Abstract
Many areas of biology and biotechnology have been revolutionized by the ability to label proteins genetically by fusion to the Aequorea green fluorescent protein (GFP). In previous fusions, the GFP has been treated as an indivisible entity, usually appended to the amino or carboxyl terminus of the host protein, occasionally inserted within the host sequence. The tightly interwoven, three-dimensional structure and intricate posttranslational self-modification required for chromophore formation would suggest that major rearrangements or insertions within GFP would prevent fluorescence. However, we now show that several rearrangements of GFPs, in which the amino and carboxyl portions are interchanged and rejoined with a short spacer connecting the original termini, still become fluorescent. These circular permutations have altered pKa values and orientations of the chromophore with respect to a fusion partner. Furthermore, certain locations within GFP tolerate insertion of entire proteins, and conformational changes in the insert can have profound effects on the fluorescence. For example, insertions of calmodulin or a zinc finger domain in place of Tyr-145 of a yellow mutant (enhanced yellow fluorescent protein) of GFP result in indicator proteins whose fluorescence can be enhanced severalfold upon metal binding. The calmodulin graft into enhanced yellow fluorescent protein can monitor cytosolic Ca2+ in single mammalian cells. The tolerance of GFPs for circular permutations and insertions shows the folding process is surprisingly robust and offers a new strategy for creating genetically encodable, physiological indicators.
The Aequorea green fluorescent protein (GFP) is a 238-aa, spontaneously fluorescent protein that has become spectacularly popular in molecular and cell biology as a transcriptional reporter, fusion tag, biosensor, or partner for fluorescence resonance energy transfer (FRET) (1). GFP has been fused to a very wide variety of proteins to render them fluorescent, but in almost every case the GFP, even if mutated to change colors or improve folding, simply has been appended to either the amino or carboxyl terminus of the host protein. In a few cases, GFP has been inserted inside the host protein, but even here the GFP-coding sequence has been kept essentially intact (2, 3). Random peptides up to 20 residues in length have been inserted at several locations within loops of GFP, but mostly with deleterious effects on GFP folding and fluorescence (4).
GFP is a seemingly monolithic, 11-stranded β-barrel that forms a nearly perfect shell around a chromophore spontaneously generated by an unusual multistep pathway involving cyclization and oxidation of residues 65–67 (1). Such a complex maturation process leading to a highly compact and rigid unit would seem very unlikely to tolerate major transpositions and insertions. Nevertheless, we now show that GFP can remain fluorescent despite a variety of circular permutations (5) and insertions of foreign proteins. When the insert itself is a conformationally sensitive receptor, ligand binding can strongly modulate the fluorescence of the GFP. Thus, insertion of receptors into GFP may offer a new strategy for generating genetically encoded indicators for important biochemical and physiological signals.
MATERIALS AND METHODS
Cloning and Gene Construction.
Enhanced yellow fluorescent proteins (EYFPs) with peptide insertions replacing Y145 were made by performing two separate PCRs on cDNA encoding EYFP, which is equivalent to GFP with the mutations S65G, V68L, Q69K, S72A, and T203Y, previously termed 10C Q69K (1, 6). The first PCR amplified the 5′ end of the EYFP cDNA and incorporated a 5′ BamHI site. The 3′ primer encoded the hexapeptide linker GGTGEL in place of Y145 and contained KpnI and SacI restriction sites for subsequent cloning. The second PCR amplified the 3′ portion of the EYFP cDNA with a 5′ primer coding for GGTGEL replacing Y145. The 3′ primer encoded an EcoRI site. These two PCR products were combined and amplified with N- and C-terminal EYFP primers to yield a full-length cDNA containing the insertion. The full-length cDNA was digested with BamHI and EcoRI, ligated, and cloned into the BamHI and EcoRI sites of pRSETB (Invitrogen) to yield the plasmid pYFPins. Next, cDNAs encoding Xenopus calmodulin (7) and the first zinc finger motif from zif268 (8) were amplified by PCR by using primers containing 5′ KpnI sites and 3′ SacI sites and then digested with KpnI and SacI. Finally, insertions into EYFPs were made by cloning cDNAs of inserted proteins between the KpnI and SacI sites of pYFPins.
Circular permutations with a new N terminus at Y145 were made by two separate PCRs. The first PCR amplified the 3′ end of enhanced cyan fluorescent protein (ECFP), enhanced green fluorescent protein (EGFP), or EYFP cDNA (5′ in the final permutated cDNA) and contained a 5′ BamHI site and a mutagenic codon for Y145M; the 3′ primer encoded the peptide linker GGTGGS and included a KpnI site. The second PCR amplified the 5′ end of the ECFP, EGFP, or EYFP cDNA with a 5′ primer coding for GGTGGS and a KpnI site; the 3′ primer contained an EcoRI site. The first PCR product was digested sequentially with BamHI and KpnI, the second PCR product was digested sequentially with EcoRI and KpnI, and the fragments were purified by agarose gel electrophoresis. The PCR fragments encoding the N and C termini then were cloned in a three-part ligation into the BamHI/EcoRI sites of pRSETB.
To construct a cameleon (7) (YC3.2) containing circularly permuted ECFP (cpECFP) instead of normal ECFP, the ECFP in YC3.1 was exchanged with cpECFP and cloned into the BamHI and EcoRI sites of pRSETB.
Random circular permutations were made after the general method of Graf and Schachman (9) with the following modifications. The cpGFP gene described above was amplified by PCR with primers that created a final PCR amplicon starting and ending at an XhoI site (ctcgag), which also coded for residues L141 and E142. The PCR product was digested with XhoI and cloned into the XhoI site of pBluescript (Invitrogen). This plasmid was amplified in bacteria, purified with a Qiagen maxi-prep, and digested with XhoI, and the ≈730-bp fragment was purified by agarose gel electrophoresis to yield a linear GFP cDNA. The linear fragment was circularized at a concentration of 5 μg/ml with 8,000 units/ml T4 DNA ligase (New England Biolabs) overnight at 16°C. After ethanol precipitation, the DNA was digested with DNase (100 units/mg DNA; Pharmacia) for 15 min at room temperature in 50 mM Tris⋅HCl, pH 7.5/1 mM MnCl2. Digestion was stopped by phenol extraction and then subsequent phenol/chloroform/isoamyl alcohol and chloroform/isoamyl alcohol extractions. The DNA was ethanol-precipitated, resuspended in 1× synthesis buffer (Stratagene), and incubated with T7 DNA polymerase (Stratagene) and T4 DNA ligase (Stratagene) at room temperature for 1 hr to repair DNA nicks and fill sticky ends. The linear, repaired, randomly permutated DNA library was purified by agarose gel electrophoresis and incorporated by blunt ligation into a triple-stop expression vector. This expression vector was made by ligating an oligonucleotide containing a 5′ EcoRV site and three downstream stop codons, one in each reading frame, between the BamHI and EcoRI sites of pRSETB. This vector (“pRSET triple stop”) was digested with EcoRV, treated with alkaline phosphatase, and purified by agarose gel electrophoresis.
Bacterial Transformation and Protein Purification.
Escherichia coli BL21(DE3) Gold (Stratagene) were transformed by electroporation in 10% glycerol with a ligation mixture (0.1-cm cuvette, 12.5 kV/cm, 200Ω, 25 μF). For protein expression, cells were grown to an OD600 of 0.6 in LB containing 100 mg/liter ampicillin, at which time they were induced with 1 mM isopropyl β-d-thiogalactoside. Bacteria were allowed to express recombinant protein for 6 hr at room temperature and then overnight at 4°C. The bacteria then were pelleted by centrifugation, resuspended in 50 mM Tris⋅HCl/300 mM NaCl, and lysed by a French press. The bacterial lysates were centrifuged at 30,000 × g for 30 min, and the proteins were purified from the supernatants with NiNTA resin (from Qiagen, used for purifying circularly permutated GFP and calmodulin insertions) or cobalt Talon resin (from CLONTECH, used for purifying zinc finger inserts).
Bacterial Colony Screening.
LB/agar plates containing ampicillin were screened by illuminating the plates with a 150-W Xe lamp and a 470 ± 20-nm excitation filter relayed through a pair of fiber optic light guides. Fluorescence was imaged by a cooled charge-coupled device camera (Sensys; Photometrics, Tucson, AZ) through a 530 ± 20-nm emission filter. Of approximately 25,000 bacterial colonies screened, about 200 became fluorescent after 24 hr at 4°C. Of these, 144 were picked for plasmid minipreps and restriction analysis. All plasmid minipreps were digested with HindIII and KpnI to analyze the site of permutated termini (HindIII cuts 3′ to the EGFP gene in pRSETB; KpnI cuts at the linker between the N and C termini of EGFP). Clones that gave restriction fragments of ≈750 bp or no visible fragments from 100 to 1,000 bp were considered to be regenerations of nearly wild-type sequence and were not investigated further. Clones that gave restriction fragments between 100 and 1,000 bp, but not 750 bp, were sequenced at their N and C termini to pinpoint the exact locations of new termini within the EGFP sequence.
Protein Titrations.
pH titrations were carried out in 125 mM KCl/20 mM NaCl/0.5 mM CaCl2/0.5 mM MgCl2/50 mM pH buffer. Buffers were chosen to span a wide pH range and included citrate (pH 4–5), 4-morpholineethanesulfonate (pH 5.5–6.5), Hepes (pH 7–8.15), glycine (pH 8.8–10.7), and phosphate (pH11.3–13.2). For each pH, a weakly buffered protein solution was mixed with an equal volume of the corresponding buffer solution and analyzed for total fluorescence in triplicate on a microplate fluorescence reader by using a 482 ± 10-nm excitation filter and a 532 ± 14-nm emission filter.
Calcium titrations of EYFP–calmodulin insertion proteins were done in a cuvette in a fluorescence spectrometer in 100 mM KCl/10 mM MOPS, pH 7.5 (buffer was run through a Chelex column to remove traces of calcium). Small aliquots of CaCl2 were added to this cuvette, and a full fluorescence emission spectrum was taken after each addition.
Zinc titrations of EYFP–zinc finger insertions were done in 50 mM Mops, pH 7.0. A fluorescence emission spectrum was taken of the protein (diluted from a stock solution stored with 1 mM DTT) with 50 μM EDTA, and then small aliquots of ZnCl2 were added as subsequent spectra were recorded.
Titration curves were generated by averaging the three intensity values for each pH (for microplate data) or by integrating the total emission intensity (for full spectra), plotting these data vs. analyte concentration, and fitting a sigmoidal curve to the data.
Mammalian Cell Expression and Imaging.
The cDNA encoding the EYFP–calmodulin insertion was cloned into the HindIII and EcoRI sites of the mammalian expression vector pCDNA3. HeLa cells were transfected with this construct through lipofection, grown overnight in DMEM at 37°C, and then grown at 28°C for 8–24 hr before imaging, which was performed at 22°C as described previously (7) but with excitation (480DF30) and emission (535DF25) filters (Omega) appropriate for single-emission-wavelength monitoring of EYFP.
RESULTS
During semirandom mutagenesis of ECFP to alter its pH sensitivity, we discovered a mutant that somehow had incorporated six new residues, FKTRHN, in place of Y145, yet remained fluorescent. This chance finding suggested that the two portions of ECFP on either side of Y145 might have some ability to fold autonomously, a hypothesis that we tested with two distinct types of manipulations: deliberate circular permutation (5) and insertion of foreign proteins. The constructs are schematized in Fig. 1.
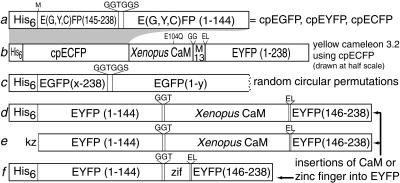
Figure 1.
Schematic structures of major new constructs. (a) Designed circular permutations of EGFP, EYFP, and ECFP starting at Y145M. His6 indicates the polyhistidine tag MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDP. Linkers and substitutions are shown above the main sequence. (b) Yellow cameleon 3.2 (YC3.2) incorporating cpECFP instead of ECFP. This sequence is drawn at half the scale of all the other constructs. M13 is the CaM-binding peptide derived from skeletal muscle myosin light chain kinase (7). (c) Random circular permutations of EGFP. The successful values of x and y are shown in Table 1. (d and e) Insertions of CaM in place of Y145 of EYFP as expressed in bacteria (d) for in vitro purification or in HeLa cells (e) for in situ monitoring of cytosolic Ca2+. kz, Kozak sequence (10) for optimal translation initiation. (f) Insertion of a zinc finger (zif), residues 334–362 of zif268 (8), in place of Y145 of EYFP.
Circular Permutations.
The original N and C termini of EGFP, ECFP, and EYFP were connected with the hexapeptide linker GGTGGS. The fluorescent protein sequences were begun at Y145, which was changed to M, and to which was prefixed an N-terminal polyhistidine tag to aid purification (Fig. 1a). N144 thus became the new C terminus. All the circularly permuted proteins had basically the same fluorescence spectra but a higher pKa (7.0, 8.3, and 8.8 for cpECFP, cpEGFP, and cpEYFP, respectively) than their native counterparts (6.4, 6.15, and 6.2 for nonpermuted ECFP, EGFP, and EYFP, respectively). The circular permutations and their parents also should differ in their spatial orientation relative to an appended protein, because amino acids 144–145 are at the opposite end of the β-barrel from the original N and C termini. To see whether the altered spatial orientation could be detected as a difference in FRET, we exploited yellow cameleons, in which ECFP and EYFP bracket calmodulin and a calmodulin target peptide so that FRET between the ECFP and EYFP is Ca2+-dependent (6, 7). Replacement of the ECFP in yellow cameleon 3.1 with Y145I-N144 cpECFP produced YC3.2 (Fig. 1b). The increase in the ratio of 528 to 476 nm emissions going from the Ca2+-free to the Ca2+-saturated states of YC3.2 was only 15%, much smaller than the ≈100% increase from YC3.1 (6). This reduction in FRET responsivity suggests that cpECFP indeed maintains a different orientation in space with respect to the rest of the cameleon than does ECFP, because other factors influencing FRET (interfluorophore distance and spectral overlap) probably have changed only very little. The lack of improvement in this case does not mean that cpGFPs will always decrease FRET efficiency. The observed effect is probably because the spacers in YC3.1 between the ECFP and calmodulin and between the M13 and YFP had been optimized extensively for maximal responsivity to Ca2+. Therefore, most alterations in orientation of the ECFP would probably be detrimental.
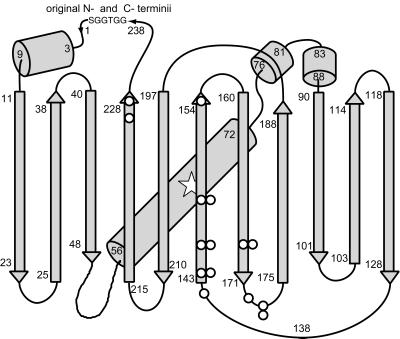
To test whether EGFP could tolerate circular permutations at sites other than Y145, we followed the systematic procedure of Graf and Schachman (9) to make random circular permutations. A circular cDNA encoding EGFP (1, 11) and the GGTGGS linker between the N and C termini was randomly nicked with DNase, made blunt-ended, cloned into the pRSETB expression vector with stop codons in all three possible reading frames, and screened for fluorescence as described in Materials and Methods. About 90% of the fluorescent colonies regenerated termini at or very near the original N and C termini, as judged by restriction fragment mapping. However, 10 nontrivial, circular permutations were found (Figs. 1c and 2), in which the original EGFP sequence was interrupted at E142, Y143, Y145, H148, D155, H169, E172, D173, A227, or I229 (Table 1). Interestingly, all these positions were in the original C-terminal half of EGFP (Fig. 2). Furthermore, some sites such as H148, H169, and A227 are in normally β-stranded segments, whereas others such as E142, Y143, and Y145 are in loops between β-strands. We think this list of functional circular permutations is likely to be nearly complete, because we examined 25,000 independent colonies, which should have been enough to saturate the 717 bp of the EGFP gene, even though only one in six of the blunt-ended DNAs would be expected to be in both the correct orientation and reading frame. Indeed, several of the new breakpoints appeared twice but with slightly different substitutions at the new termini, proving that they resulted from independent cloning events. Similarly, Y145 was recovered as a breakpoint from the random procedure, recapitulating the designed circular permutation but with a substitution proving its independent origin.
Figure 2.
Schematic drawing of the overall fold of GFP (12) modified to show starting points of fluorescent circular permutations (○), the linker (GGTGGS) connecting the original N and C termini, and the approximate location of the chromophore (open star, residues 65–67). Locations with two circles indicate where circular permutations with two different ending amino acids were isolated (Table 1).
Table 1.
Sequences and spectra of random, circular permutations of EGFP
| Starting a.a. | Ending a.a. | Excitation | Emission | 400:490 ratio |
|---|---|---|---|---|
| E142M | N144LSE | 404, 494 | 514 | 1.09 |
| Y143N | N146LSE | 410, 494 | 512 | 0.31 |
| Y143I | N144LSE | 404, 494 | 512 | 1.16 |
| Y145I | N144 | 488 | 514 | 0.20 |
| Y145M* | N144* | 487 | 512 | 0.23 |
| H148I | N149LSE | 488 | 510 | 0.08 |
| H148I | K162SE | 398, 490 | 512 | 0.35 |
| D1551 | K156SE | 400, 496 | 514 | 1.47 |
| H169H | N170LSE | 396, 490 | 514 | 1.47 |
| H169I | N170LSE | 396, 494 | 514 | 1.19 |
| E172M | I171DLSE | 398, 492 | 514 | 1.24 |
| D173I | D173LSE | 398, 494 | 514 | 1.38 |
| D173D | E172SE | 398, 492 | 514 | 1.33 |
| A227A | A227I | 400, 492 | 514 | 0.61 |
| I229I | I229 | 396, 492 | 514 | 1.21 |
Starting a.a. is the first amino acid (which may be mutated) after the constant polyhistidine tag, i.e., “x” in Fig. 1c. For example, E142M means that the cpGFP starts at amino acid 142, but the Glu has been changed to Met by the random ligation. Ending a.a. is the last GFP-derived amino acid, i.e., “y” in Fig. 1c, followed by the amino acids (if any) added by the expression vector. For example, N144LSE means that the GFP sequence ends with Asn at position 144, to which is appended the vector-encoded C-terminal tripeptide LSE. Wavelengths of fluorescence excitation and emission maxima are given in nanometers. When two excitation peaks exist, the major peak is underlined. 400:490 ratio is the ratio of the excitation amplitudes at 400 and 490 nm at pH 7.5.
Y145M–N144 is the original, rationally designed, circular permutation included for comparison.
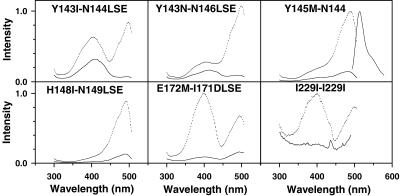
The excitation spectra of the above circular permutations usually consisted of two peaks at 396–404 and 490–496 nm (Fig. 3), closely corresponding to the two major excitation peaks for wild-type GFP. Thus, most of the circular permutations counteracted the S65T mutation (13) in the starting EGFP and generated significant populations of both the protonated and unprotonated chromophore from pH 6–8. The only exceptions were the two circular permutations at Y145 and one at H148, where the short-wavelength excitation peak from the protonated chromophore was absent or only a small shoulder. Small shifts in the position of the breakpoint or in the length of the terminations often changed drastically the ratio of the two excitation peaks, as shown by comparison of Y143I-N144LSE (Fig. 3 Upper Left) with either Y143N-N146LSE (Fig. 3 Upper Center) or Y145M-N144 (Fig. 3 Upper Right). However, only one emission at 512–514 nm was ever detectable, a property again reminiscent of wild-type GFP and presumably a result of fast proton transfer from the excited state (14).
Figure 3.
Excitation spectra of selected circular permutations of EGFP. Dotted lines (upper traces in each panel) are spectra taken at pH 8; solid lines (lower traces) are at pH 6 for the same concentration of protein. Spectra for different mutants were recorded with varying protein concentrations and gains, so amplitudes cannot be compared across permutations. The permutations are denoted by their starting and ending amino acids as in Table 1. An emission spectrum for Y145M–N144 is included in Upper Right; emission spectra for all the circular permutations were essentially identical in shape and wavelength.
All of the circular permutations were acid-quenched to a greater degree than EGFP or wild-type GFP. As an extreme case, I229–I229, which starts and ends at residue 229, lost essentially all of its fluorescence at pH 6 (Fig. 3 Lower Right). Also, most permutations showed considerable sensitivity of the ratio of 400 to 490-nm excitation amplitudes to pH, unlike the behavior of EGFP or wild-type GFP. For example, when lowering pH from 8 to 6, the circular permutations with new termini between residues 142 and 145 (Fig. 3 Upper) shifted from mostly deprotonated toward protonated chromophore spectra, as if the chromophores themselves were directly titrated by external pH. By contrast, proteins with new termini at residue 172 (Fig. 3 Lower Center) (plus 155, 169, and 173, data not shown) did the reverse; that is, their spectra corresponded to more deprotonated chromophores as pH was lowered. This paradoxical behavior was reminiscent of ratiometric “pHluorins” (15). Such reversed pH dependency could be explained if protonation of a nonchromophoric site causes a conformational change, shift in hydrogen bonding, or an electrostatic repulsion that displaces a proton from the phenolic hydroxyl of the chromophore. Finally, new termini at position 148 (Fig. 3 Lower Left) caused the excitation ratio to be relatively pH-insensitive, although the excitation amplitude remains strongly quenched by acid. Replacements of H148 by amino acids with smaller side chains in nonpermuted GFPs confer strong pH sensitivity (16).
The circularly permuted GFPs were more sensitive not only to acid but also to heat. Folding efficiency appeared to be good at 4° and 22°C in bacteria, adequate at 28–30°C in mammalian cells, but poor at 37°C (data not shown). For random screening in bacteria, we grew the colonies at 4°C to allow even poorly folding mutants to become fluorescent. A decrease in thermotolerance because of circular permutations is typical (5) and could indicate that the constant GGTGGS linker between the former N and C termini was not optimal. Recovery of folding at 37°C may require additional mutations, which need not be the same as those already known to aid nonpermutated GFPs (1).
ECFPs, EGFPs, and EYFPs with Insertions at Y145.
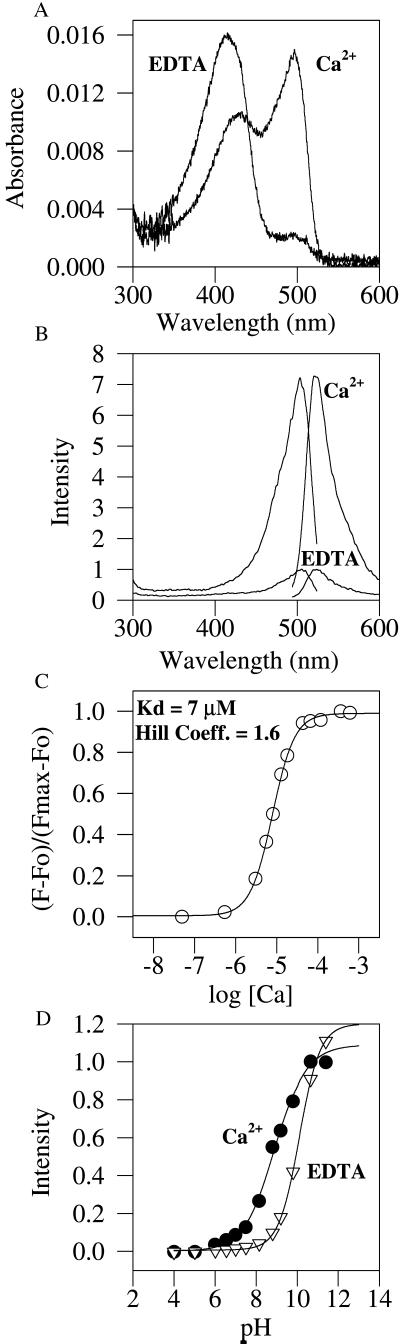
We have inserted a variety of full-length proteins at position 145 of ECFP, EGFP, or EYFP, such as Xenopus calmodulin, the S1 and S2 domains of the GluR6 glutamate receptor (17), and EYFP itself. Of these, only GluR6 led to a nonfluorescent fusion protein when inserted into EYFP (ECFP and EGFP were not tried). EYFP inserted into ECFP led to a fluorescent fusion protein in which each GFP mutant retained fluorescence and there was efficient FRET from the ECFP to the EYFP. Most interestingly, ECFP, EGFP, and EYFP all tolerated calmodulin insertion, and all resulted in Ca2+-sensitive fusion proteins. Preliminary comparisons indicated that EYFP with inserted calmodulin gave the largest responses to Ca2+, so this protein (Fig. 1d) was chosen for more detailed investigation.§ At pH 7.5, the Ca2+-free protein showed a dominant 400-nm absorbance peak (Fig. 4A) with a small shoulder at 490 nm, indicating that the YFP chromophore was predominantly protonated. Upon saturation with Ca2+ at the same pH, the 490-nm absorbance peak became dominant at the expense of the 400-nm peak, indicating that Ca2+ binding to calmodulin caused deprotonation of the chromophore. The excitation spectra at either zero or high Ca2+ (Fig. 4B) showed only the 490-nm peaks, consistent with previous findings that the 400-nm absorbing protonated species is nonfluorescent in EYFPs (18), unlike the dual-excitation peaks in many of the cpEGFPs (Fig. 3). Thus, both the excitation and emission spectra simply increased in amplitude by up to 7-fold upon saturation with Ca2+, without any significant shift in peak wavelength. Titration through intermediate values of buffered free [Ca2+] showed an apparent Kd for Ca2+ of 7 μM and a Hill coefficient of 1.6 (Fig. 4C), without any obvious inflection point separating the known low- and high-affinity sites (19). Because Ca2+ binding promoted deprotonation of the chromophore at constant pH, the Ca2+-free and Ca2+-bound proteins should show different pH titration curves. Fig. 4D confirms that they had apparent pKa values of 10.1 and 8.9, respectively.
Figure 4.
In vitro characterization of EYFP–calmodulin insert protein. (A) Absorbance spectra with EDTA and zero Ca2+, or with saturating Ca2+, both at pH 7.5 and the same protein concentration. (B) Fluorescence excitation and emission spectra obtained under the same conditions. (C) Titration at pH 7.5 with Ca2+ buffers, plotting fractional conversion to the Ca2+-bound state vs. the logarithm of the free-Ca2+ concentration in molar units. (D) Titrations of normalized fluorescence vs. pH with EDTA and zero Ca2+, or with saturating Ca2+. The midpoints of the titration curves are 10.1 in EDTA and 8.9 in excess Ca2+.
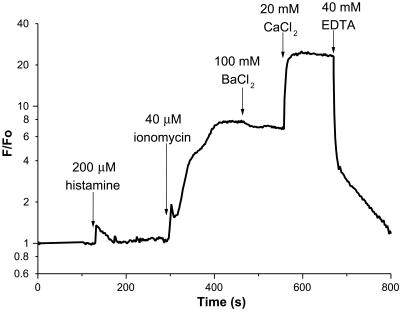
To show that the EYFP–calmodulin insertion is a viable calcium indicator within live cells, a version (Fig. 1e) without the polyhistidine tag but with an optimal eukaryotic translation initiation sequence (10) was transfected into HeLa cells and imaged in single cells. A relatively physiological stimulus such as histamine produced a brief increase in fluorescence of about 35% from baseline (Fig. 5), consistent with a low-affinity indicator reporting the histamine-induced [Ca2+]i spike (6). In one cell, we observed a brief period of oscillatory fluorescence after histamine application, with fluorescence increases of 10–40% observed on each oscillation (data not shown). The addition of ionomycin in medium containing 2 mM CaCl2 led to an 8-fold increase in brightness, in fairly good agreement with the fluorescence enhancement obtainable in vitro (Fig. 4C). BaCl2 (100 mM) was added to check for quenching by Cl− (20), which did not seem significant. Further addition of 20 mM Ca2+ led to an additional 2.5-fold increase in brightness, which was due to transient alkalinization of the cytoplasm caused by ionomycin and high calcium. Parallel measurements with BCECF indicated that cytosolic pH remained neutral during histamine and ionomycin additions but then rose to ≈8.5 upon addition of 20 mM Ca2+.
Figure 5.
Fluorescence of a typical HeLa cell expressing cytosolic EYFP–calmodulin insert as a function of time after successive additions of reagents. The fluorescence normalized by the initial fluorescence F0 is plotted on a logarithmic scale.
We also examined an insertion of the first zinc finger from zif268 into EYFP (Fig. 1f), which increased in fluorescence about 1.7-fold on binding Zn2+, again without a change in wavelengths. Although this intensity change and the Zn2+ affinity (apparent Kd ≈ 0.4 mM) were modest, they indicate that the sensitivity of EYFP fluorescence to the conformation of an inserted receptor may be of some generality.
DISCUSSION
Despite the gross rearrangements of their primary sequence described above, GFPs can still fold correctly, generate internal chromophores, and protect their excited states from vibrational deactivation. Circular permutation is known (5) to be tolerated in a variety of proteins when the original N and C termini are fairly close in space, as is true for GFPs. If a protein consists of more than one autonomous domain loosely held together by flexible linkers, it is easy to imagine that its function could be preserved despite swapping the order of those domains. However, GFPs would seem poor candidates because of their monolithic cylindrical symmetry, intricate interdigitation of the 11 strands making up their β-barrels, complexity of their maturation process, and intolerance of significant N- and C-terminal truncations (21). Nevertheless, an unbiased genetic screen located at least 10 different sites (Table 1 and Fig. 2) within the EGFP sequence at which a permuted fluorescent protein could be initiated after a constant N-terminal purification tag. Currently, we have no explanation for the locations or varying phenotypes of the permissible permutations, but they should be interesting test cases for a future understanding of protein folding. Also, we do not yet know whether the increased acid sensitivity of the fluorescence was due to the circular permutations themselves, the attachment of the constant N-terminal polyhistidine tag, or to the nature of the linker joining the former N and C termini.
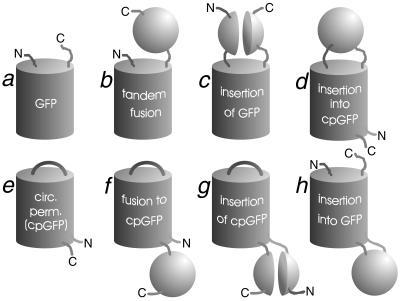
Fig. 6 summarizes the many topologies for merging the sequences of a host protein and a GFP or cpGFP. cpGFPs represent a library of mutants whose spatial orientations with respect to fusion partners should differ from those of regular GFP mutants. Thus, when conventional tandem fusions to GFPs are unsatisfactory either in the function of the host protein or in FRET to or from the GFP, a tandem fusion to a cpGFP (Fig. 6f) might be worth trying. The higher pKa values of cpGFP mutants might also be exploited to measure the pH in relatively alkaline compartments such as mitochondria. Like the pHluorins (15) and a recent nonpermuted GFP mutant, S65T/H148D (22), some of the circularly permuted EGFPs would offer excitation ratioing and might avoid the Cl− ion interference (20) and relatively easy photoisomerization (6) of the current high-pKa EYFPs.
Figure 6.
Topologies of GFP, cpGFP, and chimeras with other proteins. The other proteins are depicted schematically as spheres, when their sequences remain contiguous, or as paired hemispheres, when GFP or cpGFP is inserted within them. N and C denote amino and carboxyl termini of the full protein or chimera. Tandem fusions b and f arbitrarily show the carboxyl terminus of the GFP or cpGFP fused to the amino terminus of the other protein, but also are intended to encompass the opposite order. Topologies a, b, and c were already known; examples of e, f, and h are demonstrated in this paper.
Although we have focused on Y145 as the site for initial insertions of conformationally sensitive proteins and domains, the other nine breakpoints found by random circular permutation are also plausible sites at which proteins may likewise be inserted (Fig. 6h). In other cases, it may be preferable to insert the cpGFP sequence into the host (Fig. 6g), especially when the latter is a large, multidomain protein. The sensitivity of cpGFPs to the environment and to small changes in sequence around new termini ought to enhance the probability that their fluorescence will sense changes in conformations of proteins into which they have been spliced. Thus, we would predict that the new topologies (Fig. 6 f–h) generally will confer greater responsivity than the topologies that were known (Fig. 6 b–d).
The EYFP–calmodulin insertion (Fig. 1 d and e) has not yet been subjected to systematic mutational improvement and, therefore, is most significant as a proof that the fluorescence of GFPs can be made highly responsive to the conformation of inserted receptors. Its 7- to 8-fold enhancement of fluorescence upon Ca2+ binding is substantially larger than the ratiometric or single-wavelength dynamic range of any other genetically encodable fluorescent indicator of Ca2+ (6, 7, 23, 24). Its most fundamental limitation may be the pH sensitivity inherent in the current mechanism of modulating fluorescence via changes in pKa of the chromophore. Perhaps other insertion sites might modulate fluorescence by changing access of H2O or O2 without pH sensitivity. The lack of wavelength shifting might be remedied in an EGFP analog in which both the protonated and unprotonated species might be detectable at separate excitation wavelengths as in Fig. 3. Such excitation-ratioing indicators would complement the existing emission-ratioing cameleons and the intensity-only, EYFP-calmodulin insert. The 7-μM apparent Kd roughly matches the apparent Kd for Ca2+ binding to native calmodulin in the absence of a target protein or peptide (25) and should be tunable by further engineering.
We currently are investigating GFPs with kinase consensus sites, protease cleavage sites, glutamate-binding domains, and peptide loops containing multiple cysteines as examples of sensors for phosphorylation, protease activity, glutamate concentration, and redox potential, respectively. At present, we know that examples of all the above constructs are fluorescent, but have yet to determine optimal sequences and conditions for conformational modulation. Other tantalizing possibilities include improved membrane potential sensors, detectors of antibody–antigen interaction, and GFP-based complementation assays for protein–protein interactions analogous to previous examples based on dihydrofolate reductase (26) and β-galactosidase (27).
Acknowledgments
We thank Professor C. Zuker for helpful comments. This work was supported by the National Institutes of Health (Grant NS27177 to R.Y.T.) and the Howard Hughes Medical Institute.
ABBREVIATIONS
- GFP
Aequorea green fluorescent protein or mutant thereof
- FRET
fluorescence resonance energy transfer. ECFP, EGFP, and EYFP, enhanced cyan, green, and yellow fluorescent protein, respectively (in this paper, EYFP includes mutations V68L and Q69K in addition to the previous S65G, S72A, and T203Y)
- cp
circularly permuted
Footnotes
An apt nickname for this construct is “camgaroo1,” because it is yellowish, carries a smaller companion (calmodulin = CaM) inserted in its “pouch,” can bounce high in signal, and may spawn improved progeny.
References
- 1.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Siegel M S, Isacoff E Y. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 3.Biondi R M, Baehler P J, Reymond C D, Veron M. Nucleic Acids Res. 1998;26:4946–4952. doi: 10.1093/nar/26.21.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abedi M R, Caponigro G, Kamb A. Nucleic Acids Res. 1998;26:623–630. doi: 10.1093/nar/26.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinemann U, Hahn M. Prog Biophys Mol Biol. 1995;64:121–143. doi: 10.1016/0079-6107(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 6.Miyawaki A, Griesbeck O, Heim R, Tsien R Y. Proc Natl Acad Sci USA. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyawaki A, Llopis J, Heim R, McCaffery J M, Adams J A, Ikura M, Tsien R Y. Nature (London) 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 8.Christy B A, Lau L F, Nathans D. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graf R, Schachman H K. Proc Natl Acad Sci USA. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang T-T, Cheng L, Kain S R. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ormö M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 13.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 14.Chattoraj M, King B A, Bublitz G U, Boxer S G. Proc Natl Acad Sci USA. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miesenböck G, De Angelis D A, Rothman J E. Nature (London) 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 16.Wachter R M, Elsliger M-A, Kallio K, Hanson G T, Remington S J. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 17.Arvola M, Keinanen K. J Biol Chem. 1996;271:15527–15532. doi: 10.1074/jbc.271.26.15527. [DOI] [PubMed] [Google Scholar]
- 18.Llopis J, McCaffery J M, Miyawaki A, Farquhar M G, Tsien R Y. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjandra N, Bax A, Crivici A, Ikura M. In: Calcium as a Cellular Regulator. Carafoli E, Klee C, editors. New York: Oxford Univ. Press; 1999. pp. 152–170. [Google Scholar]
- 20.Wachter, R. M. & Remington, S. J. (1999) Curr. Biol., in press. [DOI] [PubMed]
- 21.Dopf J, Horiagon T. Gene. 1996;173:39–44. doi: 10.1016/0378-1119(95)00692-3. [DOI] [PubMed] [Google Scholar]
- 22.Elsliger M-A, Wachter R M, Hanson G T, Kallio K, Remington S J. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 23.Romoser V A, Hinkle P M, Persechini A. J Biol Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- 24.Persechini A, Lynch J A, Romoser V A. Cell Calcium. 1997;22:209–216. doi: 10.1016/s0143-4160(97)90014-2. [DOI] [PubMed] [Google Scholar]
- 25.Cox J A. Biochem J. 1988;249:621–629. doi: 10.1042/bj2490621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier J N, Campbell-Valois F-X, Michnick S W. Proc Natl Acad Sci USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]