Abstract
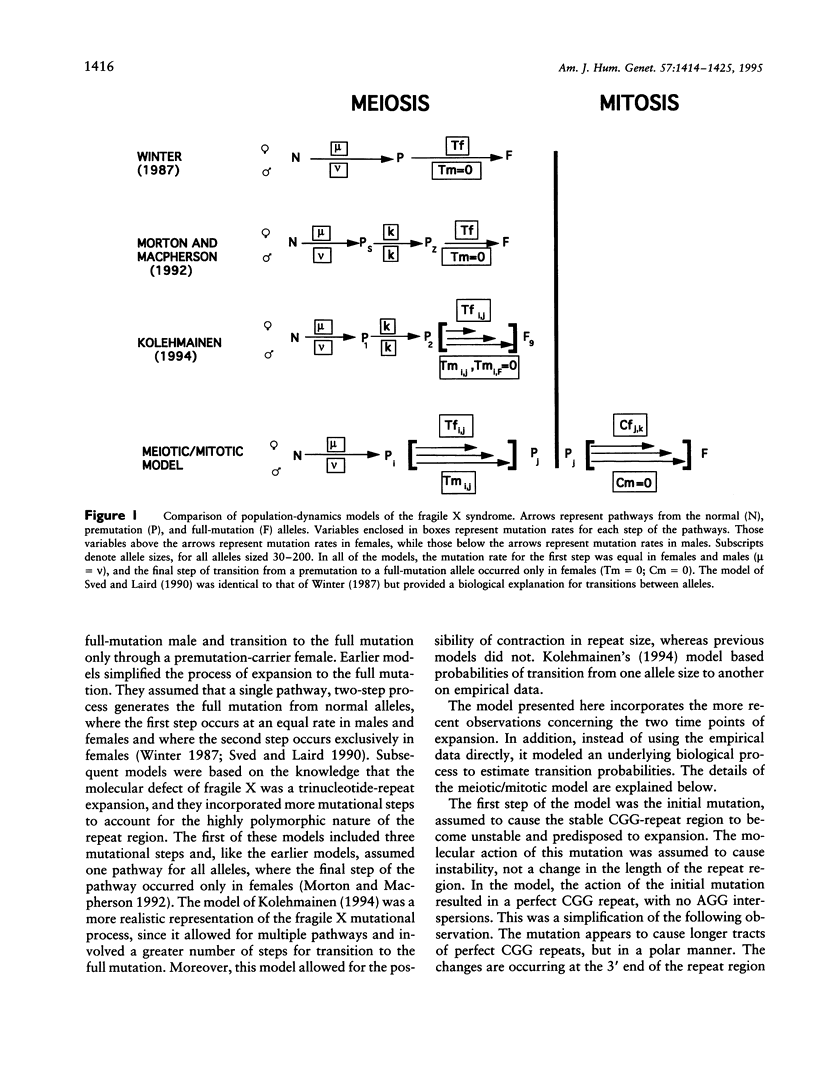
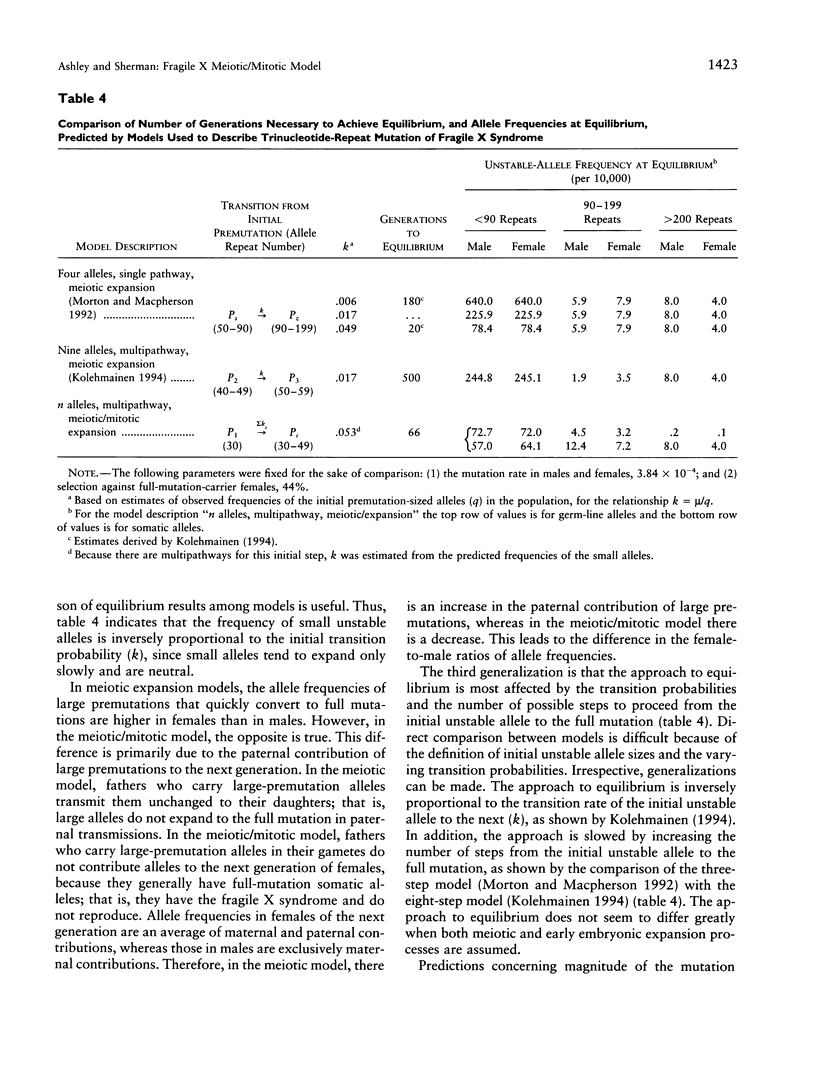
A model to explain the mutational process and population dynamics of the fragile X syndrome is presented. The mutational mechanism was assumed to be a multipathway, multistep process. Expansion of CGG repeats was based on an underlying biological process and was assumed to occur at two time points: meiosis and early embryonic development (mitosis). Meiotic expansion was assumed to occur equally in oogenesis and spermatogenesis, while mitotic expansion was restricted to somatic, or constitutional, alleles of maternal origin. Testable hypotheses were predicted by this meiotic/mitotic model. First, parental origin of mutation is predicted to be associated with the risk of a woman to have a full mutation child. Second, "contractions" seen in premutation male transmissions are predicted not to be true contractions in repeat size, but a consequence of the lack of mitotic expansion in paternally derived alleles. Third, a portion of full-mutation males should have full-mutation alleles in their sperm, due to the lack of complete selection against the full-mutation female. Fourth, a specific premutation-allele frequency distribution is predicted and differs from that based on models assuming only meiotic expansion. Last, it is predicted that approximately 65 generations are required to achieve equilibrium, but this depends greatly on the expansion probabilities.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashley C. T., Jr, Wilkinson K. D., Reines D., Warren S. T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993 Oct 22;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Chung M. Y., Ranum L. P., Duvick L. A., Servadio A., Zoghbi H. Y., Orr H. T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993 Nov;5(3):254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Wassarman P. M. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- Devys D., Biancalana V., Rousseau F., Boué J., Mandel J. L., Oberlé I. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):208–216. doi: 10.1002/ajmg.1320430134. [DOI] [PubMed] [Google Scholar]
- Edwards A., Hammond H. A., Jin L., Caskey C. T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992 Feb;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Fisch G. S., Snow K., Thibodeau S. N., Chalifaux M., Holden J. J., Nelson D. L., Howard-Peebles P. N., Maddalena A. The fragile X premutation in carriers and its effect on mutation size in offspring. Am J Hum Genet. 1995 May;56(5):1147–1155. [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Heitz D., Devys D., Imbert G., Kretz C., Mandel J. L. Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J Med Genet. 1992 Nov;29(11):794–801. doi: 10.1136/jmg.29.11.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M. C., Grewal P. K., Davies K. E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994 Sep;3(9):1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Kelly R., Bulfield G., Collick A., Gibbs M., Jeffreys A. J. Characterization of a highly unstable mouse minisatellite locus: evidence for somatic mutation during early development. Genomics. 1989 Nov;5(4):844–856. doi: 10.1016/0888-7543(89)90126-2. [DOI] [PubMed] [Google Scholar]
- Kolehmainen K. Population genetics of fragile X: a multiple allele model with variable risk of CGG repeat expansion. Am J Med Genet. 1994 Jul 15;51(4):428–435. doi: 10.1002/ajmg.1320510425. [DOI] [PubMed] [Google Scholar]
- Kruyer H., Milà M., Glover G., Carbonell P., Ballesta F., Estivill X. Fragile X syndrome and the (CGG)n mutation: two families with discordant MZ twins. Am J Hum Genet. 1994 Mar;54(3):437–442. [PMC free article] [PubMed] [Google Scholar]
- Kunst C. B., Warren S. T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994 Jun 17;77(6):853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Malmgren H., Steén-Bondeson M. L., Gustavson K. H., Seémanova E., Holmgren G., Oberlé I., Mandel J. L., Pettersson U., Dahl N. Methylation and mutation patterns in the fragile X syndrome. 1992 Apr 15-May 1Am J Med Genet. 43(1-2):268–278. doi: 10.1002/ajmg.1320430142. [DOI] [PubMed] [Google Scholar]
- McConkie-Rosell A., Lachiewicz A. M., Spiridigliozzi G. A., Tarleton J., Schoenwald S., Phelan M. C., Goonewardena P., Ding X., Brown W. T. Evidence that methylation of the FMR-I locus is responsible for variable phenotypic expression of the fragile X syndrome. Am J Hum Genet. 1993 Oct;53(4):800–809. [PMC free article] [PubMed] [Google Scholar]
- Morris A., Morton N. E., Collins A., Lawrence S., Macpherson J. N. Evolutionary dynamics of the FMR1 locus. Ann Hum Genet. 1995 Jul;59(Pt 3):283–289. doi: 10.1111/j.1469-1809.1995.tb00748.x. [DOI] [PubMed] [Google Scholar]
- Morton N. E., Macpherson J. N. Population genetics of the fragile-X syndrome: multiallelic model for the FMR1 locus. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4215–4217. doi: 10.1073/pnas.89.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet C., von Koskull H., Nordström A. M., Peippo M., Mandel J. L. Striking founder effect for the fragile X syndrome in Finland. Eur J Hum Genet. 1993;1(3):181–189. doi: 10.1159/000472412. [DOI] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Reyniers E., Vits L., De Boulle K., Van Roy B., Van Velzen D., de Graaff E., Verkerk A. J., Jorens H. Z., Darby J. K., Oostra B. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat Genet. 1993 Jun;4(2):143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Simple repeat DNA is not replicated simply. Nat Genet. 1994 Feb;6(2):114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Tarleton J., MacPherson J., Malmgren H., Dahl N., Barnicoat A., Mathew C., Mornet E., Tejada I. A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am J Hum Genet. 1994 Aug;55(2):225–237. [PMC free article] [PubMed] [Google Scholar]
- Sherman S. L., Jacobs P. A., Morton N. E., Froster-Iskenius U., Howard-Peebles P. N., Nielsen K. B., Partington M. W., Sutherland G. R., Turner G., Watson M. Further segregation analysis of the fragile X syndrome with special reference to transmitting males. Hum Genet. 1985;69(4):289–299. doi: 10.1007/BF00291644. [DOI] [PubMed] [Google Scholar]
- Sherman S. L., Morton N. E., Jacobs P. A., Turner G. The marker (X) syndrome: a cytogenetic and genetic analysis. Ann Hum Genet. 1984 Jan;48(Pt 1):21–37. doi: 10.1111/j.1469-1809.1984.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Snow K., Doud L. K., Hagerman R., Pergolizzi R. G., Erster S. H., Thibodeau S. N. Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet. 1993 Dec;53(6):1217–1228. [PMC free article] [PubMed] [Google Scholar]
- Snow K., Tester D. J., Kruckeberg K. E., Schaid D. J., Thibodeau S. N. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994 Sep;3(9):1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Sved J. A., Laird C. D. Population genetic consequences of the fragile-X syndrome, based on the X-inactivation imprinting model. Am J Hum Genet. 1990 Mar;46(3):443–451. [PMC free article] [PubMed] [Google Scholar]
- Tuckerman E., Webb T., Bundey S. E. Frequency and replication status of the fragile X, fra(X)(q27-28), in a pair of monozygotic twins of markedly differing intelligence. J Med Genet. 1985 Apr;22(2):85–91. doi: 10.1136/jmg.22.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G., Robinson H., Laing S., Purvis-Smith S. Preventive screening for the fragile X syndrome. N Engl J Med. 1986 Sep 4;315(10):607–609. doi: 10.1056/NEJM198609043151002. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Vogel F. Mutation and selection in the marker (X) syndrome. A hypothesis. Ann Hum Genet. 1984 Oct;48(Pt 4):327–332. doi: 10.1111/j.1469-1809.1984.tb00846.x. [DOI] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Winter R. M. Population genetics implications of the premutation hypothesis for the generation of the fragile X mental retardation gene. Hum Genet. 1987 Mar;75(3):269–271. doi: 10.1007/BF00281072. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Hennig I., Vogel W., Steinbach P. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet. 1993 Jun;4(2):140–142. doi: 10.1038/ng0693-140. [DOI] [PubMed] [Google Scholar]
- Wöhrle D., Hirst M. C., Kennerknecht I., Davies K. E., Steinbach P. Genotype mosaicism in fragile X fetal tissues. Hum Genet. 1992 Apr;89(1):114–116. doi: 10.1007/BF00207057. [DOI] [PubMed] [Google Scholar]
- Yu S., Mulley J., Loesch D., Turner G., Donnelly A., Gedeon A., Hillen D., Kremer E., Lynch M., Pritchard M. Fragile-X syndrome: unique genetics of the heritable unstable element. Am J Hum Genet. 1992 May;50(5):968–980. [PMC free article] [PubMed] [Google Scholar]
- Zhong N., Yang W., Dobkin C., Brown W. T. Fragile X gene instability: anchoring AGGs and linked microsatellites. Am J Hum Genet. 1995 Aug;57(2):351–361. [PMC free article] [PubMed] [Google Scholar]


