated probe (mCRE) used 5′-GCTTAACAACAGTAAGATCTCACGGACTACAGGGGAG-3′.
Glutathione S-Transferase (GST) Pull-Down.
Recombinant c-Jun and ATF-2 cDNAs were transcribed and translated in rabbit reticulocyte lysates (TNT Promega) in the presence of [35S]methionine and then incubated with GST-ER mutants as described (15).
Western Blotting.
Nuclear extracts from estradiol-stimulated MCF-7 cells were separated by SDS/PAGE in 12.5% gels and electroblotted to nitrocellulose. The membranes were incubated with a mAb against c-Jun and with polyclonal antibodies against ATF-2 (KM-1 and C19, respectively, Santa Cruz Biotechnology), and cyclin D1 (CLONTECH). The immune complexes were detected with an enhanced chemiluminescence detection kit (Amersham Pharmacia).
RESULTS
Estrogens Induce a Transient Activation of the Cyclin D1 Promoter in MCF-7 Cells.
MCF-7 cells were transfected with the D1–944 Luc plasmid, and then synchronized at G0/G1 by ICI 182,780. After stimulation by estradiol cells were harvested for assay at different times (Fig. 1A). The luciferase activity was induced by estradiol and increased progressively to reach a maximum at 7 h, then decreased progressively. In parallel experiments, we have investigated the endogenous cyclin D1 protein expression. An increase of cyclin D1 levels was observed within 2 h of treatment by estradiol. Cyclin D1 continued to increase until 9 h and then returned to reach again the control level by 24 h (Fig. 1B). ICI 182,780-arrested MCF-7 cells placed in fresh medium without hormone did not progress toward S phase.
Figure 1.
Transient activation of the cyclin D1 promoter by estradiol in MCF-7 cells. (A) MCF-7 cells were transfected by using the calcium phosphate procedure with 5 μg of D1–944 Luc and 1 μg of pCH110, then treated with 5 nM ICI 182,780 for 48 h in the presence of 5% charcoal-stripped serum (CSS). At t = 0 h, medium was changed to 5% CSS with 10 nM estradiol. At the times indicated, cells were harvested and assayed for luciferase and β-galactosidase activities. (B) Whole-cell extracts were prepared from a parallel set of transfected cells and analyzed for cyclin D1 protein by Western blotting.
Mapping the Cis-Acting Element in the Cyclin D1 Promoter that Mediates Estrogen Induction.
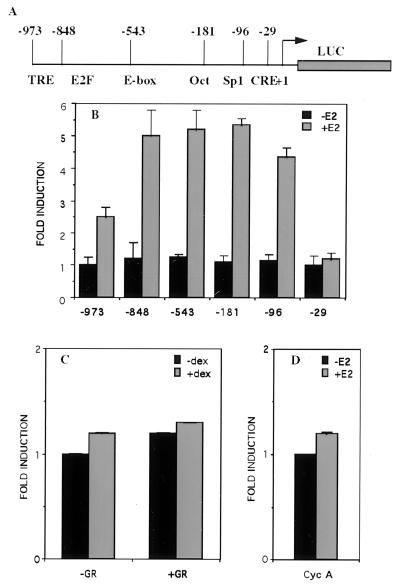
Constructs containing varying lengths of the cyclin D1 5′-flanking sequence upstream of the luciferase reporter gene were used in transient transfection studies in ER-negative HeLa cells cotransfected with an ER expression vector (Fig. 2A). The level of induction by estradiol (E2) from deleted promoters until D1–96 remains approximately constant, 4- to 5-fold (Fig. 2B). A larger deletion in which all of the regulatory elements were removed (D1–29) resulted in a promoter construct activity that was no more inducible by E2. The cyclin D1 promoter is unresponsive to activation mediated via the glucocorticosteroid receptor, whereas a control glucocorticosteroid-responsive element construct (MMTV-CAT) is activated under similar experimental conditions (Fig. 2C and data not shown). Like cyclin D1, cyclin A is a regulatory protein required for progression through the G1/S phase of the cell cycle (16). The 500-bp fragment of the human cyclin A promoter containing several sequence-specific transcriptional regulators, including Sp1, CREB/ATF, p53, E2F, NF1, and AP1 (17, 18), was unresponsive to activation mediated by the ER (Fig. 2D). In MCF-7 cells, cyclin A expression is induced late (>16 h) after E2 (11), probably indirectly as a consequence of the cell cycle progression. These results indicate that (i) the stimulation of the cyclin D1 promoter by E2 requires the presence of its receptor, and (ii) the region between −96 and −29 that encompasses a potential CRE-like element is necessary to confer regulation by estrogens. It was reported that estrogens can act via the cAMP system to regulate cAMP-mediated gene expression (19). Increasing the intracellular concentration of cAMP leads to activation of protein kinase A, resulting in phosphorylation of CREB, which is an essential step in transcriptional activation by this protein factor. Activation of adenylate cyclase by forskolin does not activate the cyclin D1 promoter. Furthermore, inhibition of protein kinase A by the specific inhibitor H89 only weakly affected the hormone response of the D1–96 construct (data not shown). These results are reminiscent of those obtained in site-directed mutagenesis studies of the CRE of the tyrosine hydroxylase gene promoter in which the mutation of the consensus CRE motif TGACGTCA to TAACGTCA diminished by more than 90% the transcription induced by forskolin (20). We therefore investigated the mechanism whereby ER activates the D1–96 promoter in the absence of any known ERE.
Figure 2.
The −96-bp region of the cyclin D1 promoter is required for activation by ER. (A) Schematic representation of the intact and 5′ deleted human cyclin D1 promoter luciferase constructs. Numbers at the top indicate the endpoints of the 5′ deletion mutants starting from the Inr element. The 3′ end of the D1 promoter in the reporter constructs is +139. Positions of the known regulatory elements are shown. HeLa cells were transfected by using Lipofectamine with: (B) 1 μg of luciferase reporter gene driven by 5′ deletion mutants of the human cyclin D1 promoter, 0.2 μg of pSG5-ER, and 0.2 μg of pCH110 as an internal standard of transfection efficiency. After 24 h, cells were incubated with 10 nM estradiol or the ethanol vehicle for 24 h. (C) 1 μg of D1–96 Luc, 0.2 μg of pSG5-GR, and 0.2 μg of pCH110. After 24 h, cells were incubated with 100 nM dexamethasone or ethanol vehicle for 24 h. (D) 1 μg of luciferase reporter gene driven by a 500-bp fragment of the human cyclin A promoter, 0.2 μg of pSG5-ER, and 0.2 μg of pCH110. After 24 h, cells were incubated with 10 nM estradiol or the ethanol vehicle for 24 h. At 48 h posttransfection, cells were harvested and luciferase activities were measured and normalized to β-galactosidase activities.
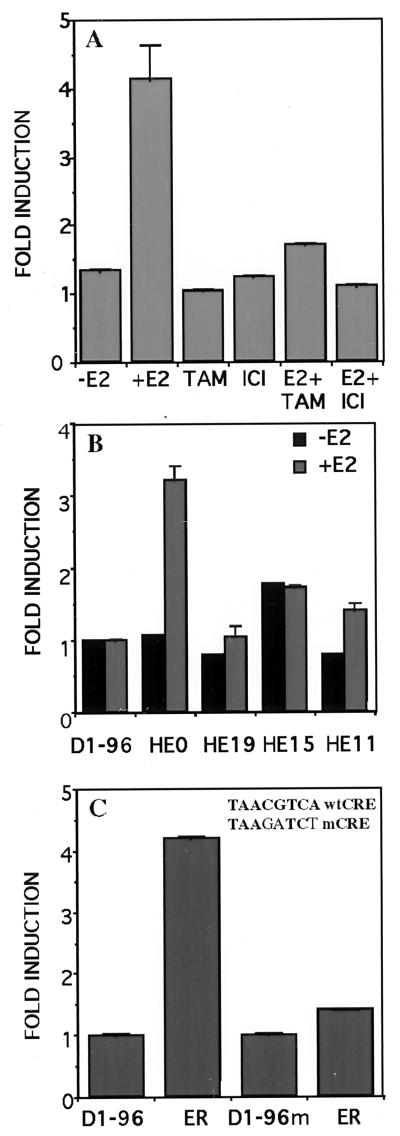
Induction of the D1–96 Promoter by Estrogen Can Be Selectively Blocked by Antagonists and Requires the Presence of Wild-Type ER.
To determine the specificity of the estrogen induction of the cyclin D1 promoter, we tested whether two estrogen antagonists, tamoxifen and ICI 182,780, could block this response. Neither of these two antagonists stimulated the cyclin D1 promoter activity (Fig. 3A). Furthermore, addition of these antagonists simultaneously with estradiol completely blocked the estrogen-induced stimulation (Fig. 3A). These results indicate that the induction of the cyclin D1 promoter requires agonist-activated ER.
Figure 3.
Induction of the D1–96 promoter by estradiol requires the presence of wild-type ER and integrity of the CRE-binding site. (A) HeLa cells were transfected with D1–96 Luc, pSG5-ER, and pCH110 as described in Fig. 2B. After 24 h, cells were incubated with 10 nM estradiol and/or 100 nM tamoxifen, and/or 5 nM ICI 182,780 or ethanol vehicle for 24 h. (B) Deletion mutants of the ER were transfected with D1–96 Luc and pCH110. After 24 h cells were incubated with 10 nM estradiol or ethanol vehicle for 24 h. (C) Effect of CRE mutation (Inset) on the activation of the D1–96 promoter by ER. The pSG5-ER was cotransfected with D1–96 Luc or with D1–96m Luc and pCH110 in HeLa cells. After 24 h, cells were incubated with 10 nM estradiol for 24 h. At 48 h posttransfection cells were harvested and luciferase activities were measured and normalized to β-galactosidase activities.
To determine the domain(s) of ER involved in the induction of the cyclin D1 promoter, mutants HE19, HE15, and HE11 were analyzed (Fig. 3B). Previous work has established that each of these variant ERs is expressed at comparable levels from these vectors (21). HE19, which contains the DBD, the ligand binding domain, and the hormone-inducible transactivating function (AF-2), was inefficient at stimulating transcription from the cyclin D1 promoter, whereas it was an efficient inducible activator from the ERE-thymidine kinase-Luc used as control (data not shown). HE15, which contains the constitutively acting transactivating domain AF-1 but lacks the ligand binding domain was 50% as efficient as the wild-type receptor, HEG0, from both promoters. The receptor DBD is also necessary for estrogen action as deletion of this domain (HE11) practically abolished induction. Thus, it appears that the DBD is required and both AF-1 and AF-2 have to cooperate for the full estrogen induction of the cyclin D1 promoter activity.
An Essential Role for CRE Motif in Estrogen Regulation of the Cyclin D1 Promoter.
To investigate whether the CRE-like motif plays a role in the estrogen-induced cyclin D1 gene activation, we compared the activity of the D1–96 construct with that of D1–96m whose CRE site was mutated in the CGTCA motif in such a way as to abolish the enhancer function of a consensus, canonical CRE (22) (Fig. 3C). Destruction of the motif caused complete loss of estrogen responsiveness, supporting an essential role of the CRE motif in mediating estrogen effects.
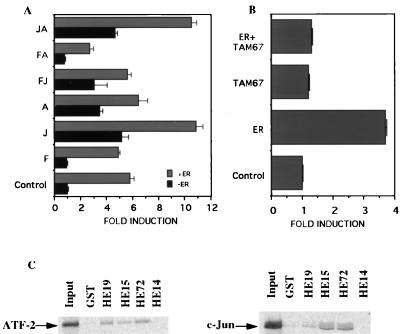
It has been suggested that the putative CRE site is the major mediator of the induction of the cyclin D1 promoter by c-Jun, presumably by binding an ATF/c-Jun heterodimeric protein complex (12). Thus we examined whether c-Jun, c-Fos, and ATF-2 overexpression affected the hormone response of the D1–96 cyclin promoter construct (Fig. 4A). In agreement with the previous results, transfection of HeLa cells with c-Jun, ATF-2, but not c-Fos, caused a 3- to 5-fold induction of the D1–96 Luc. Interestingly, a clear additional increase of the promoter activity was detected when ER was cotransfected with c-Jun or with both c-Jun and ATF-2. Control values in the absence of estrogen treatment are identical to those in the absence of ER. Cotransfection of Tam 67 (23), a dominant negative variant of c-Jun, completely abolished the induction of D1–96 construct even in the presence of ER (Fig. 4B). This result suggests that c-Jun homodimer or c-Jun/ATF-2 heterodimer contribute to the ability of ER to activate the transcription from D1–96 Luc construct.
Figure 4.
ER requires c-Jun to mediate cyclin D1 promoter activation. HeLa cells were transfected as described in Fig. 2B with: (A) D1–96 Luc together with expression vectors for ER, c-Jun, c-Fos, and ATF-2 alone or in combinations as indicated. (B) D1–96 Luc and ER and/or TAM 67. After 24 h, cells were incubated with 10 nM estradiol for 24 h. At the end of incubation cells were harvested and luciferase activities were measured and normalized to β-galactosidase activities. F, c-Fos; J, c-Jun; A, ATF-2. C. In vitro interaction of c-Jun and ATF-2 with ER. 35S-labeled c-Jun and ATF-2 were prepared by translation in vitro. Labeled protein probes were incubated with immobilized GST or GST-ER mutants. Equal amounts of 35S-labeled proteins were incubated with each resin. After extensive washing of the GST beads, the proteins were eluted, separated on PAGE, and detected by fluorography. The input lanes contained 10% of the labeled proteins.
To verify whether CRE binding proteins were required for ER-mediated induction, we have used undifferentiated F9 embryonal carcinoma cells, which lack a functional cAMP signaling pathway (24–26) and have only low levels of endogenous proteins AP-1 expression (27, 28). Transfection of HEG0 into these cells weakly supported hormone activation of the D1–96 Luc construct whereas it was efficiently inducible from ERE-thymidine kinase-Luc. Induction by transfection of c-Jun was greater than with HEG0 alone. An ≈2-fold additional increase was detected when HEG0 was cotransfected with c-Jun (data not shown). These results show that, in F9 cells, the estrogen effects at the putative CRE site require c-Jun. In this context it is noteworthy that F9 cells contain an endogenous ATF-2 activity (29).
To investigate the role of the CRE element in the regulation of cyclin D1 promoter by estrogens, a 40-bp oligonucleotide containing this element was fused to the thymidine kinase (TK) promoter in a luciferase reporter plasmid. This oligonucleotide was not able to confer inducibility by ER whereas as expected the ERE-TK-Luc construct is activated under similar experimental conditions (data not shown). These findings suggest that the CRE-mediated transcriptional regulation by ER depends on the core promoter structure.
In Vitro Interaction Between ATF-2 or c-Jun and ER.
We examined whether ER specifically interacts with ATF-2 and c-Jun. To map the interaction domain, HE15 (ABC regions, amino acids 1–282), HE19 (CDEF regions, amino acids 179–595), HE72 (a N-terminal and C-terminal deletion mutant, amino acids 160–315), and HE14 (DEF regions, amino acids 282–595) were fused to GST, expressed in Escherichia coli, and attached to glutathione-agarose beads. These immobilized mutants then were tested for their ability to interact with in vitro-translated 35S-methionine-labeled c-Jun and ATF-2 (Fig. 4C). Based on results obtained with these proteins, we propose that the region of the ER that interacts with c-Jun and ATF-2 encompasses the DBD. The control GST protein alone pelleted only background amounts of c-Jun or ATF-2.
Endogenous c-Jun and ATF-2 Proteins Specifically Bind the Cyclin D1 CRE.
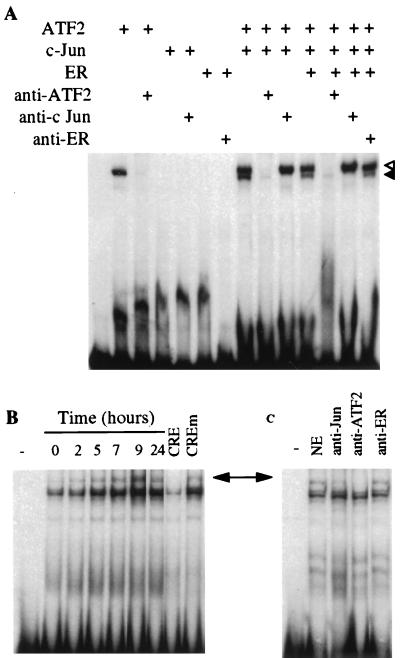
The functional properties of the cyclin D1 CRE were tested in vitro by electrophoretic mobility gel shift assay (EMSA). The cyclin D1-CRE bound in vitro- translated ATF-2 homodimer and ATF-2/c-Jun heterodimer protein complexes. The binding was specifically inhibited by the respective antibodies. In contrast c-Jun homodimer did not bind to the CRE-D1 (Fig. 5A). We also have analyzed nuclear extracts from MCF-7 cells arrested in G0/G1 by ICI 182,780 and stimulated by estradiol for different periods of time. Multiple complexes were formed with these extracts (Fig. 5B). With one exception that represented a nonspecific protein interaction, each of these complexes was specifically inhibited by a 100-fold molar excess of a cold CRE oligonucleotide but not by a mutated oligonucleotide (mCRE) identical to wild-type CRE except for three point mutations shown to abolish estrogen responsiveness in transient transfection assays (see above). We did not detect any qualitative cell cycle-dependent variations in the pattern of the complexes. However, the amounts of complexes formed increased substantially after 2–9 h of estradiol treatment and then decreased at 24 h when the cells had begun replicating their DNA. In an attempt to determine the composition of these complexes, we have used antibodies against c-Jun, ATF-2, and ER (Fig. 5C). The c-Jun and ATF-2 antibodies blocked the formation of the slower-migrating band, whereas the ER antibody had no effect, indicating that the interactions between ER and c-Jun/ATF-2 are not strong enough to withstand the conditions of EMSA. The behavior of ER in this model is reminiscent of coactivator proteins whose presence enhances the promoter activity by protein–protein interactions often undetectable by EMSA. Thus, the cyclin D1 CRE-like element-binding complex contains both ATF-2 and c-Jun proteins. The remaining bands correspond to the nonspecific band mentioned above and to a specific unidentified complex that apparently contains neither c-Jun nor ATF-2.
Figure 5.
c-Jun and ATF-2 proteins specifically bind to the cyclin D1-CRE. (A) The 32P-labeled DNA probe (≈25,000 cpm) was incubated for 30 min at room temperature with the in vitro-translated c-Jun, ATF-2 and ER proteins as indicated above each lane. For supershift assays, lysates were preincubated with the antisera for 15 min at room temperature before the addition of the radioactive probe. Open arrow indicates the ATF-2 homodimeric complexes and the closed arrow the ATF-2/c-Jun heterodimeric complexes. (B) Nuclear extracts (5 μg) of MCF-7 cells treated as described in Fig. 1A were incubated with 2 μg of poly dI-dC for 15 min at 4°C before the addition of the radioactive cyclin D1-CRE probe. For competition experiments, a 100-fold molar excess of unlabeled CRE or mutated CRE was mixed with the radioactive probe. (C) Nuclear extracts (5 μg) of MCF-7 cells treated with estradiol for 7 h were preincubated with 2 μg of poly dI-dC for 15 min at 4°C, then antisera were added for 15 min at room temperature and finally with radioactive probe. The arrow indicates the position of c-Jun/ATF-2-DNA complexes.
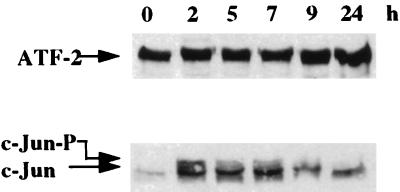
Protein extracts from synchronized MCF-7 cells were analyzed by Western blotting with the c-Jun and ATF-2 antibodies. As shown in Fig. 6, the levels of the activator ATF-2 show no significant fluctuation during the G0/G1 and G1/S phases. In contrast, c-Jun, and in particular its phosphorylated form (slower migrating band), rapidly increased and declined again as the estrogen-stimulated cell progressed through G1 phase.
Figure 6.
Estrogens induce c-Jun expression. Nuclear extracts (30 μg) from MCF-7 cells treated as described in Fig. 1A were analyzed for c-Jun and ATF-2 proteins by Western blotting. c-Jun-P indicates the phosphorylated form of c-Jun.
DISCUSSION
Induction of cyclin D1 expression is an essential part of the mitogenic action of estrogens in hormone-dependent breast cancer cells. In turn, cyclin D1 has been reported to enhance the transactivating effects of estrogens (30), independently of its role in the regulation of cyclin-dependent kinase activities.
In this work we have studied the mechanisms involved in the induction of the cyclin D1 promoter activity by estradiol-activated ER. We show that the DBD as well as both transactivating functions AF-1 and AF-2 are required for the optimal activity of the ER, and that the induction of the cyclin D1 promoter is blocked by antiestrogens. However, the cyclin D1 promoter contains no ERE-like sequence. We have mapped a region localized between positions −96 and −29 containing a putative CRE that contributes to the activation of the cyclin D1 gene promoter by estradiol. Mutation of the CRE motif causes complete loss of estrogen responsiveness, supporting an essential role of this motif in mediating estrogen effects. The action of estrogen is not significantly inhibited by H89, a specific inhibitor of cAMP-dependent protein kinase, indicating that is not a consequence of an increase in the level of intracellular cAMP. The requirement for the AF-1 and AF-2 domains implies that the ER participates at transcriptional transactivation. On the other hand, the DBD is not involved in the binding of ER to the DNA, but may be necessary for the correct spatial orientation of the receptor or for protein–protein interactions. Pull-down experiments indeed confirmed that DBD is necessary for the interactions of the ER with ATF-2 and c-Jun.
Further experiments demonstrated that c-Jun and ATF-2 proteins are required for the transcriptional regulation of the cyclin D1 promoter by estrogens. Overexpression of these proteins induced the expression of the reporter plasmid placed under the control of the D1–96 promoter. Furthermore, an additional increase in the magnitude of transcriptional activation was observed when ER was cotransfected with c-Jun or c-Jun and ATF-2. No detectable stimulation was observed with HE0 expressed in the absence of hormone. Tam 67, a dominant negative variant of c-Jun that lacks the transactivation domain (23), did not induce the expression of the indicator gene and completely abolished the response of D1–96 promoter for estrogen. Additional data indicate that the CRE-D1-dependent transactivation is mediated by the c-Jun/ATF-2 heterodimer. This conclusion follows from the observation that whereas such heterodimers bind to CRE-D1 in electrophoretic mobility shift assay, c-Jun homodimers do not. ATF-2 homodimers do bind the CRE-D1 element but their transcriptional action is not modified by estrogens in transient transfection experiments. Moreover, the binding of c-Jun/ATF-2 heterodimers to CRE-D1 is enhanced in extracts from estradiol-treated cells.
Remarkably, the CRE-D1 element was unable to induce transcription when removed from its natural environment. This result is in agreement with observations indicating that the bases adjacent to CREs exert profound influences on the transcriptional activities mediated by these elements (31). While containing no known transcriptional regulatory elements, the adjacent DNA sequences may participate in the protein-DNA interactions needed for transcriptional activation. Their role may be related to the recruitment of coactivators or of basal transcription factors after the binding of the c-Jun/ATF-2/ER complex to the CRE-D1 element.
In summary, we have demonstrated a mechanism by which ER can regulate gene transcription through a promoter that lacks canonical ERE. Taken together, these results indicate that ER plays a dual role in directly and indirectly activating the cyclin D1 promoter. In the cells blocked by ICI 182,780 in G0/G1 phase estrogen induces the c-Jun expression and phosphorylation. Subsequently, the ER directly activates the cyclin D1 promoter, where it is tethered by protein–protein interactions. In this model, the ER assumes the role of a transcriptional coactivator as it were.
Acknowledgments
We are very grateful to Dr. R. Muller for the human cyclin D1 promoter mutants. We thank Dr. M. Yaniv, Dr. M.G. Green, Dr, P. Chambon, and Dr. L. Tora for generously providing expression vectors for c-Jun, c-Fos, ATF-2 cDNA, human ER, and human ER-deleted fragments, respectively; and Dr. A. Wakeling and Zeneca pharmaceuticals for providing the antiestrogens. We thank D. Catala for technical assistance. This work was supported in part by the Centre National de la Recherche Scientifique (CNRS) and by the Association pour la Recherche sur le Cancer (No. 9835). D.C. received a fellowship from the Association pour la Recherche sur le Cancer.
ABBREVIATIONS
- ER
estrogen receptor
- GST
glutathione S-transferase
- ERE
estrogen-responsive element
- CRE
cAMP response element
- DBD
DNA binding domain
References
- 1.Clarke R, Dickson R B, Lippman M E. Crit Rev Oncol Hematol. 1992;12:1–23. doi: 10.1016/1040-8428(92)90062-u. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland R L, Hall R E, Taylor I W. Cancer Res. 1983;43:3998–4006. [PubMed] [Google Scholar]
- 3.Wakeling A E, Dukes M, Bowler J. Cancer Res. 1991;51:3867–3873. [PubMed] [Google Scholar]
- 4.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 5.Altucci L, Addeo R, Cicatiello L, Dauvois S, Parker M G, Truss M, Beato M, Sica V, Bresciani F, Weiss A. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- 6.Buckley M F, Sweeny K J E, Hamilton J A, Sini R L, Manning D L, Nicholson R I, deFazio A, Watts C K W, Musgrove E A, Sutherland R L. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 7.Beato M, Herrlich P, Schütz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 8.Gronemeyer H. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 9.Webb P, Lopez G N, Uht R M, Kushner P J. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- 10.Katzenellenbogen B S. Annu Rev Physiol. 1980;42:17–35. doi: 10.1146/annurev.ph.42.030180.000313. [DOI] [PubMed] [Google Scholar]
- 11.Prall O W J, Sarcevic B, Musgrove E A, Watts C K W, Sutherland R L. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 12.Herber B, Truss M, Beato M, Muller R. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- 13.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabbah M, Radanyi C, Redeuilh G, Baulieu E E. Biochem J. 1996;314:205–213. doi: 10.1042/bj3140205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabbah M, Kang K-I, Tora L, Redeuilh G. Biochem J. 1998;336:639–646. doi: 10.1042/bj3360639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pines J, Hunter T. Nature (London) 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 17.Heuglein B, Chenivesse X, Wang J, Eick D, Brechot C. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto M, Yoshida M, Ono K, Fujita T, Ohtani-Fujita N, Sakai T, Nikaido T. Exp Cell Res. 1994;210:94–101. doi: 10.1006/excr.1994.1014. [DOI] [PubMed] [Google Scholar]
- 19.Aronica S M, Kraus W L, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1994;91:8517–8521. doi: 10.1073/pnas.91.18.8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinti C, Yang C, Seo H, Conti B, Kim C, Joh T H, Kim K-S. J Biol Chem. 1997;272:19158–19164. doi: 10.1074/jbc.272.31.19158. [DOI] [PubMed] [Google Scholar]
- 21.Kumar V, Green S, Stack G, Berry M, Jin J-R, Chambon P. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 22.Montminy M. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 23.Brown P H, Kim S-H, Wise S C, Sabichi A L, Birrer M J. Cell Growth Differ. 1996;7:1013–1021. [PubMed] [Google Scholar]
- 24.Gonzales G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 25.Rehfuss R P, Walton K M, Loriaux M M, Goodman R H. J Biol Chem. 1991;266:18431–18434. [PubMed] [Google Scholar]
- 26.Liu F, Thompson M A, Wagner S, Greenberg M E, Green M R. J Biol Chem. 1993;268:6714–6720. [PubMed] [Google Scholar]
- 27.Yang Y H, Chambard J C, Sun Y L, Smeal T, Schmidt T J, Drouin J, Karin M. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 28.Chiu R, Boyle W J, Meek J, Smeal T, Hunter T, Karin M. Cell. 1988;54:541–552. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 29.van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb A J. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwijsen R M L, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides R J A M. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 31.Deutsch P J, Hoeffler J P, Jameson J L, Lin J C, Habener J F. J Biol Chem. 1988;263:18466–18472. [PubMed] [Google Scholar]
- 32.Nuclear Receptors Nomenclature Committee. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]