Abstract
The proteolytic cleavage of sterol regulatory element-binding proteins (SREBPs) is regulated by SREBP cleavage-activating protein (SCAP), which forms complexes with SREBPs in membranes of the endoplasmic reticulum (ER). In sterol-depleted cells, SCAP facilitates cleavage of SREBPs by Site-1 protease, thereby initiating release of active NH2-terminal fragments from the ER membrane so that they can enter the nucleus and activate gene expression. In sterol-overloaded cells, the activity of SCAP is blocked, SREBPs remain bound to membranes, and transcription of sterol-regulated genes declines. Here, we provide evidence that sterols act by inhibiting the cycling of SCAP between the ER and Golgi. We use glycosidases, glycosidase inhibitors, and a glycosylation-defective mutant cell line to demonstrate that the N-linked carbohydrates of SCAP are modified by Golgi enzymes in sterol-depleted cells. After modification, SCAP returns to the ER, as indicated by experiments that show that the Golgi-modified forms of SCAP cofractionate with ER membranes on density gradients. In sterol-overloaded cells, the Golgi modifications of SCAP do not occur, apparently because SCAP fails to leave the ER. Golgi modifications of SCAP are restored when sterol-overloaded cells are treated with brefeldin A, which causes Golgi enzymes to translocate to the ER. These studies suggest that sterols regulate the cleavage of SREBPs by modulating the ability of SCAP to transport SREBPs to a post-ER compartment that houses active Site-1 protease.
Keywords: sterol regulatory element-binding proteins, cholesterol, sterol-sensing domain, vesicular transport, glycosidases
Feedback control of cholesterol metabolism is maintained by the regulated release of sterol regulatory element-binding proteins (SREBPs) from cell membranes (1). SREBPs are a family of membrane-bound transcription factors that activate genes involved in the synthesis of cholesterol and its uptake from plasma lipoproteins. SREBPs are released proteolytically from cell membranes in a process that is regulated by a polytopic membrane protein called SCAP, which stands for SREBP cleavage-activating protein (2). SCAP forms a complex with SREBPs in endoplasmic reticulum (ER) membranes (3). Formation of this complex is required for SREBPs to be cleaved by Site-1 protease (S1P), a membrane-bound serine protease that cleaves the SREBPs at a leucine–serine bond within a hydrophilic loop that projects into the ER lumen (4). Cleavage at this site initiates a process by which the transcriptionally active fragments of SREBP leave the membrane and translocate to the nucleus (1).
When cells are overloaded with sterols, SREBP is no longer cleaved by S1P. SREBP remains membrane-bound, and sterol synthesis is suppressed. Sensitivity of SREBP cleavage to sterols is mediated by the polytopic membrane attachment domain of SCAP (2, 5, 6). This domain is believed to contain eight membrane-spanning helices, five of which comprise a region termed the sterol-sensing domain (7). Specific point mutations within this domain abolish sensitivity to sterol suppression and render cleavage of SREBP constitutive (2, 5, 6).
The mechanism by which sterols control the activity of SCAP currently is under intensive investigation. A clue emerged recently from studies of the N-linked carbohydrate chains on the polytopic membrane attachment domain of SCAP (6). When cells were overloaded with sterols, these carbohydrate chains were largely in a form that is sensitive to digestion with endoglycosidase H (endo H), which recognizes high mannose carbohydrate chains that are found on ER proteins (8). When cells were deprived of sterols, the N-linked carbohydrate chains of SCAP became resistant to endo H treatment, suggesting that SCAP had moved from the ER to the Golgi complex, where it had been acted upon by α-mannosidase II. Mutant forms of SCAP that produce constitutive cleavage of SREBPs were always endo H-resistant, even in the presence of sterols (6). These data generated the hypothesis that sterols control the exit of SCAP from the ER. In sterol-depleted cells, the SCAP/SREBP complex can move to a post-ER compartment, where it encounters active S1P. In sterol-treated cells, this movement is abolished, and SREBP is retained in the ER, where it is not susceptible to Site-1 cleavage.
The current studies were designed to test further the hypothesis that sterols regulate the movement of SCAP. We perform a series of studies with a mutant cell line and glycosidase inhibitors that show directly that the endo H resistance of SCAP is attributable to its modification by Golgi enzymes. Nevertheless, in the steady state the bulk of SCAP is contained in ER membranes as revealed by density gradient ultracentrifugation, indicating that in sterol-depleted cells SCAP cycles between ER and Golgi. These data support the hypothesis that Site 1 cleavage of SREBPs occurs normally in a post-ER compartment and that sterols regulate the ability of SCAP to escort SREBP from the ER to this compartment.
MATERIALS AND METHODS
Materials.
We obtained monoclonal anti-BiP antibody and polyclonal anticalnexin antibody from StressGen Biotechnologies (Victoria, Canada); monoclonal antitransferrin receptor antibody from Zymed; swainsonine from Genzyme; kifunensine from Toronto Research Chemicals (Downsview, ON, Canada); brefeldin A from Calbiochem; endo D from Boehringer Mannheim; endo H, peptide N-glycosidase F (PNGase F), and Clostridium perfringens neuraminidase from New England Biolabs; and Nycodenz from Sigma. Other reagents were obtained from previously reported sources (6, 9).
Cell Culture.
All cells were grown in monolayer at 37°C in an atmosphere of 8–9% CO2. Chinese hamster ovary (CHO)-7 cells, a clone of CHO-K1 cells adapted to growth in lipoprotein-deficient serum (9), were grown in medium A (a 1:1 mixture of Ham’s F-12 medium and DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 5% (vol/vol) newborn calf lipoprotein-deficient serum. Clone 15B cells, a mutant CHO cell line deficient in N-acetylglucosamine (GlcNAc) transferase I activity (10, 11) (kindly provided by Stuart Kornfeld, Washington University, St. Louis, MO), were grown in medium A supplemented with 5% FCS. Cells were set up for experiments on day 0 at a density of 5 × 105 cells per 100-mm dish, refed on day 2, and harvested on day 3.
Isolation of Cell Membranes and Trypsin Treatment.
Cell monolayers were harvested, and membrane fractions were prepared and treated with trypsin as described previously (6). Briefly, cell pellets were resuspended in 0.4 ml of buffer B (10 mM Hepes⋅KOH, pH 7.4/10 mM KCl/1.5 mM MgCl2/5 mM sodium EDTA/5 mM sodium EGTA/250 mM sucrose), passed through a 22-gauge needle, and centrifuged at 1,000 × g for 5 min. The postnuclear supernatants then were centrifuged at 15,000 × g for 10 min, and the resulting membrane pellets were resuspended in 0.1 ml of buffer C (buffer B containing 100 mM NaCl). Equal amounts of protein then were incubated in the absence or presence of 1 μg of trypsin in a total volume of 58 μl for 30 min at 30°C. Reactions were stopped by addition of 2 μl of soybean trypsin inhibitor (400 units).
Glycosidase Treatments.
Cells were harvested, and membrane fractions were prepared and treated with trypsin as described above. For subsequent treatment with endo H, individual samples received 10 μl of solution containing 3.5% (wt/vol) SDS and 7% (vol/vol) 2-mercaptoethanol. After heating at 100°C for 10 min, each sample received sequential additions of 9 μl of 0.5 M sodium citrate (pH 5.5), 5 μl of solution containing 17× protease inhibitors (a concentration of 1× corresponding to 10 μg/ml leupeptin, 5 μg/ml pepstatin A, and 2 μg/ml aprotinin), followed by 1 μl of endo H (0.05 units). For treatment with PNGase F, trypsin-treated samples were denatured in the presence of SDS and 2-mercaptoethanol as described above and then received sequential additions of 7 μl of 0.5 M sodium phosphate (pH 7.5), 7 μl of solution containing 10% (vol/vol) Nonidet P-40 and 12× protease inhibitors, followed by 1 μl of PNGase F (7.7 × 10−3 units). For treatment with neuraminidase or endo D, membranes were incubated with trypsin as described above and then received sequential additions of 5 μl of solution containing 17× protease inhibitors and 8.5 μl of 10% (vol/vol) Triton X-100. After rocking at 4°C for 1 hr, the samples received 9 μl of 0.5 M sodium citrate (pH 5.5) and 1 μl of neuraminidase (50 units) or endo D (10−3 units). All reactions were carried out overnight at 37°C and stopped by addition of 20 μl of buffer D [0.25 M Tris⋅HCl, pH 6.8/2% SDS/10% (vol/vol) glycerol/0.05% (wt/vol) bromophenol blue/4% 2-mercaptoethanol]. The mixtures then were heated at 100°C for 5 min and subjected to SDS/PAGE (5–12% gradient gels).
Density Gradient Centrifugation.
Culture dishes with monolayers of CHO-7 cells were placed on ice and washed once with 5 ml of PBS and once with 5 ml of buffer E (10 mM triethanolamine⋅acetic acid, pH 7.4/0.25 M sucrose/1 mM sodium EDTA/1× protease inhibitors). Pooled cells from 40 dishes then were scraped into 0.8 ml of buffer E, followed by homogenization and cell fractionation on preformed Nycodenz gradients as described by Hammond and Helenius (12). The gradients were centrifuged for 45 min in an SW 41 rotor (Beckman) at 4°C at 37,000 × g, deceleration was performed without brake, and 19 fractions (600 μl/fraction) were collected from each gradient. The bottom three fractions contained aggregated material and were not analyzed further. Aliquots of each fraction (80 μl) received 20 μl of buffer F (150 mM Tris⋅HCl, pH 6.8/15% SDS/25% glycerol/12.5% 2-mercaptoethanol/0.02% bromophenol blue), were heated at 100°C for 5 min, and then were subjected to 7% SDS/PAGE. Additional aliquots from each fraction (50 μl) were used to determine Golgi α-mannosidase II activity as described (13).
For combined trypsin and endo H treatments of gradient fractions, 120-μl aliquots from each of two consecutive fractions were pooled and incubated with 5 μg of trypsin in a total volume of 250 μl for 30 min at 30°C. Reactions were stopped by the addition of 5 μl of soybean trypsin inhibitor (1,000 units). Aliquots (0.2 ml) from each reaction were then diluted by the addition of 1 ml of buffer E and centrifuged at 2 × 105 g for 45 min in a Beckman TLA 100.2 rotor at 4°C. The resulting pellets were dissolved in 0.1 ml of solution containing 0.5% SDS and 1% 2-mercaptoethanol, heated at 100°C for 10 min, and supplemented with 12 μl of 0.5 M sodium citrate (pH 5.5) and 8 μl of solution containing 15× protease inhibitors. Subsequently, each sample was split into two 60-μl aliquots and incubated overnight at 37°C in the absence or presence of 0.05 units of endo H. Reactions were stopped by the addition of 20 μl of buffer D. The mixtures then were heated at 100°C for 5 min and subjected to SDS/PAGE (5–12% gradient gels).
Immunoblot Analysis.
mAb IgG-9D5 directed against amino acids 540–707 of hamster SCAP was prepared as described previously (3). All other antibodies were obtained from commercial sources as described above. Gels were calibrated with molecular weight markers (Amersham or Bio-Rad). After SDS/PAGE, proteins were transferred to Hybond C-extra nitrocellulose sheets (Amersham) and incubated with antibodies at the indicated concentration. Bound antibodies were visualized by chemiluminescence and exposed to Kodak X-Omat Blue XB-1 film at room temperature as described previously (6). To quantify the distribution of calnexin and SCAP after density gradient centrifugation, immunoblots were scanned with a Hewlett–Packard ScanJet 4c/T page scanner and quantified by densitometry by using nih image 1.60 software. Signals from simultaneously exposed standard curves were used for calibration.
RESULTS
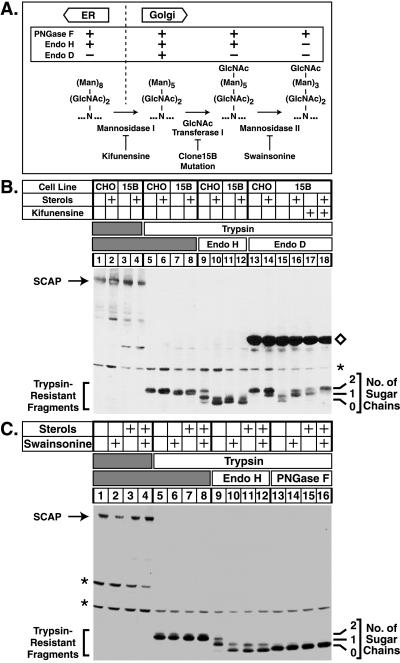
The experiments in Fig. 1 were designed to determine whether the sterol-regulated modifications of the N-linked oligosaccharides on SCAP from endo H-sensitive to endo H-resistant structures are attributable to the actions of Golgi enzymes. N-linked oligosaccharides of proteins residing in the ER have a configuration that is sensitive to endo H. As glycoproteins are transported to the Golgi, their sugars are successively modified by α-mannosidase I, GlcNAc transferase I, and α-mannosidase II (8) (see Fig. 1A). The reaction catalyzed by the latter enzyme gives rise to an oligosaccharide structure that is resistant to digestion by endo H (14). If SCAP were transported to the Golgi, inhibition of any of these steps should prevent SCAP from becoming resistant to endo H.
Figure 1.
Sterol-regulated modifications of SCAP oligosaccharides are catalyzed by Golgi-specific enzymes. (A) Schematic representation of enzymatic steps involved in conversion of high mannose to complex N-linked oligosaccharides. The table above the flow chart indicates sensitivities of individual N-linked sugars to hydrolysis by PNGase F, endo H, and endo D. Kifunensine is an inhibitor of Golgi α-mannosidase I, and swainsonine is an inhibitor of Golgi α-mannosidase II (15). Clone 15B cells lack GlcNAc transferase I activity (11). N, asparagine; Man, mannose. (B) On day 0, CHO-7 cells (CHO) or Clone 15B cells (15B) were set up in medium A supplemented with 10% FCS and 50 μg protein/ml human low density lipoprotein. On day 2, cells were switched to medium A containing 10% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% (vol/vol) ethanol in the absence or presence of sterols (1 μg/ml 25-hydroxycholesterol plus 10 μg/ml cholesterol) and kifunensine (2.5 μg/ml) as indicated. After incubation at 37°C for 16 hr, cells were harvested, and membrane fractions were prepared as described under Materials and Methods. Aliquots of membrane fractions (45 μg protein) were incubated in the absence (lanes 1–4) or presence (lanes 5–18) of trypsin. Proteolysis was stopped, the samples were incubated in the absence (lanes 1–8) or presence (lanes 9–18) of the indicated glycosidase, subjected to SDS/PAGE (5–12% gel), transferred to nitrocellulose, blotted with 10 μg/ml IgG-9D5 (anti-SCAP), and exposed to film for 45 sec. (C) On day 0, CHO-7 cells were set up as described above. On day 2, cells were switched to medium A containing 10% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence or presence of sterols and swainsonine (5 μg/ml) as indicated. After incubation for 16 hr, cells were harvested as above. Aliquots of membrane fractions (55 μg protein) were incubated in the absence (lanes 1–4) or presence (lanes 5–16) of trypsin. Proteolysis was stopped, the samples were incubated in the absence (lanes 1–8) or presence (lanes 9–16) of the indicated glycosidase, subjected to SDS/PAGE, transferred to nitrocellulose, and blotted as above. The filter was exposed to film for 20 sec. Numbers on the right denote the number of N-linked sugar chains present on protease-protected SCAP fragments. ⋄, Endo D protein, which cross-reacts with IgG-9D5; ∗, cross-reacting proteins of unknown identity.
The experimental approach is based on our previous observation that SCAP contains a glycosylated luminal loop of ≈170 aa that is protected from proteolysis when intact membranes are treated with trypsin. This loop carries two N-linked oligosaccharides (6, 7). After digestion with trypsin, the trypsin-resistant fragment can be visualized by SDS/PAGE and immunoblotting as a 36-kDa band (6, 7). The structures of the two sugar chains on this fragment can be analyzed by their susceptibility to hydrolysis by specific endoglycosidases (see Fig. 1A).
In the experiment in Fig. 1B, we compared the structures of the sugar chains on SCAP from wild-type CHO-7 cells and Clone 15B cells, a CHO-derived cell line that lacks GlcNAc transferase I activity (10, 11). These cells cannot produce endo H-resistant sugars; instead, they accumulate secretory glycoproteins carrying N-linked oligosaccharides of the (Man)5-(GlcNAc)2-Asn form (11), a structure that is uniquely susceptible to hydrolysis by endo D (14). Cells were grown in the absence or presence of sterols, and membrane fractions were treated successively with trypsin and glycosidases. In the absence of trypsin, SCAP migrated as a 150-kDa band in both cell lines and there was no effect of sterols (lanes 1–4). Trypsin treatment generated a resistant fragment of ≈36 kDa that was not affected by genetic background or sterols (lanes 5–8). This fragment is known to contain two N-linked sugar chains (6). In cells grown without sterols, endo H treatment left one or two of these chains intact, indicating that all of the SCAP fragments contained at least one endo H-resistant chain (lane 9). When wild-type cells were grown in the presence of sterols, endo H treatment produced SCAP fragments that had either zero or one sugar chain, indicating that all of the fragments contained at least one endo H-sensitive chain (lane 10). By contrast, the sugar chains on SCAP from GlcNAc transferase I-deficient Clone 15B cells were predominantly endo H-sensitive in cells grown either in the absence or presence of sterols (lanes 11 and 12). This result demonstrates that processing of SCAP from an endo H-sensitive to an endo H-resistant form requires the activity of GlcNAc transferase I. To rule out the possibility that SCAP from Clone 15B cells is not transported normally to the Golgi, we treated these membranes with endo D (lanes 13–18). SCAP from Clone 15B cells grown in the absence of sterols acquired endo D-sensitive sugars (lane 15), whereas SCAP from cells grown in the presence of sterols remained in an endo D-resistant form (lane 16). Processing to an endo D-sensitive form was inhibited when cells were incubated with kifunensine, a specific inhibitor of α-mannosidase I (lane 17) (ref. 15).
In the experiment in Fig. 1C, wild-type CHO-7 cells were grown in the absence or presence of sterols and swainsonine, a specific inhibitor of α-mannosidase II (15). In cells grown in the absence of sterols, SCAP was found in an endo H-resistant form, and this processing was inhibited by swainsonine (Fig. 1C, compare lanes 9 and 10). In this experiment, we also treated the trypsin-resistant SCAP fragment from wild-type cells with PNGase F, which hydrolyzes N-linked carbohydrates whether or not they have been modified in the Golgi. PNGase F removed both N-linked sugar chains whether or not the cells were incubated with sterols (lanes 13–16). Taken together, the results of Fig. 1 B and C demonstrate that the processing of SCAP to an endo H-resistant form requires the successive action of α-mannosidase I, GlcNAc transferase I, and α-mannosidase II, all of which are Golgi enzymes (16, 17).
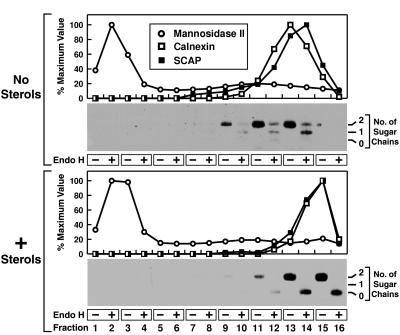
We next sought to determine the subcellular localization of SCAP through density gradient ultracentrifugation. In Fig. 2 postnuclear supernatants from CHO-7 cells grown in the absence or presence of sterols were subjected to Nycodenz density gradient centrifugation, and individual fractions were analyzed for the presence of SCAP as well as Golgi and ER-specific markers. Surprisingly, SCAP colocalized with the ER-specific marker calnexin (12, 18) in cells grown either in the absence or presence of sterols (Fig. 2 Upper and Lower, respectively). Immunoblots of the same samples treated with trypsin and endo H showed that SCAP from cells grown without sterols carried endo H-resistant sugar chains even though the protein cofractionated with ER markers. Thus, in the steady state most SCAP appears to reside in the ER both in the absence and in the presence of sterols. This suggests that sterol deprivation allows a transient translocation of SCAP from the ER to the Golgi, whereupon the protein quickly recycles back to the ER.
Figure 2.
Subcellular localization of SCAP as determined by density gradient centrifugation. On day 0, CHO-7 cells were set up in medium A supplemented with 10% FCS. On day 2, cells were switched to medium A containing 10% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence (no sterols) or presence (+ sterols) of 1 μg/ml 25-hydroxycholesterol plus 10 μg/ml cholesterol as indicated. After incubation at 37°C for 16 hr, cells were harvested, and postnuclear supernatants were fractionated by Nycodenz gradient centrifugation as described in Materials and Methods. Fractions were collected from top to bottom. Graphs show aliquots from individual fractions that were analyzed for α-mannosidase II activity. Additional aliquots were subjected to SDS/PAGE (5–12% gel), transferred to nitrocellulose, and blotted with 2 μg/ml anticalnexin antibody and 10 μg/ml IgG-9D5 (anti-SCAP). Immunoreactivity was detected by chemiluminescence and quantified by densitometry. Distributions are plotted as “% maximum value,” with 100% corresponding to the highest value of each measured parameter. Immunoblots show aliquots from consecutive fractions that were pooled and treated with trypsin as described in Materials and Methods. Reactions then were divided into two equal aliquots, which were incubated in the absence or presence of endo H as indicated, subjected to SDS/PAGE, and immunoblotted with 10 μg/ml IgG-9D5. Filters were exposed to film for 6 min (Upper) and 1 min (Lower), respectively. The apparent difference in the density of the ER fractions in the nontreated vs. sterol-treated panels is a result of variation in sample collection.
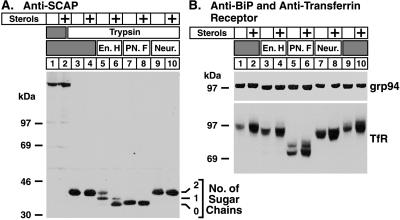
To determine how far SCAP moves into the Golgi in the absence of sterols, we analyzed the sugar chains on SCAP for their sensitivity to neuraminidase, a glycosidase that removes sialic acid residues from oligosaccharides (14). Addition of sialic acid occurs in the trans-Golgi (19). Sensitivity to neuraminidase thus is used as a marker to determine whether a protein has reached the trans-cisternae of the Golgi. As shown in Fig. 3A, SCAP became resistant to endo H in the absence of sterols (lane 5). However, the protein remained resistant to neuraminidase (lanes 9), indicating that it did not reach the trans-Golgi. To confirm that the neuraminidase was active under the conditions of our in vitro assay, we treated membrane extracts from CHO cells with neuraminidase, performed SDS/PAGE, and blotted with an antibody against the transferrin receptor, a sialylated glycoprotein of the plasma membrane (Fig. 3B Lower, lanes 7–10). The mobility of this receptor increased after neuraminidase treatment whether the cells were incubated in the absence or presence of sterols.
Figure 3.
Specificity of the sterol-regulated oligosaccharide modifications of SCAP. On day 0, CHO-7 cells were set up in medium A supplemented with 10% FCS. On day 2, cells were switched to medium A containing 10% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence or presence (+ sterols) of 1 μg/ml 25-hydroxycholesterol plus 10 μg/ml cholesterol as indicated. After incubation at 37°C for 16 hr, cells were harvested, and membrane fractions were prepared as described in Materials and Methods. (A) Aliquots (60 μg of protein) were incubated in the absence or presence of trypsin as indicated. Proteolysis was stopped, and the samples were incubated either in the absence (lanes 1–4) or presence (lanes 5–10) of the indicated glycosidase, subjected to SDS/PAGE (5–12% gel), and transferred to nitrocellulose. The filter was blotted with 10 μg/ml IgG-9D5 (anti-SCAP) and exposed to film for 25 sec. (B) Aliquots of the same membrane fractions used in A (16 μg of protein) were incubated without prior trypsin treatment either in the absence (lanes 1, 2, 9, and 10) or presence (lanes 3–8) of the indicated glycosidase, subjected to SDS/PAGE (6.5% gel), and transferred to nitrocellulose. Filters were blotted with 2 μg/ml anti-BiP antibody (Upper) and 1 μg/ml antitransferrin receptor antibody (Lower) and exposed to film for 5 sec and 2 sec, respectively. En. H, endo H; PN. F, PNGase F; Neur., neuraminidase; TfR, transferrin receptor.
To exclude the unlikely possibility that sterol deprivation caused Golgi enzymes to move to the ER, we analyzed the endo H sensitivity of grp94, an abundant luminal ER glycoprotein (20). As shown in Fig. 3B Upper, grp94 was sensitive to endo H both in the absence and presence of sterols. That grp94 is not intrinsically resistant to modification by Golgi enzymes was demonstrated by Lippincott-Schwartz et al. (21), who showed that grp94 acquires endo H-resistant sugars upon treatment of cells with brefeldin A, a fungal metabolite that causes rapid redistribution of lipids and proteins of the Golgi to the ER.
In principle, sterols could regulate the modification of SCAP by Golgi enzymes in two ways. According to one model, SCAP recycles between the ER and the Golgi both in the absence and presence of sterols, but its oligosaccharides become resistant to enzymatic modification in the presence of sterols. In a second model, SCAP remains in the ER in the presence of sterols. Upon induction by sterol deprivation, SCAP exits the ER, transiently moves to the Golgi, where its carbohydrates are modified, and then recycles back to the ER.
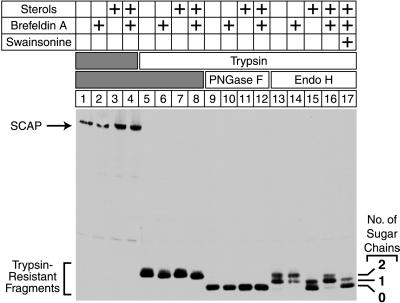
To distinguish between these two possibilities, we analyzed the sugar chains on SCAP after treatment of cells with brefeldin A. As shown in Fig. 4, SCAP was endo H-sensitive when cells were grown in the presence of sterols (lane 15). However, when cells were grown in the presence of both sterols and brefeldin A, SCAP was processed to the endo H-resistant form (lane 16), and this effect was inhibited by swainsonine (lane 17), indicating that brefeldin A-induced modification of SCAP required the action of the Golgi enzyme α-mannosidase II (see Fig. 1A).
Figure 4.
Brefeldin A induces endo H-resistance of SCAP in the presence of sterols. On day 0, CHO-7 cells were set up in medium A supplemented with 10% FCS. On day 2, cells were switched to medium A containing 10% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence or presence (+ sterols) of 1 μg/ml 25-hydroxycholesterol plus 10 μg/ml cholesterol as indicated. After incubation at 37°C for 10.5 hr, cultures received either no addition (lanes 1–16) or 5 μg/ml swainsonine (lane 17). After a 30-min preincubation, cells were incubated for an additional 5 hr with 0.1% (vol/vol) methanol in the absence or presence of 10 μg/ml brefeldin A as indicated. Cells were harvested, and membrane fractions were prepared as described in Materials and Methods. Aliquots (54 μg of protein) were incubated in the absence (lanes 1–4) or presence (lanes 5–17) of trypsin. Proteolysis was stopped, and the samples were incubated either in the absence (lanes 1–8) or presence (lanes 9–17) of the indicated glycosidase, subjected to SDS/PAGE (5–12% gel), and transferred to nitrocellulose. The filter was blotted with 10 μg/ml IgG-9D5 (anti-SCAP) and exposed to film for 2 min.
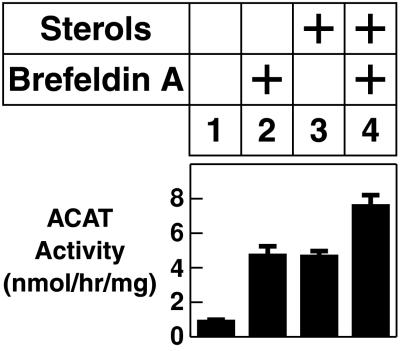
A trivial explanation for the ability of brefeldin A to block sterol suppression of SCAP processing would be that brefeldin A blocks the transport of the inhibitory sterols to the ER. To rule out this possibility, we measured the effect of brefeldin A on the ability of sterols to stimulate the conversion of cholesterol to cholesteryl esters. This reaction is catalyzed by the ER enzyme acyl-CoA:cholesterol acyltransferase, and its stimulation requires that cholesterol is transported to the ER (22). To make these measurements, CHO cells were incubated with or without sterols, and then they were labeled for 2 hr with [14C]oleate. As shown in Fig. 5, the rate of [14C]oleate incorporation into cholesteryl [14C]oleate was low when the cells were incubated in the absence of sterols (lane 1), and it rose 5-fold when sterols were present (lane 3). Brefeldin A alone led to an increase in cholesteryl [14C]oleate synthesis (lane 2), and there was a further increase when sterols were added (lane 4). These results demonstrate that the ER of brefeldin A-treated cells contains abundant sterols as shown earlier by Ridgway and Lagace (24).
Figure 5.
Effect of brefeldin A on sterol-regulated acyl-CoA:cholesterol acyltransferase activity. On day 0, CHO-7 cells were set up at 2.5 × 105 cells per 60-mm dish in medium A supplemented with 5% FCS. On day 2, cells were switched to medium A containing 5% newborn calf lipoprotein-deficient serum, 50 μM compactin, 50 μM sodium mevalonate, and 0.2% ethanol in the absence or presence (+ sterols) of 1 μg/ml 25-hydroxycholesterol plus 10 μg/ml cholesterol as indicated. After incubation at 37°C for 11 hr, the cells were incubated for an additional 5 hr with 0.1% methanol in the absence or presence of 10 μg/ml brefeldin A as indicated. During the final 2 hr, each monolayer was pulse-labeled with 0.2 mM sodium [14C]oleate (7,600 dpm/pmol), after which the cells were harvested for measurement of the rate of [14C]oleate incorporation into cholesteryl [14C]oleate as described previously (23). Error bars denote the SD of triplicate values.
DISCUSSION
The current experiments provide evidence that SCAP cycles between the ER and the medial-Golgi in sterol-deprived cells and that sterols block the exit of SCAP from the ER, thereby preventing SCAP from reaching the Golgi. That SCAP reaches the Golgi in sterol-deprived cells was suggested by the earlier demonstration that the N-linked carbohydrates of SCAP become resistant to endo H in sterol-deprived cells (6). In the current paper, this suggestion is supported by extensive studies of the N-linked carbohydrates of SCAP in wild-type CHO cells and in Clone 15B cells, which lack Golgi GlcNAc transferase I. Further documentation is provided by experiments with kifunensine (an inhibitor of α-mannosidase I), swainsonine (an inhibitor of α-mannosidase II), and endo D [specific for (Man)5-(GlcNAc)2-Asn-containing glycopeptides] (see Fig. 1A). The conclusion that sterols prevent this movement is based on the observation that the N-linked carbohydrates of SCAP are not fully modified by any Golgi-associated enzymes in sterol-treated cells. It is supported by the finding that SCAP becomes endo H-resistant in sterol-treated cells when the cells are treated with brefeldin A, which causes Golgi glycosidases to move to the ER.
In sterol-deprived cells, the endo H-resistant forms of SCAP cofractionate with ER markers and are well separated from Golgi markers on density gradients (Fig. 2), a result that is consistent with an earlier study showing ER localization of SCAP by immunofluorescence (3). These findings indicate that SCAP does not remain in the Golgi, but, rather, it cycles back to the ER. Endo H-resistant SCAP is also resistant to neuraminidase, suggesting that the protein returns to the ER before reaching the sialyltransferases in the trans-Golgi.
The finding that sterols control the exit of SCAP from the ER raises the crucial question as to whether this is the mechanism by which sterols control the proteolytic processing of SREBPs by S1P. Such control might be possible if the active form of S1P is located in a post-ER compartment and if SCAP escorts SREBP to this compartment in a sterol-regulated fashion. This escort mechanism is made more feasible by the previous finding that most cellular SCAP is present in a complex with SREBPs (3, 25), and, therefore, it should carry SREBPs with it when it leaves the ER.
The compartment that houses active S1P currently is unknown, but certain clues exist. S1P is synthesized as an inactive precursor designated S1P-A (26). This precursor undergoes autocatalytic cleavage to release an NH2-terminal propeptide, thereby converting itself to an active form designated S1P-B. S1P-B undergoes a further autocatalytic cleavage to generate S1P-C, which is also active. S1P-A and S1P-B are endo H-sensitive, indicating that they have not reached α-mannosidase II in the medial-Golgi. S1P-C is endo H-resistant, suggesting that it may be formed in the medial-Golgi (26).
Previous evidence indicates that SREBPs are cleaved in a pre-Golgi compartment. This conclusion was based on studies in which signals for N-linked glycosylation were inserted into the luminal loop of SREBP-2 (27). Cleavage by S1P occurred while the N-linked carbohydrates of SREBP-2 were still in an endo H-sensitive state. This pre-Golgi compartment might be the ER or it might be a post-ER compartment that is proximal to the medial-Golgi. Even though S1P-B is endo H-sensitive, it may also exist in a post-ER, pre-Golgi compartment. SCAP may transport SREBPs to this compartment in a sterol-regulated fashion. After SREBP is cleaved, SCAP must dissociate from the COOH-terminal fragment of SREBP so that it can return to the ER and form a new complex with another SREBP molecule. Studies to test this hypothesis are now underway.
Acknowledgments
We thank Mark Lehrman and Michael Roth for helpful discussions, Peter Espenshade for critical review of the manuscript, Kara Robinson and Deborah Morgan for excellent technical assistance, and Lisa Beatty for invaluable help with cultured cells. This work was supported by research funds from the National Institutes of Health (HL20948) and the Perot Family Foundation. R.A.D.-B. is the recipient of a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund. S.S. is the recipient of a fellowship from the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- SREBP
sterol regulatory element-binding protein
- SCAP
SREBP cleavage-activating protein
- CHO
Chinese hamster ovary
- endo
endoglycosidase
- ER
endoplasmic reticulum
- GlcNAc
N-acetylglucosamine
- PNGase F
peptide N-glycosidase F
- S1P
Site-1 protease
References
- 1.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 3.Sakai J, Nohturfft A, Cheng D, Ho Y K, Brown M S, Goldstein J L. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 4.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 5.Nohturfft A, Hua X, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1996;93:13709–13714. doi: 10.1073/pnas.93.24.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nohturfft A, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nohturfft A, Brown M S, Goldstein J L. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 9.Metherall J E, Goldstein J L, Luskey K L, Brown M S. J Biol Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- 10.Gottlieb C, Baenziger J, Kornfeld S. J Biol Chem. 1975;250:3303–3309. [PubMed] [Google Scholar]
- 11.Li E, Kornfeld S. J Biol Chem. 1978;253:6426–6431. [PubMed] [Google Scholar]
- 12.Hammond C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storrie B, Madden E A. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 14.Maley F, Trimble R B, Tarentino A L, Plummer T H., Jr Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 15.Kaushal G P, Elbein A D. Methods Enzymol. 1994;230:316–329. doi: 10.1016/0076-6879(94)30021-6. [DOI] [PubMed] [Google Scholar]
- 16.Dunphy W G, Brands R, Rothman J E. Cell. 1985;40:463–472. doi: 10.1016/0092-8674(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 17.Velasco A, Hendricks L, Moremen K W, Tulsiani D R P, Touster O, Farquhar M G. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochstenbach F, David V, Watkins S, Brenner M B. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth J, Taatjes D J, Lucocq J M, Weinstein J, Paulson J C. Cell. 1985;43:287–295. doi: 10.1016/0092-8674(85)90034-0. [DOI] [PubMed] [Google Scholar]
- 20.Wearsch P A, Nicchitta C V. Protein Expression Purif. 1996;7:114–121. doi: 10.1006/prep.1996.0015. [DOI] [PubMed] [Google Scholar]
- 21.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein J L, Dana S E, Brown M S. Proc Natl Acad Sci USA. 1974;71:4288–4292. doi: 10.1073/pnas.71.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 24.Ridgway N D, Lagace T A. J Biol Chem. 1995;270:8023–8031. doi: 10.1074/jbc.270.14.8023. [DOI] [PubMed] [Google Scholar]
- 25.Sakai J, Nohturfft A, Goldstein J L, Brown M S. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 26.Espenshade P J, Cheng D, Goldstein J L, Brown M S. J Biol Chem. 1999;274:22795–22804. doi: 10.1074/jbc.274.32.22795. [DOI] [PubMed] [Google Scholar]
- 27.Duncan E A, Brown M S, Goldstein J L, Sakai J. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]