Abstract
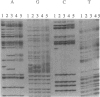
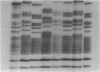
The first hypervariable segment of the human mtDNA control region contains a homopolymeric tract of cytosines between nt 16184 and 16193, interrupted at position 16189 by a thymine, according to the Cambridge reference sequence. A variant commonly found in population screening is a T-to-C transition at nt 16189, resulting in an uninterrupted homopolymeric tract. Direct sequencing of individuals with this variant produces a characteristic blurred sequence in nucleotides beyond the tract. Sequencing clones from these individuals revealed that this is caused by high levels of length heteroplasmy in the homopolymeric tract and low levels of length heteroplasmy in the four adenines following the tract. We have developed a rapid method involving densitometry of sequencing gels to quantify the relative proportions of different length variants present in an individual. We have used this to study the proportions of length variants in individuals from three twin pairs and two maternal lineages. While unrelated individuals usually have different proportions of length variants, all maternally related individuals studied have the same proportions, even if they are only distantly related. It is not obvious how identical heteroplasmic profiles are maintained in maternally related individuals, but some possible mechanisms are suggested.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashley M. V., Laipis P. J., Hauswirth W. W. Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res. 1989 Sep 25;17(18):7325–7331. doi: 10.1093/nar/17.18.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Casane D., Dennebouy N., de Rochambeau H., Mounolou J. C., Monnerot M. Genetic analysis of systematic mitochondrial heteroplasmy in rabbits. Genetics. 1994 Oct;138(2):471–480. doi: 10.1093/genetics/138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghivizzani S. C., Mackay S. L., Madsen C. S., Laipis P. J., Hauswirth W. W. Transcribed heteroplasmic repeated sequences in the porcine mitochondrial DNA D-loop region. J Mol Evol. 1993 Jul;37(1):36–37. doi: 10.1007/BF00170460. [DOI] [PubMed] [Google Scholar]
- Harrison R. G., Rand D. M., Wheeler W. C. Mitochondrial DNA size variation within individual crickets. Science. 1985 Jun 21;228(4706):1446–1448. doi: 10.1126/science.228.4706.1446. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Di Rienzo A., Kocher T. D., Wilson A. C. Toward a more accurate time scale for the human mitochondrial DNA tree. J Mol Evol. 1993 Oct;37(4):347–354. doi: 10.1007/BF00178865. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Clayton D. A. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985 Nov 25;13(22):8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Laipis P. J. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Van de Walle M. J., Laipis P. J., Olivo P. D. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Hoelzel A. R., Lopez J. V., Dover G. A., O'Brien S. J. Rapid evolution of a heteroplasmic repetitive sequence in the mitochondrial DNA control region of carnivores. J Mol Evol. 1994 Aug;39(2):191–199. doi: 10.1007/BF00163807. [DOI] [PubMed] [Google Scholar]
- Horai S., Hayasaka K. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet. 1990 Apr;46(4):828–842. [PMC free article] [PubMed] [Google Scholar]
- Howell N., Halvorson S., Kubacka I., McCullough D. A., Bindoff L. A., Turnbull D. M. Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Hum Genet. 1992 Sep-Oct;90(1-2):117–120. doi: 10.1007/BF00210753. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Lindberg G. L., Brown D. R., Beitz D. C., Freeman A. E., Mayfield J. E., Myers A. M. Replacement of bovine mitochondrial DNA by a sequence variant within one generation. Genetics. 1991 Sep;129(1):247–255. doi: 10.1093/genetics/129.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikó L., Matsumoto L. Number of mitochondria and some properties of mitochondrial DNA in the mouse egg. Dev Biol. 1976 Mar;49(1):1–10. doi: 10.1016/0012-1606(76)90253-0. [DOI] [PubMed] [Google Scholar]
- Rand D. M., Harrison R. G. Molecular population genetics of mtDNA size variation in crickets. Genetics. 1989 Mar;121(3):551–569. doi: 10.1093/genetics/121.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Simple repeat DNA is not replicated simply. Nat Genet. 1994 Feb;6(2):114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- Robin E. D., Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988 Sep;136(3):507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M., Monnerot M., Mounolou J. C. Mitochondrial DNA heteroplasmy in Drosophila mauritiana. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6942–6946. doi: 10.1073/pnas.80.22.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Veltri K. L., Espiritu M., Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990 Apr;143(1):160–164. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- Vilkki J., Savontaus M. L., Nikoskelainen E. K. Segregation of mitochondrial genomes in a heteroplasmic lineage with Leber hereditary optic neuroretinopathy. Am J Hum Genet. 1990 Jul;47(1):95–100. [PMC free article] [PubMed] [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991 Apr;10(4):506–513. [PubMed] [Google Scholar]
- de Stordeur E., Solignac M., Monnerot M., Mounolou J. C. The generation of transplasmic Drosophila simulans by cytoplasmic injection: effects of segregation and selection on the perpetuation of mitochondrial DNA heteroplasmy. Mol Gen Genet. 1989 Dec;220(1):127–132. doi: 10.1007/BF00260866. [DOI] [PubMed] [Google Scholar]