Abstract
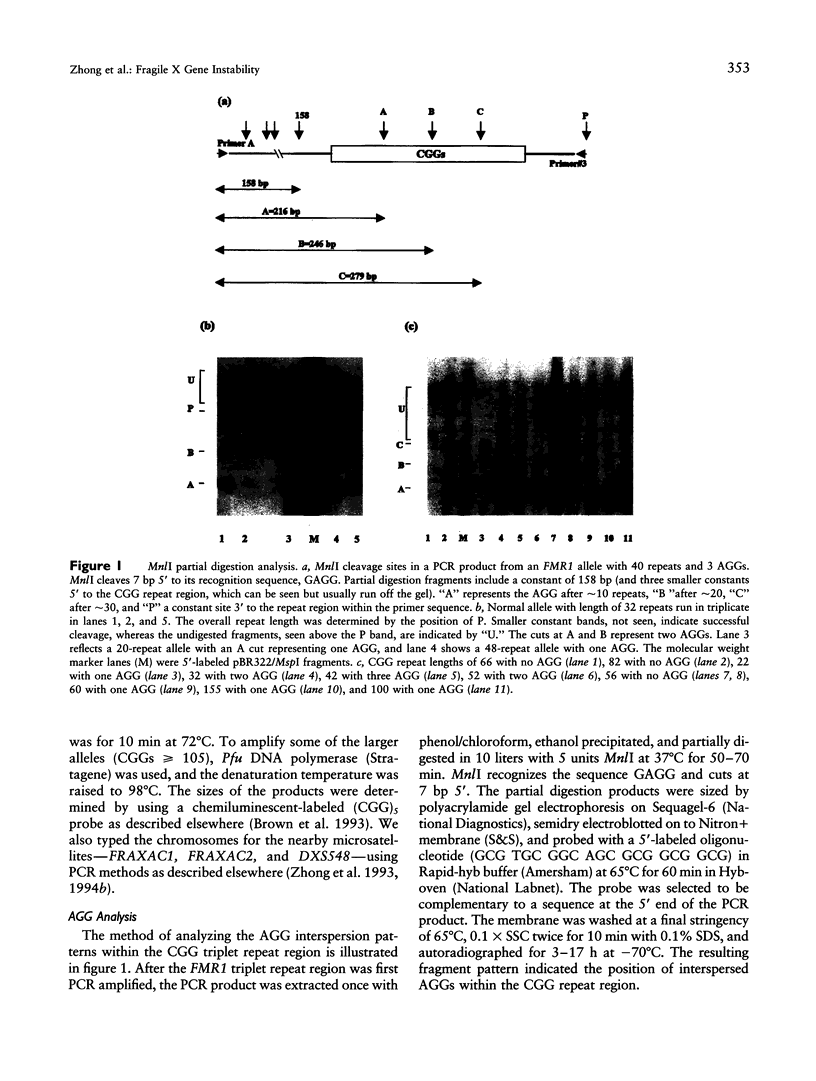
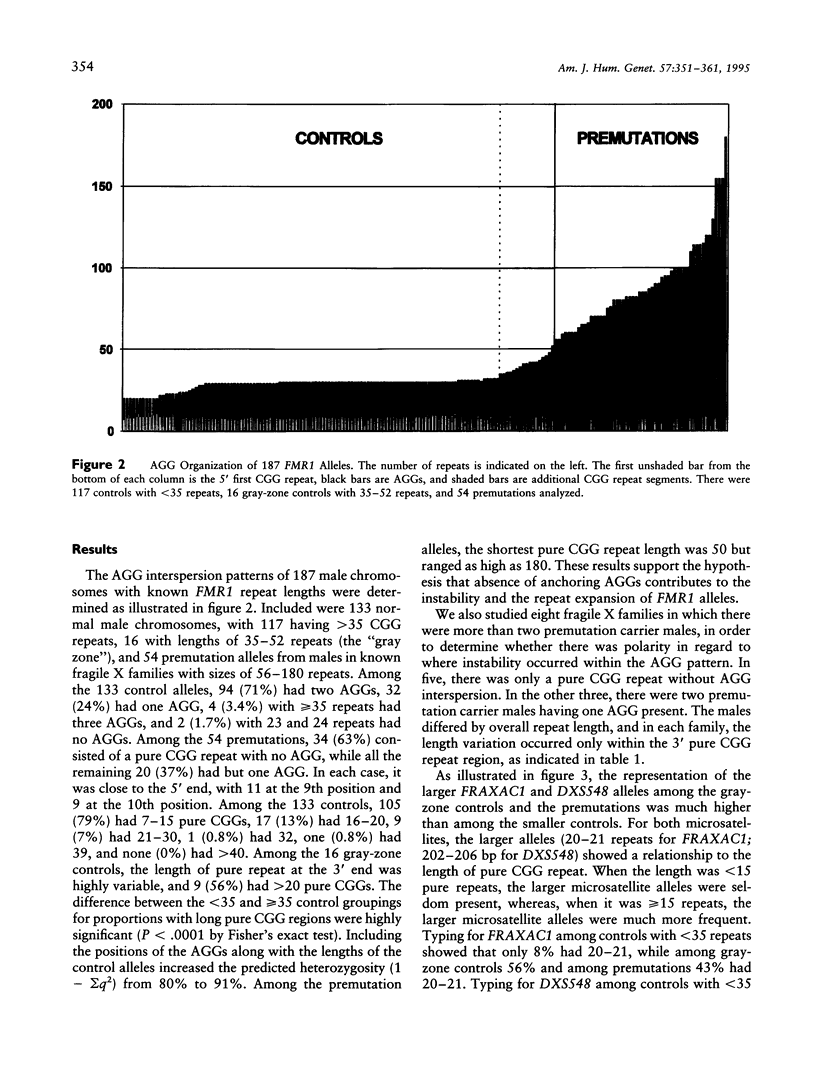
Interspersed AGGs within the FMR1 gene CGG repeat region may anchor the sequence and prevent slippage during replication. In order to detect the AGG position variations, we developed a method employing partial MnlI restriction analysis and analyzed X chromosomes from 187 males, including 133 normal controls (117 with 20-34 and 16 with 35-52 repeats), plus 54 fragile X premutations with 56-180 repeats. Among controls, the interspersed AGG positions were highly polymorphic, with a heterozygosity of 91%. Among the control samples, 1.5% had no AGG positions, 25% had one, 71% had two, and 3% had three. Among the fragile X premutation samples, 63% had no AGG, while 37% had only one AGG. Analysis of premutation samples within fragile X families showed that variation occurred only within the 3' end of the region. Thus, the instability was polar. Controls with > or = 15 pure CGG repeats were associated with the longest alleles of two nearby microsatellites, FRAXAC1 with 20-21 repeats and DXS548 with 202-206 bp and with increased microsatellite heterozygosity. The association of long pure CGG regions, as with fragile X chromosomes, with the longer and more heterozygous microsatellite alleles suggests they may be related mechanistically. Further, our results do not support a recent suggestion that the frequency of fragile X alleles may be increasing. Finally, analysis of a set of nonhuman primate samples showed that long pure CGG tracks are variable in size and are located within the 3' region, which suggests that polar instability within FMR1 is evolutionarily quite old.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashley C. T., Sutcliffe J. S., Kunst C. B., Leiner H. A., Eichler E. E., Nelson D. L., Warren S. T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat Genet. 1993 Jul;4(3):244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Bowcock A. M., Ruiz-Linares A., Tomfohrde J., Minch E., Kidd J. R., Cavalli-Sforza L. L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994 Mar 31;368(6470):455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992 Feb 21;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Brown W. T., Houck G. E., Jr, Jeziorowska A., Levinson F. N., Ding X., Dobkin C., Zhong N., Henderson J., Brooks S. S., Jenkins E. C. Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA. 1993 Oct 6;270(13):1569–1575. [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Buyle S., Reyniers E., Vits L., De Boulle K., Handig I., Wuyts F. L., Deelen W., Halley D. J., Oostra B. A., Willems P. J. Founder effect in a Belgian-Dutch fragile X population. Hum Genet. 1993 Oct 1;92(3):269–272. doi: 10.1007/BF00244471. [DOI] [PubMed] [Google Scholar]
- Chung M. Y., Ranum L. P., Duvick L. A., Servadio A., Zoghbi H. Y., Orr H. T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993 Nov;5(3):254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- Deka R., Chakraborty R., DeCroo S., Rothhammer F., Barton S. A., Ferrell R. E. Characteristics of polymorphism at a VNTR locus 3' to the apolipoprotein B gene in five human populations. Am J Hum Genet. 1992 Dec;51(6):1325–1333. [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Grewal P. K., Davies K. E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994 Sep;3(9):1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Knight S. J., Christodoulou Z., Grewal P. K., Fryns J. P., Davies K. E. Origins of the fragile X syndrome mutation. J Med Genet. 1993 Aug;30(8):647–650. doi: 10.1136/jmg.30.8.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hästbacka J., de la Chapelle A., Kaitila I., Sistonen P., Weaver A., Lander E. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet. 1992 Nov;2(3):204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- Imbert G., Kretz C., Johnson K., Mandel J. L. Origin of the expansion mutation in myotonic dystrophy. Nat Genet. 1993 May;4(1):72–76. doi: 10.1038/ng0593-72. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Tamaki K., MacLeod A., Monckton D. G., Neil D. L., Armour J. A. Complex gene conversion events in germline mutation at human minisatellites. Nat Genet. 1994 Feb;6(2):136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994 Nov;8(3):221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Knight S. J., Flannery A. V., Hirst M. C., Campbell L., Christodoulou Z., Phelps S. R., Pointon J., Middleton-Price H. R., Barnicoat A., Pembrey M. E. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993 Jul 16;74(1):127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- Koide R., Ikeuchi T., Onodera O., Tanaka H., Igarashi S., Endo K., Takahashi H., Kondo R., Ishikawa A., Hayashi T. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nat Genet. 1994 Jan;6(1):9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Kunst C. B., Warren S. T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994 Jun 17;77(6):853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Macpherson J. N., Bullman H., Youings S. A., Jacobs P. A. Insert size and flanking haplotype in fragile X and normal populations: possible multiple origins for the fragile X mutation. Hum Mol Genet. 1994 Mar;3(3):399–405. doi: 10.1093/hmg/3.3.399. [DOI] [PubMed] [Google Scholar]
- Nancarrow J. K., Kremer E., Holman K., Eyre H., Doggett N. A., Le Paslier D., Callen D. F., Sutherland G. R., Richards R. I. Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science. 1994 Jun 24;264(5167):1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- Orr H. T., Chung M. Y., Banfi S., Kwiatkowski T. J., Jr, Servadio A., Beaudet A. L., McCall A. E., Duvick L. A., Ranum L. P., Zoghbi H. Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993 Jul;4(3):221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Oudet C., Mornet E., Serre J. L., Thomas F., Lentes-Zengerling S., Kretz C., Deluchat C., Tejada I., Boué J., Boué A. Linkage disequilibrium between the fragile X mutation and two closely linked CA repeats suggests that fragile X chromosomes are derived from a small number of founder chromosomes. Am J Hum Genet. 1993 Feb;52(2):297–304. [PMC free article] [PubMed] [Google Scholar]
- Oudet C., von Koskull H., Nordström A. M., Peippo M., Mandel J. L. Striking founder effect for the fragile X syndrome in Finland. Eur J Hum Genet. 1993;1(3):181–189. doi: 10.1159/000472412. [DOI] [PubMed] [Google Scholar]
- Parrish J. E., Oostra B. A., Verkerk A. J., Richards C. S., Reynolds J., Spikes A. S., Shaffer L. G., Nelson D. L. Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat Genet. 1994 Nov;8(3):229–235. doi: 10.1038/ng1194-229. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Friend K., Kremer E., Hillen D., Staples A., Brown W. T., Goonewardena P., Tarleton J., Schwartz C. Evidence of founder chromosomes in fragile X syndrome. Nat Genet. 1992 Jul;1(4):257–260. doi: 10.1038/ng0792-257. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Kondo I., Holman K., Yamauchi M., Seki N., Kishi K., Staples A., Sutherland G. R., Hori T. Haplotype analysis at the FRAXA locus in the Japanese population. Am J Med Genet. 1994 Jul 15;51(4):412–416. doi: 10.1002/ajmg.1320510422. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992 Sep 4;70(5):709–712. doi: 10.1016/0092-8674(92)90302-s. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D. C., Leggo J., Amos W., Barton D. E., Ferguson-Smith M. A. Myotonic dystrophy CTG repeats and the associated insertion/deletion polymorphism in human and primate populations. Hum Mol Genet. 1994 Nov;3(11):2031–2035. [PubMed] [Google Scholar]
- Snow K., Doud L. K., Hagerman R., Pergolizzi R. G., Erster S. H., Thibodeau S. N. Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet. 1993 Dec;53(6):1217–1228. [PMC free article] [PubMed] [Google Scholar]
- Snow K., Tester D. J., Kruckeberg K. E., Schaid D. J., Thibodeau S. N. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994 Sep;3(9):1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Squitieri F., Andrew S. E., Goldberg Y. P., Kremer B., Spence N., Zeisler J., Nichol K., Theilmann J., Greenberg J., Goto J. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994 Dec;3(12):2103–2114. doi: 10.1093/hmg/3.12.2103. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Amirhaeri S., Kang S., Wells R. D., Griffith J. D. Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science. 1994 Jul 29;265(5172):669–671. doi: 10.1126/science.8036515. [DOI] [PubMed] [Google Scholar]
- Warren S. T., Nelson D. L. Advances in molecular analysis of fragile X syndrome. JAMA. 1994 Feb 16;271(7):536–542. [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Gyapay G., Dib C., Vignal A., Morissette J., Millasseau P., Vaysseix G., Lathrop M. A second-generation linkage map of the human genome. Nature. 1992 Oct 29;359(6398):794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., Holman K., Mulley J. C., Warren S. T., Schlessinger D. Fragile X genotype characterized by an unstable region of DNA. Science. 1991 May 24;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- Zerylnick C., Torroni A., Sherman S. L., Warren S. T. Normal variation at the myotonic dystrophy locus in global human populations. Am J Hum Genet. 1995 Jan;56(1):123–130. [PMC free article] [PubMed] [Google Scholar]
- Zhong N., Dobkin C., Brown W. T. A complex mutable polymorphism located within the fragile X gene. Nat Genet. 1993 Nov;5(3):248–253. doi: 10.1038/ng1193-248. [DOI] [PubMed] [Google Scholar]
- Zhong N., Liu X., Gou S., Houck G. E., Jr, Li S., Dobkin C., Brown W. T. Distribution of FMR-1 and associated microsatellite alleles in a normal Chinese population. Am J Med Genet. 1994 Jul 15;51(4):417–422. doi: 10.1002/ajmg.1320510423. [DOI] [PubMed] [Google Scholar]
- Zhong N., Ye L., Dobkin C., Brown W. T. Fragile X founder chromosome effects: linkage disequilibrium or microsatellite heterogeneity? Am J Med Genet. 1994 Jul 15;51(4):405–411. doi: 10.1002/ajmg.1320510421. [DOI] [PubMed] [Google Scholar]
- van den Ouweland A. M., Deelen W. H., Kunst C. B., Uzielli M. L., Nelson D. L., Warren S. T., Oostra B. A., Halley D. J. Loss of mutation at the FMR1 locus through multiple exchanges between maternal X chromosomes. Hum Mol Genet. 1994 Oct;3(10):1823–1827. doi: 10.1093/hmg/3.10.1823. [DOI] [PubMed] [Google Scholar]



