Abstract
Numerous species of bacteria use an elegant regulatory mechanism known as quorum sensing to control the expression of specific genes in a cell-density dependent manner. In Gram-negative bacteria, quorum sensing systems function through a cell-to-cell signal molecule (autoinducer) that consists of a homoserine lactone with a fatty acid side chain. Such is the case in the opportunistic human pathogen Pseudomonas aeruginosa, which contains two quorum sensing systems (las and rhl) that operate via the autoinducers, N-(3-oxododecanoyl)-l-homoserine lactone and N-butyryl-l-homoserine lactone. The study of these signal molecules has shown that they bind to and activate transcriptional activator proteins that specifically induce numerous P. aeruginosa virulence genes. We report here that P. aeruginosa produces another signal molecule, 2-heptyl-3-hydroxy-4-quinolone, which has been designated as the Pseudomonas quinolone signal. It was found that this unique cell-to-cell signal controlled the expression of lasB, which encodes for the major virulence factor, LasB elastase. We also show that the synthesis and bioactivity of Pseudomonas quinolone signal were mediated by the P. aeruginosa las and rhl quorum sensing systems, respectively. The demonstration that 2-heptyl-3-hydroxy-4-quinolone can function as an intercellular signal sheds light on the role of secondary metabolites and shows that P. aeruginosa cell-to-cell signaling is not restricted to acyl-homoserine lactones.
Bacteria must constantly monitor the environment in which they live for changes that require an adaptive response. In the case of cell density, many species are able to react to the achievement of a critical density through a cell-to-cell signaling mechanism known as quorum sensing. In Gram-negative bacteria, quorum sensing systems consist of an acylated homoserine lactone signal molecule (referred to as an autoinducer) and an autoinducer-dependent transcriptional activator protein (referred to as an “R protein”) (reviewed in ref. 1). Bacteria at a low cell density produce a basal level of autoinducer, and, as a population grows, autoinducer concentration increases concomitantly with cell density. On reaching a threshold concentration, autoinducer binds to and thereby activates an R protein, which then induces or ceases to repress specific target genes. In this manner, intercellular signals enable a bacterial population to control the expression of specific genes in response to cell density. Such is the case with the ubiquitous environmental microbe Pseudomonas aeruginosa. This opportunistic pathogen has at least two homologous quorum sensing systems, the las and rhl systems, which control a battery of virulence genes (reviewed in ref. 2). The P. aeruginosa quorum sensing systems have been shown to function through two autoinducers, N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) (Fig. 1A), and N-butyryl-l-homoserine lactone (C4-HSL) (Fig. 1B) (3, 4). In the las system, LasI catalyzes the synthesis of the 3-oxo-C12-HSL signal molecule, which binds to and activates the transcriptional activator protein LasR (5, 6). Similarly, in the rhl system, RhlI catalyzes the synthesis of the C4-HSL signal molecule, which binds to and activates the transcriptional activator protein RhlR (7, 8).
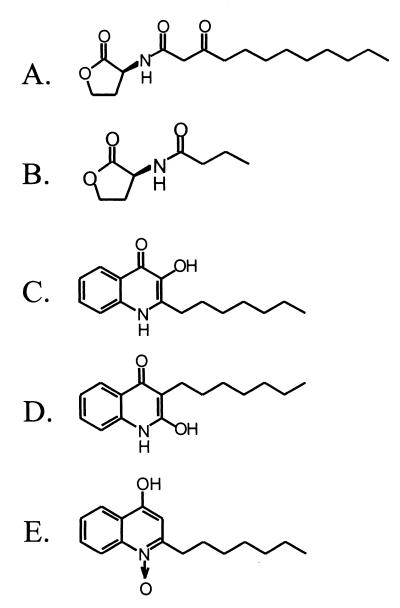
Figure 1.
Chemical structures of relevant compounds. (A) 3-oxo-C12-HSL. (B) C4-HSL. (C) 2-heptyl-3-hydroxy-4-quinolone (PQS). (D) 2-hydroxy-3-heptyl-4-quinolone. (E) 2-heptyl-4-hydroxy-quinoline-N-oxide.
Quorum sensing in P. aeruginosa first was discovered because of the ability of 3-oxo-C12-HSL and LasR to induce the expression of the lasB gene, which encodes for the elastin-hydrolyzing protease, LasB (3, 5, 6, 9). Subsequently, the expression of lasB also was shown to be controlled by C4-HSL and RhlR, indicating that both known P. aeruginosa cell-to-cell signals were involved in the regulation of this major virulence factor (4, 10–12). During our analysis of C4-HSL production, it was found that P. aeruginosa was capable of producing a third unknown signal that activated lasB. This observation led to the discovery and identification of a P. aeruginosa cell-to-cell signal molecule. This molecule belongs to the 4-quinolone chemical family, which is best known for the antibiotic activity of many of its members. We proved that this signal was 2-heptyl-3-hydroxy-4-quinolone and have designated it as the Pseudomonas quinolone signal (PQS). We also showed that this compound induced lasB in P. aeruginosa and depended on the P. aeruginosa quorum sensing systems for its production and bioactivity.
MATERIALS AND METHODS
Strains, Plasmids, and Reagents.
The following bacterial strains were used in this study: Escherichia coli strain DH5α (13); the wild-type P. aeruginosa strain PAO1 (14); and the PAO1 mutant strains PAO-JP2 (lasI, rhlI) (12), PAO-JP3 (lasR, rhlR) (12), and PAO-R1 (lasR) (6). Unless otherwise indicated, bacterial cultures were grown in peptone trypticase soy broth (15) supplemented with 200 μg/ml carbenicillin where appropriate. Plasmid pECP39, which encodes a truncated form of LasR (ΔLasR) that is capable of activating LasR-controlled genes in the absence of an autoinducer, was constructed as follows. First, the intermediate plasmid pKDT39 was constructed by ligating the lasR DNA that encodes LasR amino acids 160–239 in-frame to the histidine fusion site of the expression vector pTRCHisC (Invitrogen). This resulted in the trcp-ΔlasR fusion, which encodes for a truncated, autoinducer-independent form of LasR, the expression of which is controlled by the trc promoter. The trcp-ΔlasR-containing DNA fragment from pKDT39 then was ligated into the P. aeruginosa cloning vector pUCP22 (16) to form pECP39 (bla, lacIq, trcp-ΔlasR, oriP, oriC). Plasmid pTS400 (5) contains a lasB′-lacZ translational fusion, and plasmid pECP62.5 (12) contains tacp-rhlR and a lasB′-lacZ translational fusion. Transformations and other molecular techniques were completed with standard procedures (17). The PQS analog 2-hydroxy-3-heptyl-4-quinolone (Fig. 1D) was synthesized as described (18), and 2-heptyl-4-hydroxy-quinoline-N-oxide (Fig. 1E) was purchased from Sigma.
The PQS Bioassay.
To monitor PQS bioactivity, 1-ml cultures of P. aeruginosa strain PAO-R1 (pTS400) were grown for 18 h at 37°C with shaking (260 rpm; initial optical density was 0.02 at 660 nm) in the presence of culture extract or synthetic PQS. This was followed by measuring activation of the lasB′-lacZ fusion with the use of β-galactosidase (β-gal) assays as described by Miller (19). For culture extracts, bacterial culture fluid was extracted with ethyl acetate and was prepared as described (3). Extracts that were resuspended in ethyl acetate were added to bioassay tubes, and ethyl acetate was removed by drying under a stream of nitrogen. Unless otherwise specified, the amount of material assayed was extracted from 10 ml of culture fluid.
Thin Layer Chromatography (TLC) and High Performance Liquid Chromatography (HPLC).
Preparative TLC plates were obtained by making 1-mm layers of a slurry of 55-g silica gel G (Machery & Nagel) in a solution of 5 g of KH2PO4 in 95 ml of water on 20- × 20-cm glass plates. Plates then were air-dried and activated at 100°C for 1 h. The solvent for TLC was a 17:2:1 mixture of dichloromethane-acetonitrile-dioxane (vol/vol). HPLC was performed on a Beckman–Altex Ultrasphere 10-mm × 25-cm C18 reverse phase column. Extracts were resuspended in 0.25 ml of methanol and were loaded onto the column that then was eluted (2 ml/min) with an acetonitrile/water gradient (10 - 100% over 120 min). Fractions were collected at the indicated intervals and were dried by rotary evaporation at room temperature. Evaporated samples were stored at −20°C before being dissolved in methanol and were assayed for PQS activity.
Purification of PQS.
PQS was purified from 1.2 liters of P. aeruginosa strain PAO-JP2 (pECP39) that was grown for 24 h at 37°C with shaking (260 rpm; initial optical density was 0.05 at 660 nm). Cultures were centrifuged for 10 min at 10,000 × g, and spent supernatant was removed and extracted twice with ethyl acetate as described (3). The ethyl acetate extract was dried with sodium sulfate and was concentrated by rotary evaporation at room temperature. The concentrated material then was extracted and concentrated three times with progressively smaller volumes of a 1:1 mixture of ethyl acetate and acetonitrile (final volume, 0.75 ml). This concentrated extract was fractionated by using Short Body C18 Sep-Pak Plus cartridges (Waters). After loading the extract onto a cartridge (0.25 ml per cartridge), it was washed with 10, 30, and 40% acetonitrile in water (3 ml per cartridge). The active material then was eluted with 55% acetonitrile in water. This partially purified extract was dissolved in 0.3 ml of 90% dioxane in water and was further purified by preparative TLC. After loading the extract, the TLC plate was eluted twice with 17:2:1 dichloromethane/acetonitrile/dioxane. A fluorescent blue band extending from Rf 0.30–0.48 was removed and eluted with 1:1 acetonitrile/ethyl acetate (3 × 3 ml). The eluate, which contained 2-heptyl-3-hydroxy-4-quinolone (Fig. 1C), was concentrated and analyzed as described below.
Chemical Analysis of Natural and Synthetic PQS.
1H and 13C NMR spectra in DMSO-d6 were recorded on a Bruker (Bellerica, MA) AM400 NMR spectrometer operating at 400 MHz for 1H or 100.5 MHz for 13C. The chemical shifts are reported in δ (ppm) relative to residual DMSO-d5 (2.49 ppm) for 1H or relative to DMSO-d6 (39.43 ppm) for 13C, and coupling constants are given in hertz. Infrared (IR) spectra were recorded on a Perkin–Elmer 1600 series Fourier Transformed-IR. Melting points were recorded on a Mel-Temp melting point apparatus and are uncorrected. The ultraviolet spectra were recorded on a Shimadzu UV-1601 PC spectrophotometer. Low-resolution MS were recorded on a Hewlett–Packard 5973 mass selective detector fitted with an SIS direct insertion probe, and high-resolution electron impact spectra were recorded at the University of California-Riverside Mass Spectrometry Facility.
Synthesis of PQS.
A mixture of 2-heptylquinolone (2.00 g, 8.22 mmol), hexamine (0.58 g, 4.11 mmol), and trifluoroacetic acid (12.3 ml) was stirred at reflux under argon for 27 h. Methanol (20 ml) and water (20 ml) were added, and heating was continued for 50 min. Hydrochloric acid (2.5 M, 10 ml) was added, and heating was continued for 30 min. The mixture was allowed to cool, and the precipitate was removed by filtration and was washed with water. The solid was triturated with acetone (10 ml) and then was removed by filtration to give 3-formyl-2-heptylquinolone (0.99 g, 44%), which crystallized from methanol/ethyl acetate as colorless needles: mp 244–247°C (dec) (Found: C, 75.55; H, 7.95; N, 5.09%. C17H21NO2 requires C, 75.24; H, 7.80; N, 5.16%). 1H NMR 12.11 (br s, 1H), 10.37 (s, 1H), 8.12 (d, J 8.0, 1H), 7.70 (dd, J 8.2, 7.1, 1H), 7.57 (d, J 8.2, 1H), 7.38 (dd, J 8.0, 7.1, 1H), 3.01 (t, J 7.7, 2H), 1.56 (quin., J 7.4, 2H), 1.39–1.16 (m, 8H), 0.82 (t, J 6.4, 3H). 13C NMR 190.7 (d), 178.0 (s), 159.9 (s), 139.1 (s), 132.9 (d), 126.1 (s), 124.8 (d), 124.9 (d), 118.6 (d), 113.3 (s), 31.5 (t), 31.1 (t), 28.9 (t), 28.8 (t), 28.3 (t), 22.0 (t), 13.8 (q). m/z 271 (5%), 253 (8%), 242 (5%), 228 (3%), 214 (8%), 200 (13%), 187 (20%), 172 (16%), 159 (100%), 130 (8%).
Aqueous hydrogen peroxide (1.05 M, 1.49 ml, 1.56 mmol) was added to a solution of 3-formyl-2-heptylquinolone (0.41 g, 1.49 mmol) in ethanol (4.5 ml) and aqueous sodium hydroxide (1.08 M, 1.49 ml, 1.6 mmol) under argon, and the mixture was stirred at room temperature for 6 h. The precipitate was removed by filtration, was air dried, and was crystallized from ethyl acetate to give 2-heptyl-3-hydroxy-4-quinolone (0.29 g, 74%), as off-white needles: mp 196–198°C (dec) (Found: C, 74.30; H, 8.42; N, 5.21%. C16H21NO2 requires C, 74.10; H, 8.16; N, 5.40%). λmax (log ɛ) methanol 217 (4.43), 250 (4.56), 292 (3.34), 343 (4.08), 351 nm (4.08). νmax (KBr) 3,275, 2,929, 1,639, 1,599, 1,558, 1,488, 1,467, 1,416, 1,367, 1,265, 1,115, 1,073, 1,025, 954, 934, 863, 756, 710, 895, 574 cm−1. 13C NMR 168.8 (s), 137.7 (s), 137.3 (s), 135.4 (s), 129.8 (d), 124.4 (d), 122.1 (s), 121.4 (d), 117.7 (d), 31.1 (t), 28.7 (t), 28.4 (t), 28.0 (t), 27.7 (t), 21.9 (t), 13.8 (q). 1H NMR and MS of synthetic PQS are presented in Results.
RESULTS
Discovery of a Novel Cell-to-Cell Signal.
The LasB elastase is a significant P. aeruginosa virulence factor that is controlled by both the las and rhl quorum sensing systems (6, 8). It has been shown that the transcription of lasB is greatly reduced in either a P. aeruginosa lasI or rhlI mutant, indicating the importance of 3-oxo-C12-HSL and C4-HSL, respectively (11, 12). Consistent with this finding, strain PAO-R1 (pTS400), which contains a LasR null mutation and a lasB′-lacZ fusion, does not express elastase or β-gal (6, 7). This phenotype results from the absence of LasR, which positively regulates genes controlled by the las quorum sensing system including rhlR, which is required for the rhl quorum sensing system to function (8, 20, 21). As one would expect, the absence of lasR also renders the lasB′-lacZ fusion in strain PAO-R1 (lasR−) (pTS400) unresponsive to 3-oxo-C12-HSL (4). Additionally, lasB′-lacZ in this strain is mildly activated by exogenously added C4-HSL, which is probably attributable to the presence of low amounts of RhlR that can be produced in the absence of LasR (4, 20). Therefore, we were surprised to find that the addition of a spent culture media extract from P. aeruginosa strain PAO1 (wild type) to strain PAO-R1 (lasR−) (pTS400) caused a major induction of lasB′-lacZ (Fig. 2A). This induction could not be mimicked with the addition of synthetic 3-oxo-C12-HSL and/or C4-HSL (data not shown), indicating that an unknown third signal was present in the media from the wild-type strain PAO1. HPLC separation of a strain PAO1 spent media extract resulted in two fractions that activated the lasB promoter in strain PAO-R1 (lasR−) (pTS400) (data not shown). One fraction co-eluted with C4-HSL, and the other eluted near 3-oxo-C12-HSL. We presumed the material that eluted near 3-oxo-C12-HSL was a novel cell-to-cell signal.
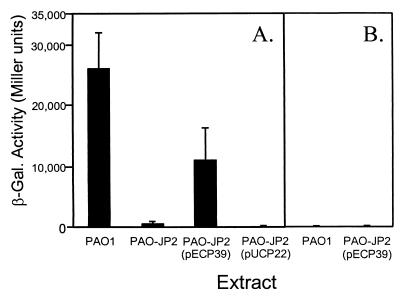
Figure 2.
A novel signal induces lasB in P. aeruginosa. Strain PAO-R1 (lasR−) (pTS400) (A) or strain PAO-JP3 (lasR−, rhlR−) (pTS400) (B) was grown in the presence of culture supernatant extracts from the following P. aeruginosa strains: PAO1 (wild type), PAO-JP2 (lasI−, rhlI−), PAO-JP2 (pECP39), or PAO-JP2 (pUCP22). After 18 h of growth, β-gal activity was assayed and is presented in Miller units +/− SDn−1. (Note: A control culture with no extract added produced 8 Miller units of β-gal activity.) Data represent the means of duplicate assays performed during three separate experiments.
The lack of lasB′-lacZ induction in strain PAO-R1 (lasR−) (pTS400) without the addition of extract (5) showed that production of the novel signal required LasR and presumably 3-oxo-C12-HSL. Thus, HPLC purification of the novel signal from wild-type strain PAO1 was hampered by the fact that it was not readily separated from 3-oxo-C12-HSL, which is required for LasR activity. This presented a technical difficulty that was overcome by construction of a gene that encoded an autoinducer-independent form of LasR. This truncated LasR, referred to as ΔLasR, has a large N-terminal deletion and is encoded on plasmid pECP39. The ΔLasR protein, which was based on a similar truncated construct of the Vibrio fischeri LuxR protein (a LasR homolog) (22), can activate LasR-controlled genes in the absence of 3-oxo-C12-HSL (data not shown). Therefore, ΔLasR was used to induce the production of the novel signal in strain PAO-JP2 (lasI−, rhlI−). The double autoinducer mutant, strain PAO-JP2, was transformed with plasmid pECP39, and it was found that a spent culture media extract from this strain contained the novel signal that activated lasB′-lacZ in strain PAO-R1 (lasR−) (pTS400) (Fig. 2A). There were no lasB′-lacZ activating signals in spent media extracts from cultures of strain PAO-JP2 (lasI−, rhlI−) alone or containing the control vector pUCP22 (Fig. 2A). This confirmed that the synthesis of the new signal depended on LasR and indicated that the signal was not produced by a known autoinducer synthase because strain PAO-JP2 does not have a functional LasI or RhlI.
An initial analysis of the novel signal activity showed that, when the lasR, rhlR double mutant, strain PAO-JP3, containing pTS400 replaced strain PAO-R1(lasR−) (pTS400) in our bioassay, the signal in culture extracts of strains PAO1 and PAO-JP2 (lasI−, rhlI−) (pECP39) was no longer able to induce lasB′-lacZ (Fig. 2B). This suggested that rhlR was required for the bioactivity of the new signal. With this information, we attempted to define the requirements for signal activity by developing an E. coli bioassay for the signal. E. coli strain DH5α was transformed with plasmid pECP62.5, which contains the lasB′-lacZ reporter and the inducible tacp-rhlR gene. If the signal worked directly through RhlR, then the induction of tacp-rhlR in the presence of exogenously added signal should result in the activation of lasB′-lacZ in E. coli strain DH5α (pECP62.5). We found that the signal did not induce lasB′-lacZ when RhlR was expressed in E. coli strain DH5α (pECP62.5) (data not shown). This suggested that the signal’s requirement for rhlR may be attributable to an additional P. aeruginosa component that is controlled by RhlR or, alternatively, that E. coli may be less permeable to this signal.
Purification and Identification of the Novel Cell-to-Cell Signal.
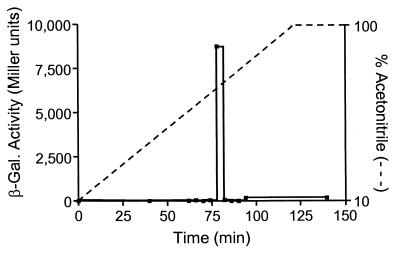
HPLC analysis of a spent media extract from a culture of strain PAO-JP2 (lasI−, rhlI−) (pECP39) showed that a single peak of bioactivity was eluted from an acetonitrile/water gradient (Fig. 3). This indicated that, if multiple signals capable of activating lasB′-lacZ were produced, they would be similar with regard to their hydrophobicity. The eluted signal was further purified by preparative TLC (see Materials and Methods) and was chemically analyzed to determine its structure.
Figure 3.
HPLC analysis of a novel signal extracted from P. aeruginosa culture medium. A culture medium extract from strain PAO-JP2 (lasI−, rhlI−) (pECP39) was separated by reverse-phase HPLC, and fractions were collected between the times indicated by each point on the graph. The extract was loaded onto the column at time 0. Fractions were assayed for our bioactive signal, and results are presented as β-gal activity in Miller units. These data are the average of duplicate β-gal assays from one experiment that was representative of multiple repeated experiments. The dashed line (----) indicates acetonitrile concentration.
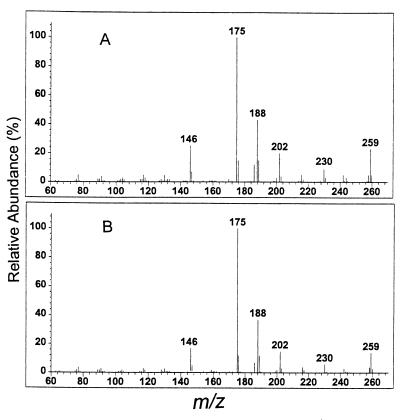
Chemical analysis of the purified signal indicated that, unlike most other Gram-negative autoinducers, it was not an acylated homoserine lactone. Low-resolution mass spectroscopy analysis showed a molecular ion of m/z 259 and a fragmentation pattern consistent with an alkylquinolone that had an additional oxygenation on the heteroaromatic core (Fig. 4A). The ultraviolet spectrum (data not shown) and downfield signals of the 1H NMR (Fig. 5A) were similar to those of the known 3-hydroxy-2-methyl-4-quinolone (23, 24). However, the molecular ion and upfield signals of the 1H NMR (Fig. 5A) suggested the presence of a heptyl rather than a methyl chain in position 2. High-resolution mass spectroscopy gave a molecular ion with an m/z of 259.1571, consistent with the chemical composition C16H21NO2 (calculated = 259.1572). Taken together, these data indicated that the novel signal was 2-heptyl-3-hydroxy-4-quinolone (Fig. 1C), which we have designated as the Pseudomonas quinolone signal (PQS).
Figure 4.
Electron impact mass spectra of purified natural PQS (A) and synthetic PQS (B). Natural PQS was purified from strain PAO-JP2 (lasI−, rhlI−) (pECP39). Comparable peaks are labeled with their respective m/z.
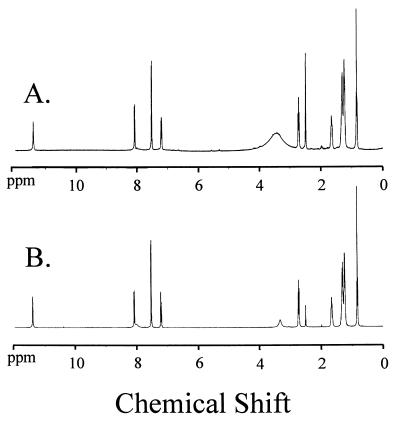
Figure 5.
1H NMR spectra of purified, natural PQS (A) and synthetic PQS (B) in DMSO-d6. 1H NMR spectra for natural and synthetic PQS are identical and can be described as follows: 11.42 (br s, 1H), 8.08 (d, J 8.0, 1H), 8.05 (br s, 1H), 7.55–7.48 (m, 2H), 7.23–7.17 (m, 1H), 2.72 (t, J 7.5, 2H), 1.65 (quin., J 6.4, 2H), 1.35–1.18 (m, 8H), 0.82 (t, J 6.7, 3H).
Analysis of Synthetic PQS.
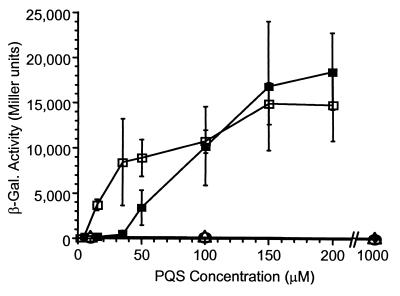
To confirm our identification of PQS, we prepared synthetic 2-heptyl-3-hydroxy-4-quinolone by sequential Duff reaction (25) and Dakin oxidation (26) of 2-heptylquinolone (27) (see Materials and Methods). The low- resolution mass spectrum (Fig. 4B), high-resolution mass spectrum (m/z of 259.1578), and 1H NMR spectrum (Fig. 5B) of synthetic 2-heptyl-3-hydroxy-4-quinolone were indistinguishable from those of purified natural PQS. Synthetic and natural PQS were also identical with regard to ultraviolet spectroscopy and TLC analysis (data not shown). Furthermore, the synthetic material was active in the PQS bioassay. In the presence of increasing concentrations of synthetic or natural PQS, there was a dose-dependent induction of lasB′-lacZ in strain PAO-R1 (lasR−) (pTS400) (Fig. 6).
Figure 6.
PQS bioassay with synthetic or natural PQS. Synthetic PQS (open squares), natural PQS (closed squares), 2-hydroxy-3-heptyl-4-quinolone (circles), or 2-heptyl-4-hydroxy-quinoline-N-oxide (triangles) were added to bioassay cultures at the indicated concentrations. Data represent the means of duplicate assays performed during three separate experiments and are presented as β-gal activity in Miller units +/− SDn−1.
The purification of PQS from a 1.2-liter culture of strain PAO-JP2 (lasI−, rhlI−) (pECP39) yielded an average of 1.8 mg of PQS, which is equivalent to a concentration of ≈6 μM in the culture fluid. This concentration, which is likely to be an underestimate of the actual amount in culture fluid (assuming the total recovery in the purification was <100%), was within the range required for activation of the lasB promoter in P. aeruginosa strain PAO-R1(lasR−) (pTS400) (Fig. 6; 5 μM synthetic PQS = 148 Miller units). It is also interesting to note here that two PQS analogs were not capable of activating lasB′-lacZ in strain PAO-R1 (lasR−) (pTS400). Despite their structural similarities to PQS, the compounds 2-hydroxy-3-heptyl-4-quinolone (Fig. 1D) and 2-heptyl-4-hydroxy-quinoline-N-oxide (Fig. 1E) were not active when tested in our PQS bioassay (Fig. 6). These results suggested that the PQS target may be specific with regard to the compounds by which it is activated.
DISCUSSION
We have discovered that P. aeruginosa produces a cell-to-cell signal that is unlike any previously reported intercellular signal molecule. This molecule was determined to have a 4-quinolone base structure and therefore has been designated as the Pseudomonas quinolone signal (PQS). Our results showed that exogenously added PQS induced a lasB′-lacZ fusion in the P. aeruginosa lasR mutant, strain PAO-R1, containing the plasmid pTS400 (Fig. 2A). This indicates that there is a third cell-to-cell signal, in addition to the autoinducers 3-oxo-C12-HSL and C4-HSL, that is involved in lasB induction by P. aeruginosa. Despite the ability of PQS to activate lasB′-lacZ in the absence of lasR, the production of PQS required an active LasR protein (Fig. 2A). This suggested that a gene (or genes) required for PQS synthesis is controlled through the las quorum sensing system. We also found that at least RhlR was required for PQS to act as a signal because lasB′-lacZ was not induced by PQS in the lasR, rhlR double mutant, strain PAO-JP3, containing the plasmid pTS400 (Fig. 2B). Although the basis of the requirement for rhlR is not known, we suggest that RhlR may serve to regulate the expression of a P. aeruginosa gene that is involved in the response to PQS. This idea stems from our inability to replicate the PQS response by adding PQS to a recombinant E. coli strain expressing RhlR in the presence of lasB′-lacZ (data not shown). These initial experiments on PQS indicated that this signal depended on both P. aeruginosa quorum sensing systems and that it was involved in the regulation of the major virulence factor, LasB elastase.
The addition of a third molecule to the P. aeruginosa intercellular signal repertoire increases the complexity of the quorum sensing hierarchy. With regard to the relationship of these cell-to-cell signals, our data imply that 3-oxo-C12-HSL is required for the production of PQS, and previous results suggested that 3-oxo-C12-HSL also positively regulates the production of C4-HSL (20, 21). These findings mean that 3-oxo-C12-HSL can be considered the dominant P. aeruginosa cell-to-cell signal that is responsible, with LasR, for initiating the start of the quorum sensing response. The relationship between PQS and C4-HSL is difficult to determine from our data. To act as signals, both molecules require RhlR. The C4-HSL signal has been shown to interact with RhlR (12), but it is not clear whether PQS acts directly or indirectly through RhlR. In either case, it is possible that PQS and C4-HSL produce an additive effect on the induction of lasB. Determining the relative position of PQS within the P. aeruginosa quorum sensing hierarchy will provide an interesting insight as to its role in the regulation of specific genes.
To facilitate the purification of PQS, we constructed a mutated lasR gene that encoded a truncated protein (ΔLasR) capable of activating LasR-controlled genes in the absence of 3-oxo-C12-HSL. PQS was purified by sequential reverse phase column chromatography and TLC from the double autoinducer mutant, strain PAO-JP2, that was expressing the ΔLasR protein. Based primarily on the spectral properties of the purified material (Figs. 4 and 5), we concluded that the novel signal molecule was 2-heptyl-3-hydroxy-4-quinolone (Fig. 1C). This compound was synthesized, and all chemical analysis (including high and low resolution MS, 1H NMR spectra, UV spectra, and TLC) showed that synthetic 2-heptyl-3-hydroxy-4-quinolone was identical to natural PQS.
As a confirmation of the structural assignment, we showed that synthetic PQS, but not other similar quinolones, had biological activity (Fig. 6). Based on the activity profile of synthetic PQS and the amount of signal we purified from P. aeruginosa culture fluid, we conservatively estimated that the concentration of PQS in culture fluid was ≈6 μM, which is in the active concentration range (Fig. 6). This concentration is similar to the approximate concentrations of 3-oxo-C12-HSL (1 to 5 μM; refs. 3 and 4) and C4-HSL (10 μM; ref. 4) found in culture supernatants. The amount of PQS required to activate our bioassay to half of its maximum activity was ≈30 μM (Fig. 6). Similar P. aeruginosa bioassays for 3-oxo-C12-HSL and C4-HSL showed that a concentration of ≈1 μM was required for these signals to activate their respective assays to half of their maximum activity (4). These differences suggest that, with regard to lasB′-lacZ induction, the relative potency of PQS may not be as high as the potencies of 3-oxo-C12-HSL and C4-HSL. However, we do not yet fully understand the biological significance of this 4-quinolone signal in P. aeruginosa. The conditions under which PQS may be involved in lasB induction and whether lasB or another gene is the primary target of PQS are some of the questions that remain to be answered.
It has been well established that 4-quinolones are secondary metabolites that can have potent antibiotic activity (28). It is now evident that at least one member of this family of molecules, 2-heptyl-3-hydroxy-4-quinolone, has a role in cell-to-cell signaling. The modern 4-quinolone antibiotics are halogenated at the 6 or 8 position and are commonly used to treat a variety of both Gram-positive and Gram-negative infections (29). These molecules are bactericidal and are believed to target DNA gyrase, which can be naturally mutated in P. aeruginosa to produce resistant strains (30, 31). Despite the development of this resistance phenotype, some 4-quinolone antibiotics have a secondary effect in resistant strains. The synthesis of a number of virulence factors, many of which (including elastase) are controlled by quorum sensing, is reduced by 4-quinolone antibiotics (32). This finding is supported by in vivo studies in which 4-quinolone antibiotic therapy has been shown to reduce lung damage in rats without decreasing the bacterial population (33). The mechanisms of such activities are unknown; however, our results lead us to theorize that 4-quinolone antibiotics may interfere with 4-quinolone signaling and gene activation. Confirmation of such a hypothesis would suggest that 4-quinolone signal molecules may be a viable target for the development of antibacterial therapies. It is also interesting to note that many synthetic 4-quinolones are exported by active efflux (reviewed in ref. 34) and recent reports show that active efflux is involved in the export of the P. aeruginosa autoinducer 3-oxo-C12-HSL (35, 36). This leads to the speculation that PQS may be exported by active efflux and that this transport mechanism could be involved in multiple cell-to-cell signaling pathways.
P. aeruginosa produces a myriad of secondary metabolites, most of which (including 4-quinolones) were discovered because they possess an observable bioactivity such as antibacterial or phytotoxic activity (28). PQS does not have detectable anti-Staphylococcus aureus or anti-E. coli activity (data not shown). However, this molecule showed activity as an intercellular signal that induced the P. aeruginosa virulence gene lasB. These findings lead to the conclusion that acyl-homoserine lactones represent one type of cell-to-cell signal in P. aeruginosa but that there are also other types of intercellular signals involved in the regulation of virulence gene expression. It has previously been shown that other Gram-negative bacteria produce quorum sensing signals that are not acyl-homoserine lactones. Ralstonia solanacearum has been shown to produce a fatty acid methyl ester that acts as a signal (37), and both E. coli and Salmonella typhimurium produce an unidentified intercellular signal that appears to not be an acyl-homoserine lactone (38). The discovery that 4-quinolones can act as intercellular signals demonstrates that bacterial cell-to-cell signals are an increasingly diverse group of molecules. The elucidation of the mechanism by which PQS regulates lasB should be an important first step toward understanding the role of this type of signaling in P. aeruginosa virulence.
Acknowledgments
We thank K. D. Tucker for the construction of pKDT39 and C. J. Smith, C. Van Delden, and C. S. Pesci for help in manuscript preparation and thoughtful insight. This work was supported by National Institutes of Health Grant R01-AI33713 (to B.H.I. and A.S.K.), Cystic Fibrosis Foundation Research Fellowship Grants PESCI96FO and PESCI99I0 (to E.C.P.), and National Institutes of Health Predoctoral Training Grant 5-T32AI07362 (to J.P.P.). E.P.G. was supported by grants from the Cystic Fibrosis Foundation (GREENB97ZO) and the National Science Foundation (MCB-9808308).
ABBREVIATIONS
- PQS
Pseudomonas quinolone signal
- 3-oxo-C12-HSL
N-(3-oxododecanoyl)-l-homoserine lactone
- C4-HSL
N-butyryl-l-homoserine lactone
- β-gal
β-galactosidase
References
- 1.Fuqua W C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Pesci E C, Iglewski B H. In: Cell-Cell Signaling in Bacteria. Dunny G, Winans S C, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 147–155. [Google Scholar]
- 3.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson J P, Passador L, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 6.Gambello M J, Iglewski B H. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsek M R, Schaefer A L, Greenberg E P. Mol Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 8.Ochsner U A, Koch A K, Fiechter A, Reiser J. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morihara K. J Bacteriol. 1964;88:745–757. doi: 10.1128/jb.88.3.745-757.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, et al. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brint J M, Ohman D E. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson J P, Pesci E C, Iglewski B H. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway B W, Krishnapillai V, Morgan A F. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohman D E, Cryz S J, Iglewski B H. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 18.Laschober R, Stadlbauer W. Liebigs Ann. Chem. 1990. 1083–1086. [Google Scholar]
- 19.Miller J A. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 352–355. [Google Scholar]
- 20.Pesci E C, Pearson J P, Seed P S, Iglewski B H. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 22.Stevens A M, Dolan K M, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behrman E J, Kiser R L, Garas W F, Behrman E C, Pitt B M. J. Chem. Res. 1995. M 1051–1063. [Google Scholar]
- 24.Spence T W M, Tennant G. J Chem Soc. 1971;C:3712–3719. [Google Scholar]
- 25.Smith W E. J Org Chem. 1972;37:3973–3974. [Google Scholar]
- 26.Morgan L R, Jr, Schunior R J, Boyer J H. J Org Chem. 1963;28:260–261. [Google Scholar]
- 27.Somanathan R, Smith K M. J Heterocyclic Chem. 1981;18:1077–1079. [Google Scholar]
- 28.Leisinger T, Margraff R. Microbiol Rev. 1979;43:422–442. doi: 10.1128/mr.43.3.422-442.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith J T, Zeiler H-J. In: Quinolone Antibacterials. Kuhlmann J, Dalhoff A, Zeiler H-J, editors. Berlin: Springer; 1998. pp. 1–11. [Google Scholar]
- 30.Maxwell A, Critchlow S E. In: Quinolone Antibacterials. Kuhlmann J, Dalhoff A, Zeiler H-J, editors. Berlin: Springer; 1998. pp. 119–166. [Google Scholar]
- 31.Piddock L V J. Drugs. 1995;49, Suppl. 2:29–35. doi: 10.2165/00003495-199500492-00006. [DOI] [PubMed] [Google Scholar]
- 32.Grimwood K, To M, Rabin H R, Woods D E. Antimicrob Agents Chemother. 1989;33:41–47. doi: 10.1128/aac.33.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimwood K, To M, Rabin H R, Woods D E. J Antimicrob Chemother. 1989;24:937–945. doi: 10.1093/jac/24.6.937. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido H. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans K L, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson J P, Van Delden C, Iglewski B H. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flavier A B, Clough S J, Schell M A, Denny T P. Mol Microbiol. 1997;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- 38.Surrette M G, Miller M B, Bassler B L. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]