Abstract
Based on the belief that marine larvae, which can spend days to months in the planktonic stage, could be transported considerable distances by ocean currents, it has long been assumed that populations of coastal species with a planktonic larval stage are demographically open and highly “connected.” Such assumptions about the connectivity of coastal populations govern approaches to managing marine resources and shape our fundamental understanding of population dynamics and evolution, yet are rarely tested directly due to the small size and high mortality of marine larvae in a physically complex environment. Here, we document a successful application of elemental fingerprinting as a tracking tool to determine sources of settled invertebrates and show that coastal mussel larvae, previously thought to be highly dispersed, can be retained within 20–30 km of their natal origin. We compare two closely related and co-occurring species, Mytilus californianus and Mytilus galloprovincialis, and determine that, despite expected similarities, they exhibit substantially different connectivity patterns. Our use of an in situ larval culturing technique overcomes the previous challenge of applying microchemical tracking methods to species with completely planktonic development. The exchange of larvae and resulting connectivities among marine populations have fundamental consequences for the evolution and ecology of species and for the management of coastal resources.
Keywords: elemental fingerprinting, in situ larval culturing, larval retention, larval transport, Mytilus
Because many benthic marine species have larvae that can spend days to months in the planktonic stage, it has long been assumed that they are transported great distances and are widely dispersed (1). This assumption has led marine ecologists for much of the 20th century to presume that most coastal benthic populations were demographically open and highly “connected” through larval transport (2). Recent technological advances coupled with a recognition of the importance of the behavior, mortality, physical variability, and oceanographic retention features of larvae have led to a paradigm shift in recent years that has forced researchers to focus on the evidence of and mechanisms leading to closed populations, in which “self-seeding” occurs at local spatial scales (2, 3). Quantitative knowledge of larval connectivities can revolutionize understanding of a broad range of topics, including marine population dynamics (1), processes of local extinction and recolonization (4), scales of adaptation (5), marine reserve design (6), spread of invasive species (7), and species response to climate change (8). Despite the importance of quantifying connectivity of coastal populations, it has rarely been accomplished directly (9, 10) because of the small size and high mortality of marine larvae in a highly complex marine environment. In this study, elemental fingerprinting and in situ larval culturing were used to directly determine and compare the patterns of connectivity of two closely related species of mussel, Mytilus californianus and Mytilus galloprovincialis, in San Diego County, California, across medium spatial scales (<30 km). We specifically sought to answer these questions: Can we identify specific sources of mussel recruits settling in San Diego, CA? How much self-seeding occurs at intermediate (20–30 km) spatial scales, and how does the degree of self-seeding vary along the coastline? How similar are the connectivity patterns of two closely related, co-occurring species? By answering these questions, we lay the groundwork for large-scale exploration of connectivity patterns in open-coast species of ecological and economic significance.
Elemental fingerprinting takes advantage of location-specific chemical signatures recorded in hard parts of marine organisms at the time of their formation, which can serve as geographic “tags” (10). The microchemistry of larval structures that are retained after settlement can be used to identify the location where the hard part was formed, thus allowing the reconstruction of the larval origin of settled juveniles. This approach has been successfully applied to address issues related to movement of juvenile and adult fish (11, 12) as well as to dispersal of larval fish (10, 13) by examining the chemistry of their relatively large and well studied otoliths.
Although the use of chemical signatures of invertebrate hard parts (e.g., shells and statoliths) as a larval tracking tool shows great promise, prior studies have either tracked only newly released larvae (14), documented the existence the spatial variation in elemental signatures of adults (15), or generated an elemental “map” of location-specific shell chemistry necessary to compare and predict origins of unknown samples (16). Prior exploratory studies have used adult shell (15) to create this reference map but have met with limited success because of ontogenetic changes in shell formation found in many species. For example, mytilid mussel adult shell (dissoconch) is composed of both aragonite and calcite (17), whereas larval shell (prodissoconch) is mostly aragonitic (18), leading to demonstrable differences in how they incorporate various elements. Other studies have focused on a limited number of species that retain their embryos at the site of natal origin before release into the plankton stage (e.g., benthic encapsulation) (16). Our study takes the next step with invertebrates by collecting new recruits and using elemental fingerprinting to determine their natal origins.
In this study, we developed a method to generate larval reference signatures through in situ larval culturing. This approach is applicable to a broad number of invertebrate species with diverse life-history strategies, including many commercially important species of shellfish (oysters, scallops, clams, and mussels). Location-specific reference chemical signatures needed to interpret recruit origins were determined by raising larvae of two species of mytilid mussel, M. californianus and M. galloprovincialis, in situ at 13 sites spanning 75 km of exposed shoreline and three embayments (Fig. 1) and by analyzing resulting larval shells for trace element composition. Using this regional reference map, we compared the expected larval chemistry for each region to the prodisscoconch shell chemistry of newly settled recruits, with the assumption that they had developed in the water column during the period of in situ larval culturing. We predicted the natal origins of these wild-caught juvenile mussels by using this chemical reference map.
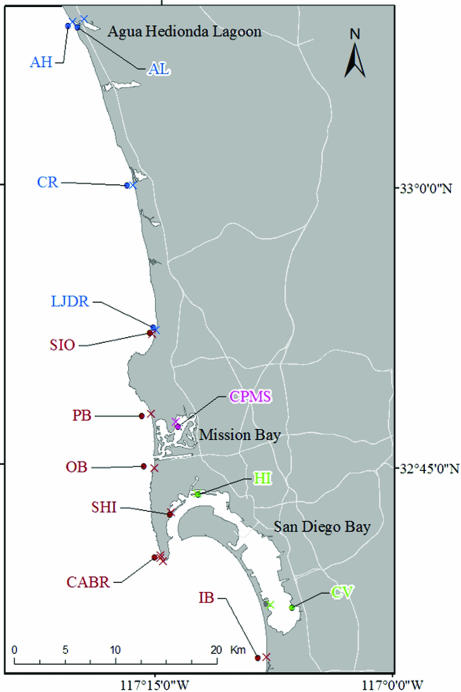
Fig. 1.
Map of larval outplanting and juvenile collection sites. Northern coastal region (blue): AL, Agua Hedionda Lagoon; AH, Agua Hedionda; CR, Cardiff Reef; LJDR, La Jolla Dike Rock. Southern coastal region (red): OB, Ocean Beach Pier; CABR, Cabrillo National Monument; IB, Imperial Beach Pier; SHI, Shelter Island. Outer bay region (green): HI, Harbor Island. Inner bay region (pink): CV, Chula Vista; CPMS, Crown Point Mitigation Site. The dark circles correspond to offshore stations, whereas paired intertidal stations are represented by ×.
Results
Evaluation of Outplanting Success.
The in situ larval culture experiment yielded larval shells that were mostly >100 μm long (108.5 ± 12.7 μm long × 77.6 ± 10.1 μm wide, mean ± 1 SD) that were formed entirely in the field at known locations. Widdows (19) estimated natural mortality rates to be ≈10–20% per day (20–50% survival after 7 days). We observed survival rates of the field-raised larvae (1.8 ± 3.6% SD) to be somewhat lower than the laboratory-raised control larvae (8.1 ± 7.7% SD in larval “homes”). However, there was a large range of survival rates among our sites (from 0% to 15.95%) and higher survival for M. californianus (2.58 ± 4.27% SD) than M. galloprovincialis (0.34 ± 0.47% SD). To improve sample size and site representation for origin assessment of M. galloprovincialis juveniles, elemental fingerprints were developed by using chemical signatures of larvae of both species combined. The element ratio (Co/Ca) that led to the greatest difference between the two species was not used in this analysis. Only outplanted M. californianus larvae were considered when evaluating M. californianus juveniles (see Tables 1 and 2 for sample sizes). The classification success of the determined elemental fingerprints was greater when considering M. californianus alone compared with both species.
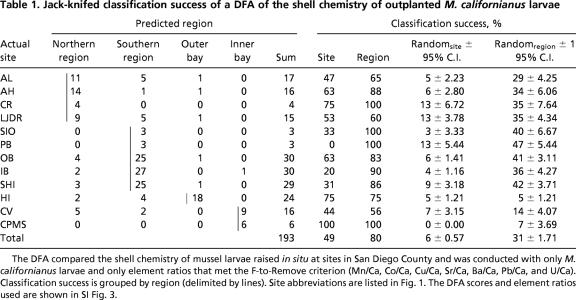
Table 1.
Jack-knifed classification success of a DFA of the shell chemistry of outplanted M. californianus larvae
The DFA compared the shell chemistry of mussel larvae raised in situ at sites in San Diego County and was conducted with only M. californianus larvae and only element ratios that met the F-to-Remove criterion (Mn/Ca, Co/Ca, Cu/Ca, Sr/Ca, Ba/Ca, Pb/Ca, and U/Ca). Classification success is grouped by region (delimited by lines). Site abbreviations are listed in Fig. 1. The DFA scores and element ratios used are shown in SI Fig. 3.
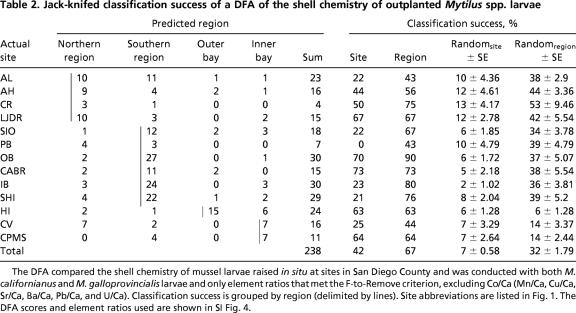
Table 2.
Jack-knifed classification success of a DFA of the shell chemistry of outplanted Mytilus spp. larvae
The DFA compared the shell chemistry of mussel larvae raised in situ at sites in San Diego County and was conducted with both M. californianus and M. galloprovincialis larvae and only element ratios that met the F-to-Remove criterion, excluding Co/Ca (Mn/Ca, Cu/Ca, Sr/Ca, Ba/Ca, Pb/Ca, and U/Ca). Classification success is grouped by region (delimited by lines). Site abbreviations are listed in Fig. 1. The DFA scores and element ratios used are shown in SI Fig. 4.
Creation of a Larval Reference Model.
Differences in the shell chemistry of outplanted larvae were sufficient to discriminate among study areas [using discriminant function analysis (DFA)]. Site-specific chemistry-based classification success (jack-knifed) was highly variable, with a mean of 49% for M. californianus (0–100%) (Table 1) and a mean of 42% (0–73%) for both species combined (Table 2). However, our past studies have indicated that the appropriate scale on which trace elemental signals of mussel juvenile shell can be resolved along the San Diego County open coast is on the order of 20 km (15). Larger regional groupings (northern coastal, southern coastal, outer bays, and inner bays) (Fig. 1) yielded much higher classification success for larval shell [80% for M. californianus (Table 1) and 67% for both species combined (Table 2)] than grouping by site. Therefore, these regional elemental fingerprints appear to be the most reliable and were used to determine natal origins.
To determine the robustness of these classifications relative to random chance, the data were randomized (i.e., the element ratios for a single larval mussel shell were kept together but assigned a different site by using a random number generator) 10 times, and the same DFA was run to generate averages and 95% confidence intervals. Average classification success was considerably higher than the randomly generated value, whether considered by site or region (Tables 1 and 2) (e.g., by region: M. californianus, 31 ± 1.71%; M. galloprovincialis, 32 ± 1.79%) classification success. An exception was Pacific Beach (Crystal) Pier (PB), which had very low sample sizes (total, seven larvae) (Tables 1 and 2).
The element ratios most important in discriminating among reference signatures varied among regions and between species. Due to the large number of sites considered, the resulting elemental fingerprinting models were complex (96% and 93% of the variability attributed to the first four DFA scores in M. californianus and the combined species analysis, respectively) [supporting information (SI) Figs. 3 and 4]. M. californianus larvae raised in inner bays were the most distinct from those raised in the other regions due to higher Co/Ca values (score 1) (SI Fig. 3). The southern region and the outer bay M. californianus larvae were distinguished by score 2 (mostly because of high Pb/Ca in the southern coastal region and high Cu/Ca in the outer bay) and score 3 (mostly because of high Mn/Ca in the outer bay). The northern region M. californianus larvae were best distinguished from the southern and outer bay regions by score 4 because of higher U/Ca and lower Ba/Ca and Sr/Ca in larval shells from the north. The most important elemental ratios contributing to regional discrimination in the two-species analysis were somewhat similar to those in the M. californianus case (SI Fig. 4). Higher Mn/Ca and Cu/Ca in the bay sites and higher Pb/Ca and Ba/Ca in the southern coastal sites accounted for most, but not all, of the variation between the regions (74% of dispersion attributed to DFA scores 1 and 2). The higher U/Ca and Mn/Ca in larval shells outplanted at Cabrillo National Monument (Fig. 1) led to the distinctness of the elemental fingerprint from this site.
The Scripps Institution of Oceanography Pier (SIO) and PB, which in a previous study were assigned to the northern and southern coastal regions, respectively, based on juvenile dissoconchs collected in 2001 (15), were both assigned to the southern coastal region in this study. In the previous study, these two sites were considered “transition” areas, and the classification success for both was quite low. It is, therefore, not surprising that their regional classification would vary, especially given the lower sample sizes of M. californianus at these two sites (Table 1). The temperature at SIO was more similar to sites farther north throughout the outplanting period (unpublished data), and the SIO and La Jolla Dike Rock sites are <1 km apart. The assignment of SIO in the southern region is uncertain; however, there were very few juveniles assigned to this site of origin (1 of 125 M. californianus and 9 of 108 M. galloprovincialis), so this uncertainty had little influence on the connectivity patterns.
Determination of the Natal Origins of Juveniles.
Newly recruited individuals of M. californianus and M. galloprovincialis are very difficult to identify to species visually. A PCR-based assay (15) was used to determine the species of mussel in 99 individuals (43%). The assay does not discriminate between M. galloprovincialis and the similar Mytilus trossulus; thus, some of the San Diego Bay mussels could be M. trossulus or hybrids between the two species, although they are rare (20). To identify the remaining 131 individuals that did not amplify using the molecular genetic assay, a DFA (Systat 9) was conducted with the PCR-identified mussels as known grouping variables and with site, dissoconch shell chemistry, shell length/width ratio, and the angle of the hinge relative to the ventral margin (21) as predicting variables. The accuracy of this combination of variables (as determined by using identified mussels as unknowns and predicting species identities) was 93% (54 of 58) for M. californianus and 85% (28 of 33) for M. galloprovincialis. M. californianus recruits were found only at open coast sites, whereas M. galloprovincialis recruits were found at all sites but were especially abundant in the south and in bays. This pattern was consistent with our observations that, in southern California, M. californianus adults were found mainly on exposed rocky shores, whereas M. galloprovincialis adults were found in embayments and on open coasts. The total number of juveniles of each species is shown in Fig. 2. At least one M. galloprovincialis was found at every site (Fig. 2B), whereas no M. californianus were found in bays.
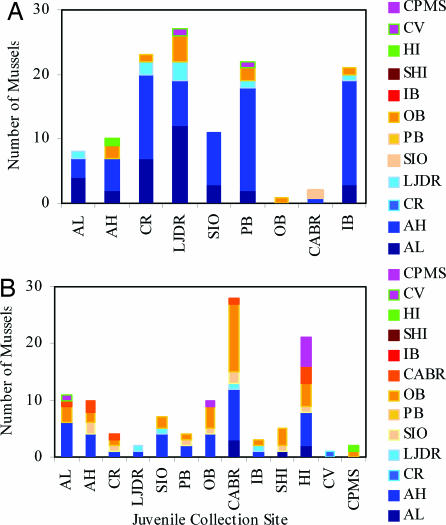
Fig. 2.
Natal origins of juvenile mussels. Sites on the x axis represent collection locations of juvenile mussels, and the colors of the bars indicate predicted natal origins determined by using larval shell chemistry for M. californianus (A) and M. galloprovincialis (B). The northern coastal region is represented by various shades of blue. The southern coastal region is represented by various shades of red and orange. The outer-bay region is represented by green, and the inner-bay region is represented by two shades of pink. Site abbreviations are listed in Fig. 1. Sites where no M. californianus juveniles were found are not shown in A.
The natal origins of the juveniles of the two species were surprisingly different, although regional self-seeding was found in both (Fig. 2). For M. californianus, the majority (88%) of individuals settling at all sites originated in the northern region, with high self-recruitment in the north (87% of juveniles collected in the north region originated in that region) and high larval importation in the south (91% of juveniles collected in the southern region originated outside of that region) (Fig. 2A). This pattern reflects asymmetric mixing (22) at the regional scale, with most larval transport from north to south. M. galloprovincialis recruits exhibited more diverse origins, with a mix of northern (45%), southern (47%), and bay (7%) sources (Fig. 2B). In this case, a multisource, near-random mixing model applies, with low site-specific self-recruitment but average regional self-seeding rates of nearly 40%.
There were a number of sources of error in assigning these origins, most notably an error associated with species identification of individuals that did not amplify with PCR and an error associated with the DFA reference signatures. When the analysis was repeated with only individuals that were positively identified with PCR, the results were almost identical to the whole sample set for M. californianus (87% northern, 11% southern, 0% outer bay, and 2% inner bay for PCR only vs. 88%, 10%, 1%, and 2%, respectively, for the total sample) and almost identical for M. galloprovincialis (41% northern, 43% southern, 0% outer bay, and 15% inner bay for PCR only vs. 45%, 47%, 0%, and 7%, respectively, for the total sample). Use of both species to create a reference elemental fingerprint for M. galloprovincialis led to reduced power to discriminate among regions and must be considered when interpreting the assignment of natal origins to M. galloprovincialis settlers.
Discussion
By using elemental fingerprinting and in situ larval culturing, specific sources of mussel recruits settling in San Diego were identified. Most M. californianus originated from the northern part of the study area, suggesting in May 2003 that these populations followed a “single source” model of larval replenishment. M. galloprovincialis populations in San Diego County appeared to originate from a larger number of sources, including bays and southern sites, although a smaller sample size led to more equivocal results. Self-seeding on intermediate spatial scales did occur but was species- and location-dependent.
This study revealed unexpected differences in connectivity and rates of self-seeding between two adjacent segments of coastline (north and south San Diego County) and between two congeneric species developing in the same waters at the same time. The mechanism behind the observed differences between the two species is unknown. We explored the possibility that these connectivity differences were caused by differences in larval release sites dictated by the distribution of adult stock. However, whole-coastline estimates of mussel cover for both M. californianus (15,320 m2, with 50% in the north and 50% in the south) and for M. galloprovincialis (15,124m2, with 45% in the north and 55% in the south) (unpublished data)‡ suggest that north–south differences in supply are unlikely to cause observed between-species variation in connectivity. Notably, most M. galloprovincialis are released from large, southern-region embayments (San Diego Bay and Mission Bay) and from a mussel farm in a small embayment to the north (Agua Hedionda Lagoon) (Fig. 1). Because M. californianus larvae are released along the open coast and M. galloprovincialis larvae are released mainly into bays, the susceptibility to influence by the prevailing southward surface current just offshore of this region (23) might vary between these species.
Local differences in life-history parameters interacting with ocean currents may also cause differences in connectivity patterns. Variations in larval duration, delay of metamorphosis, initiation of spawning, local mortality, and vertical position of the larvae within the water column may affect connectivities. For both species, the range of larval duration appears to be broad, given that mytilid mussels can demonstrate considerable developmental plasticity (24). Duration estimates for M. galloprovincialis planktonic development range from 14 days (25) to 6 weeks (26), and estimates for M. californianus range from 9 (27) to 45 days (28). Further study of the mechanisms responsible for the observed differences in larval connectivity patterns in two seemingly similar species will deepen our understanding of the consequences of various life-history strategies for larval connectivity.
The challenge remains to integrate our understanding of connectivity and elemental fingerprinting over appropriate temporal and spatial scales to effectively use this high-resolution information for the management and conservation of coastal resources. The temporal stability of the elemental fingerprints on seasonal and interannual scales and in an ontogenetic context will need to be determined with time series studies (e.g., ref. 15). Mussel connectivity patterns themselves are likely to vary temporally, because seasonal and interannual changes in circulation have been documented for the study region (23). Repeated fingerprinting activities, paired with simultaneous monitoring of settlement, and demographic and physical modeling should help determine not only the origin of settlers but also the relative importance of specific cohorts and larval sources to the persistence of populations (e.g., refs. 29 and 30).
One powerful benefit of elemental fingerprinting as a tool for studying population connectivity derives from the direct determination of natal origins in a discrete period. In contrast, genetic approaches to determining population connectivity tend to allow for more generalized conclusions about the contribution of different natal regions to the total population across longer time scales and are biased toward rare mixing events (although see ref. 6 for exceptions). For example, a recent large-scale examination of the genetic population structure of M. californianus throughout its range on the west coast of North America found little geographic structure and a large amount of chaotic small-scale variability (31). This result implies that, on evolutionary time scales and throughout its biogeographic range, M. californianus populations are panmictic and open but that on smaller scales the connectivity between populations can vary. The elemental fingerprinting approach, which is much more sensitive to self-recruitment and short-term patterns, indicates that mussel larval retention does occur on smaller spatial scales (tens of kilometers) and on time scales relevant to conservation (years and shorter) and implies that a single source can contribute the majority of new offspring to a single cohort across a broader region. By examining connectivity on different scales using multiple approaches, such as genetics and fingerprinting, a more complete and realistic understanding of the system emerges (3).
A few recent studies, most focusing on fish and in island environments, have contradicted the paradigm that self-recruitment in coastal organisms is a rare event (reviewed in refs. 2, 3, and 6), although direct evidence of larval retention in mostly continuous coastal populations has been difficult to obtain (32). In this study, we successfully documented medium-scale (20–30 km) self-recruitment within coastal mussel populations. Although mussels were previously assumed to be highly dispersed (19, 33) due to their poor swimming abilities (34), our result is consistent with two recent studies that opportunistically tracked the rate of spread of single, well marked populations of mussels: an invasive population of M. galloprovincialis in South Africa, which spread <100 km per year [90% of individuals were found within 5 km of the original population after four years (35)], and a population of Mytilus edulis and M. galloprovincialis hybrids in southwest England (typical dispersal per generation, 30 km) (36). Across a 75-km expanse of coastline, we found unexpected spatial heterogeneity in mytilid connectivity patterns, with a high degree of self-seeding in northern M. californianus populations and high larval importation of the same species in the south. Using in situ larval culturing and elemental fingerprinting, we were able to move beyond a simple determination of open or closed populations and toward a spatially specific, directly observed, and applicable model of larval connectivity.
We have successfully applied trace elemental fingerprinting to determine natal origins and quantify connectivity patterns of settled marine invertebrates. As direct determination of natal origins is applied to an increasing number of species, our general paradigms about how marine populations are connected are likely to be further tested and challenged. This result will affect our understanding of how species evolve, interact, and are distributed, and it will alter our strategies to protect them effectively.
Materials and Methods
Natal origins of early settlers for two mussel species were evaluated by (i) spawning and culturing larvae of both species at 13 sites, (ii) determining the chemical signature of larval shells, and (iii) comparing these elemental fingerprints to the retained larval shells (prodissoconchs) of “wild” juveniles from the same areas. All tools and containers used for handling mussel larvae were made with nonmetal materials that had been previously washed in nitric acid. M. californianus and M. galloprovincialis adults were collected from SIO on May 11, 2003 and induced to spawn by exposure to mechanical disturbance and heat (20–22°C; ambient ocean water, ≈17°C). Gametes were obtained from one male and female M. galloprovincialis and four male and one female M. californianus, and fertilization was carried out separately.
Larvae were outplanted in 215-ml “homes,” which consisted of a poly(vinyl chloride) pipe coupler 3.8 cm in diameter and a 14-cm-long piece of pipe with an open cap of 35-μm nitex mesh on either end. Before this experiment, the homes were leached of contaminants in flowing seawater for ≈3 months and acid washed. The mesh was rinsed well with Milli-Q water, soaked in 1% nitric acid for 24 h, and then soaked in Milli-Q water for ≈1 week. Two homes of M. californianus embryos and one home of M. galloprovincialis embryos, each containing ≈100,000 larvae, were outplanted at each site. Homes were filled from 7 to 9.5 h after fertilization. As a control for larval home effects, larvae of each species were raised in larval homes in the laboratory (identical to those used in situ) and loose in buckets of sterilized seawater. These laboratory cultures were fed Isochrysis sp. (Instant Algae Premium 1800; Aquatic Eco-Systems, Apopka, FL) when the water was changed once per day.
Control larvae raised in the laboratory were analyzed to determine whether there was a caging or species effect on shell chemistry. There was a detectable chemical effect between the larvae raised loose in buckets and in poly(vinyl chloride) larval homes (73% classification success in DFA just comparing homes and buckets, regardless of species). This difference was mostly due to higher Sr/Ca and lower U/Ca in the larval shells raised in homes, whereas all other element ratios did not differ greatly between them (i.e., all failed to meet the “F-to-Remove” criteria used to select variables when creating DFA in the rest of this study). However, it is important to place these differences in context with differences in the field. When compared with sites where both species were analyzed (Agua Hedionda Lagoon, Crown Point Mitigation Site, PB, and SIO), M. californianus raised in the laboratory was never misclassified as coming from a field site, but 9 of 33 laboratory-raised M. galloprovincialis were misclassified as coming from PB or SIO. Because the laboratory larvae were raised with seawater from the SIO pier, this misclassification is not surprising.
To assess the species difference in shell chemistry, sites with large sample sizes of both species were compared. The shell chemistry of Crown Point Mitigation Site larvae was particularly different depending on species. This difference was attributed to very high Co/Ca levels in M. californianus from this site. When this analysis was repeated without Co/Ca considered within the DFA, the differences between the species were somewhat lessened.
Larval homes were outplanted on moorings at 13 sites in San Diego County, California, each located offshore of a known source of mytilid mussels (Fig. 1). Moorings were in 10 m of water at open coast stations and 5–6 m of water in embayments. Larval homes were outplanted 2 m below mean lower low water, along with a temperature logger, on one buoy per site. By using predicted tides during this period, it was estimated that the larvae remained between 1.5 and 4 m below the surface, which is the approximate depth at which mytilid larvae have been found in nature (M. edulis in the White Sea) (37). The total length of time from fertilization to deployment ranged from 8.5 to 15 h.
After 7 days of in situ exposure to seawater at each site, the homes were retrieved. The contents of the homes were filtered in the laboratory by using the existing mesh and water collected from the corresponding site, then stored in an acid-washed, 50-ml centrifuge tube at −20°C. Because the larvae were added as shell-less embryos, it was assumed that all larval shells found at the end of the deployment were formed in situ at the deployment site.
Newly recruited juvenile mussels, assumed to have been planktonic larvae during the second to third weeks in May 2003, were obtained from the intertidal zone by collecting adult mussels and turf-forming algae just inshore from the 13 outplanting sites on June 3–6, 2003. Samples were frozen (−20°C) in Ziploc bags within 2 h of collection, and individuals with a maximum length of <3 mm were removed under a dissecting microscope and stored separately in acid-washed vials at −20°C.
All preparation of larval and juvenile shells for chemical analysis was done in a clean room with acid-washed nonmetal supplies, trace-metal-free reagents, and Milli-Q or quartz-distilled (QD) water. Larval shells were separated and visually sorted into a Petri dish with a pipette under a dissecting microscope, then treated with 15% Suprapur hydrogen peroxide [EMD Chemicals (Carlsbad, CA) through VWR (West Chester, PA)] buffered in 0.05 M Suprapur NaOH (EMD Chemicals through VWR) for 10–11 h to remove all organic material, rinsed in QD water three times, transferred to a petrographic slide covered in double-sided tape (Scotch Brand), and allowed to dry under a Class-100 laminar-flow hood.
Juvenile shells were manually split, and the soft parts were removed and frozen for molecular genetic identification. The right valve was placed in individual vials, to which 15% H2O2 buffered in 0.05 M NaOH was added for ≈18 h. Shells were then rinsed in QD water, 1% HNO3 (OPTIMA grade; Fisher Scientific, Hampton, NJ) was added for <10 s, and the acid was removed. The samples were rinsed three times in QD water and then mounted.
A total of 233 juvenile mussels were used in this analysis, with an average of 18 juveniles per site. More than 10 juveniles were analyzed for all sites except three, where too few settlers were found. Mussels ranged in length from 663 to 3,009 μm (1,595 ± 544 μm, mean ± 1 SD). In addition, one very large individual (6 mm) from Chula Vista was used because it was the only individual found at that site.
Larval and juvenile shells were analyzed by using a New Wave UP 213-nm laser ablation unit with an Element 2 double-focusing, single collector, magnetic sector inductively coupled plasma-mass spectrometer (ThermoQuest Finnigan LA-ICP-MS; Thermo Scientific, Waltham, MA). Our methods were similar to those reported in Becker et al. (15). The LA-ICP-MS system was modified so that 2% HNO3 (OPTIMA) was aspirated through the nebulizer, and this aerosol was mixed with the sample gas (He) in the spray chamber. Larvae were analyzed individually with a single 75-μm laser line (25% power, 10 Hz, 40-μm spot size, 15 μm/s). Samples from a single home were mounted together, and six homes were mounted on a single slide; an individual from each home was sampled in rotating order in an effort to reduce bias. The larval shells on newly recruited juveniles were analyzed with a 75-μm laser line (30% power, 10 Hz, 20-μm spot size, 15 μm/s) located on the early prodissoconch perpendicular to the axis of growth. The isotope menu for larvae and juveniles consisted of 48Ca, 55Mn, 59Co, 88Sr, 138Ba, 208Pb, and 238U. To correct for interferences in 138Ba (and as a marker for the slide material), 118Sn was also included. Glass standards spiked with trace elements (National Institute of Standards and Technology Standard Reference Material 612, 614, and 616) were analyzed at the beginning, middle, and end of each run day to account for machine drift and convert isotope intensities to “absolute” ratios. The slide and tape were sampled to test for contamination.
To determine isotope intensities, a chromatogram was generated for each element in each sample by using Element Software. Resulting peaks (i.e., peaks having a maximum value greater than three standard deviations above the mean of the background) were analyzed individually, and background levels were subtracted from peaks using linear regression of nonpeak values. Raw counts per second (cps) were calculated by determining the area under each peak for each isotope in each sample. The background-corrected cps values were then multiplied by a correction factor generated by the standard (National Institute of Standards and Technology), using recorded run times and linear estimations of machine drift. The sample cps values were then divided by the counts of 48Ca, a rare isotope of Ca, which was used as an internal standard for the amount of shell ablated. Because of a lack of matrix-matched standards, the absolute ratios determined by this method, although consistent within studies using these standards, might not be directly comparable with studies using different standards (15).
Resulting element ratios (X:48Ca) were analyzed with linear DFA to examine differences between the chemistry of larval shells raised at the various sites and to determine the effects of species and control treatment on the elemental signatures. A DFA model to define elemental fingerprints for each site was created by using shell elemental ratios of the outplanted larvae. The resulting jack-knifed classification matrix indicated that 20-km regions (northern coastal, southern coastal, outer bays, and inner bays) (Fig. 1) provided the best classification success. Once a satisfactory DFA model was created by using outplanted larval shell chemistry, the prodissoconch element ratios were assessed as “unknowns” to determine the site of origin for each recruited individual.
Supplementary Material
Acknowledgments
We thank E. Kisfaludy, L. Fajardo Mellor, K. Riser, and many volunteers who assisted with field and laboratory work. Analyses were conducted in the SIO Analytical Facility with assistance from A. Deyhle, B. Deck, C. Mahn, and P. Castillo. Genetic identification was done in the laboratory of R. Burton by K. Gruenthal. P. Dayton, E. Sala, and two anonymous reviewers provided useful comments. This study was supported by the Cabrillo National Monument Foundation, the University of California Marine Science Council through a California Environmental Quality Initiative Graduate Support Fellowship and University of California Office of the President Grant 02 T CEQI 08 0105, U.S. Office of Naval Research Grants N00014-00-0174 and N00014-00-1-0473, and National Science Foundation Grant OCE 03-27209.
Abbreviations
- DFA
discriminant function analysis
- PB
Pacific Beach (Crystal) Pier
- SIO
The Scripps Institution of Oceanography Pier
- QD
quartz-distilled.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611651104/DC1.
Adult mussel cover was approximated by determining an average percent cover with 20–40 point estimates at each site and multiplying by the length and height of the coastline at that site (measured on-site and calculated with ArcView software). Pier cover was estimated at each pier by determining the average cover per piling and multiplying by the number of pilings.
References
- 1.Caley MJ, Carr MH, Hixon MA, Hughes TP, Jones GP, Menge BA. Annu Rev Ecol Syst. 1996;27:477–500. [Google Scholar]
- 2.Swearer SE, Shima JS, Hellberg ME, Thorrold SR, Jones GP, Robertson DR, Morgan SG, Selkoe KA, Ruiz GM, Warner RR. Bull Mar Sci. 2002;70:251–271. [Google Scholar]
- 3.Levin LA. Integr Comp Biol. 2006;46:282–297. doi: 10.1093/icb/icj024. [DOI] [PubMed] [Google Scholar]
- 4.Strathmann RR, Hughes TR, Kuris AM, Lindeman KC, Morgan SG, Pandolfi JM, Warner RR. Bull Mar Sci. 2002;70:377–396. [Google Scholar]
- 5.Warner RR. Coral Reefs. 1997;16:S115–S120. [Google Scholar]
- 6.Palumbi SR. Ann Rev Environ Resources. 2004;29:31–68. [Google Scholar]
- 7.Neubert MG, Caswell H. Ecology. 2000;81:1613–1628. [Google Scholar]
- 8.Harley CDG, Hughes AR, Hultgren KM, Miner BG, Sorte CJB, Thornber CS, Rodriguez LF, Tomanek L, Williams SL. Ecol Lett. 2006;9:228–241. doi: 10.1111/j.1461-0248.2005.00871.x. [DOI] [PubMed] [Google Scholar]
- 9.Levin LA. Ophelia. 1990;32:115–144. [Google Scholar]
- 10.Thorrold SR, Jones GP, Hellberg ME, Burton RS, Swearer SE, Neigel JE, Morgan SG, Warner RR. Bull Mar Sci. 2002;70:291–308. [Google Scholar]
- 11.Campana SE. Mar Ecol Prog Ser. 1999;188:263–297. [Google Scholar]
- 12.Campana SE, Thorrold SR. Can J Fish Aquat Sci. 2001;58:30–38. [Google Scholar]
- 13.Swearer SE, Caselle JE, Lea DW, Warner RR. Nature. 1999;402:799–802. [Google Scholar]
- 14.DiBacco C, Levin LA. Limnol Oceanogr. 2000;45:871–880. [Google Scholar]
- 15.Becker BJ, Fodrie FJ, McMillan PA, Levin LA. Limnol Oceanogr. 2005;50:48–61. [Google Scholar]
- 16.Zacherl DC. Mar Ecol Prog Ser. 2005;290:145–163. [Google Scholar]
- 17.Dodd JR. J Paleontol. 1964;38:1065–1071. [Google Scholar]
- 18.Fuller SC, Lutz RA. Malacologica. 1988;29:363–371. [Google Scholar]
- 19.Widdows J. Aquaculture. 1991;94:147–163. [Google Scholar]
- 20.Suchanek TH, Geller JB, Kreiser BR, Mitton JB. Biol Bull. 1997;193:187–194. doi: 10.2307/1542764. [DOI] [PubMed] [Google Scholar]
- 21.Martel AL, Robles C, Beckenbach K, Smith MJ. Invertebr Bio. 1999;118:149–164. [Google Scholar]
- 22.Wares JP, Gaines SD, Cunningham CW. Evolution (Lawrence, Kans) 2001;55:295–306. doi: 10.1111/j.0014-3820.2001.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 23.Winant CD, Bratkovich AW. J Phys Oceanogr. 1981;11:71–86. [Google Scholar]
- 24.Bayne BL. Ophelia. 1965;2:1–47. [Google Scholar]
- 25.Satuito CG, Natoyama K, Yamazaki M, Fusetani N. Fisheries Sci (Tokyo) 1994;60:65–68. [Google Scholar]
- 26.Chícharo LMZ, Chicharo MA. J Exp Mar Bio Ecol. 2000;243:81–94. [Google Scholar]
- 27.Strathmann MF. Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast. Seattle: Univ of Washington Press; 1987. p. 314. [Google Scholar]
- 28.Trevelyan GA, Chang ES. J World Maricult Soc. 1983;14:137–148. [Google Scholar]
- 29.Cowen RK, Paris CB, Srinivasan A. Science. 2006;311:522–527. doi: 10.1126/science.1122039. [DOI] [PubMed] [Google Scholar]
- 30.Hastings A, Botsford LW. Proc Natl Acad Sci USA. 2006;103:6067–6072. doi: 10.1073/pnas.0506651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel JD. PhD Dissertation. Santa Cruz: Univ of California; 2004. [Google Scholar]
- 32.Sponaugle S, Cowen RK, Shanks A, Morgan SG, Leis JM, Pineda JS, Boehlert GW, Kingsford MJ, Lindeman KC, Grimes C, et al. Bull Mar Sci. 2002;70:341–375. [Google Scholar]
- 33.Bayne BL. Marine Mussels, Their Ecology and Physiology. Cambridge, UK: Cambridge Univ Press; 1976. pp. 118–120. [Google Scholar]
- 34.Chia F-S, Buckland-Nicks J, Young CM. Can J Zool. 1984;62:1205–1222. [Google Scholar]
- 35.McQuaid CD, Phillips TE. Mar Ecol Prog Ser. 2000;201:211–220. [Google Scholar]
- 36.Gilg MR, Hilbish TJ. Ecology. 2003;84:2989–2998. [Google Scholar]
- 37.Dobretsov SV, Miron G. Mar Ecol Prog Ser. 2001;218:179–187. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.