Abstract
Amplification or overexpression of growth factor receptors is a frequent occurrence in malignant gliomas. Using both expression profiling and in situ hybridization, we identified insulin-like growth factor 2 (IGF2) as a marker for a subset of glioblastomas (GBMs) that lack amplification or overexpression of EGF receptor. Among 165 primary high-grade astrocytomas, 13% of grade IV tumors and 2% of grade III tumors expressed IGF2 mRNA levels >50-fold the sample population median. IGF2-overexpressing tumors frequently displayed PTEN loss, were highly proliferative, exhibited strong staining for phospho-Akt, and belonged to a subclass of GBMs characterized by poor survival. Using a serum-free culture system, we discovered that IGF2 can substitute for EGF to support the growth of GBM-derived neurospheres. The growth-promoting effects of IGF2 were mediated by the insulin-like growth factor receptor 1 and phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3), a regulatory subunit of phosphoinositide 3-kinase that shows genomic gains in some highly proliferative GBM cases. PIK3R3 knockdown inhibited IGF2-induced growth of GBM-derived neurospheres. The current results provide evidence that the IGF2–PIK3R3 signaling axis is involved in promoting the growth of a subclass of highly aggressive human GBMs that lack EGF receptor amplification. Our data underscore the importance of the phosphoinositide 3-kinase/Akt pathway for growth of high-grade gliomas and suggest that multiple molecular alterations that activate this signaling cascade may promote tumorigenesis. Further, these findings highlight the parallels between growth factors or receptors that are overexpressed in GBMs and those that support in vitro growth of tumor-derived stem-like cells.
Keywords: astrocytoma, brain tumor, expression profiling, glioma, gliomagenesis
Malignant gliomas are the most common primary brain tumors in adults. In glioblastomas (GBMs), the most aggressive glioma subtype, tumor formation and growth appear to be driven by amplification or overexpression (OE) of gene products involved in growth factor-initiated signal transduction acting in cooperation with genetic alterations disrupting cell-cycle control (1). Parallels between growth factor signaling elements implicated in GBM growth and those that regulate key stages in neural development are consistent with recent evidence suggesting neural stem and/or progenitor cells as the cell type of origin for GBM (2, 3).
EGF receptor (EGFR) amplifications, often accompanied by the activating mutation EGFRvIII, have been reported in 30–50% of human GBMs (4). Alterations in other growth factor-induced signaling cascades include amplification and/or OE of FGF, PDGFRα, PDGFRβ, PDGF, and c-Met receptor and have typically been described in gliomas with unamplified EGFR (4). Of the genomic alterations described in GBM, PTEN mutation and/or deletion is the most common, with an estimated frequency of 70–90% (4). These findings, along with the prognostic value of PTEN status in GBM cases (see ref. 5), suggest the importance of the phosphoinositide 3-kinase (PI3K)/Akt pathway in promoting highly aggressive glial malignancies.
Mouse models provide compelling evidence for the ability of EGFR or PDGF expression in neural progenitors to cooperate with inactivation of p53 or INK4A/ARF to drive the formation of lesions that closely resemble the histopathology of human gliomas (6, 7). Both neural stem cells derived from human brain and stem-like cells derived from human GBMs can be supported and expanded in the presence of EGF or PDGF (2, 3, 8, 9), and PDGF plays an instructive role in neural development (10). The demonstration that PTEN loss increases the pool of self-renewing neural stem cells and induces loss of homeostatic control of proliferation (11) is reminiscent of the cell-cycle dysregulation that occurs during gliomagenesis. Taken together, this growing body of evidence indicates that the PI3K/Akt signaling axis, engaged downstream of growth factor receptors, functions as a “master regulator” during both neurogenesis and glioma formation.
In the current study, we used gene expression profiling to identify additional growth factor signaling elements that may contribute to GBM formation and reveal evidence that insulin-like growth factor 2 (IGF2) might provide an alternative to EGFR activation for supporting tumor growth.
Results
IGF2 Is Overexpressed in a Subgroup of GBMs Distinct from EGFR-Overexpressing (EGFR-OE) GBMs.
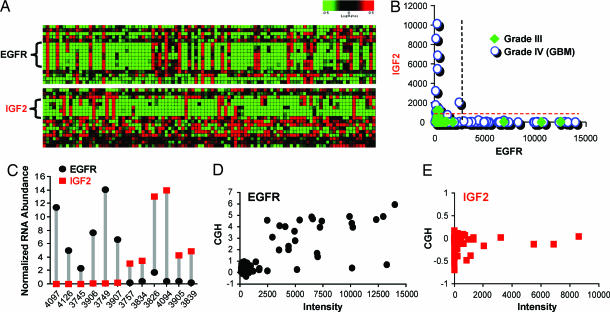
Expression profiles from 194 high-grade astrocytomas (representing 165 cases) and 13 normal brain samples were used for analysis. Genes known to be amplified in GBM such as EGFR (Fig. 1A Upper) or PDGFRα (data not shown) could be readily identified by strong expression in a discrete subset of samples. Screening revealed a separate group of samples that overexpressed IGF2 (Fig. 1A Lower). Among primary grade III tumors (n = 44), EGFR OE was seen in three cases (7%), whereas one case (2%) overexpressed IGF2, with no overlap between them (Fig. 1B). A substantially larger proportion of grade IV tumors displayed OE of either IGF2 or EGFR. Of 121 primary GBM samples, 34 (28%) overexpressed EGFR and a nonoverlapping set of 16 (13%) overexpressed IGF2 (Fig. 1B). Among GBM cases, the mutual exclusivity between EGFR-OE and IGF2-overexpressing (IGF2-OE) tumors is statistically significant (P < 0.05; Fisher's exact test).
Fig. 1.
EGFR-OE and IGF2-OE are nonoverlapping across human GBM samples. (A) Heat map displaying microarray data for EGFR (Upper) and IGF2 (Lower) in a set of GBMs. Z score-normalized intensity values are depicted for Affymetrix probes to EGFR or IGF2 as mapped to chromosomes 7p and 11p, respectively. (B) Plot representing the Affymetrix intensity values for EGFR and IGF2 in grade III gliomas (filled symbols) and GBMs (open circles). No overlap occurs between EGFR-OE and IGF2-OE cases. Dashed lines correspond to cut-off values for IGF2-OE (red) and EGFR-OE (black). (C) Normalized mRNA levels (abundance relative to that of Rab14) for EGFR and IGF2 measured by Taqman in 12 selected cases. (D and E) Comparison of log2 CGH ratios vs. expression values for EGFR (D) and IGF2 (E).
Among cases for which additional RNA was available, those identified by microarray as most strongly IGF2-OE or EGFR-OE were selected for determination of IGF2 and EGFR mRNA abundance by Taqman RT-PCR. Results validate the observation of nonoverlapping EGFR and IGF2 OE in human GBMs and demonstrate that relative RNA abundance of EGFR and IGF2 within strongly positive samples is comparable (Fig. 1C). Consistent with microarray data, EGFR was detected in all samples, whereas IGF2 levels were near the limit of detection in non-IGF2-OE samples.
Among the cases of high-grade astrocytoma profiled by microarray, 27 pairs of matched primary and recurrent tumors were available for analysis [see supporting information (SI) Table 2]. All 19 primary tumors negative for both IGF2 OE and EGFR OE gave rise to recurrent tumors that did not show OE of either element (SI Fig. 7). Of the six instances of EGFR OE primary tumors, five recurrences maintained strong expression (three of which met our criteria for EGFR OE) and one showed a dramatic reduction of EGFR signal to baseline with a concomitant appearance of strong IGF2 OE. Regarding IGF2 OE, one positive primary tumor and one borderline positive primary tumor both showed IGF2 OE at recurrence (SI Fig. 7).
Comparative genomic hybridization (CGH) of 88 GBM specimens used for expression profiling revealed EGFR amplification in 25% of the cases. Good concordance was seen between cases demonstrating EGFR amplification and OE (Fig. 1D and Table 1): 20 of 21 amplified cases showed EGFR OE, whereas, conversely, 21 of 25 EGFR-OE cases showed amplification. No copy number gains or losses were detected near the IGF2 locus (Fig. 1E). CGH analysis of other copy number alterations frequently reported in GBMs is summarized in Table 1. Amplification of PDGFRα was seen in five cases, all of which lacked OE of IGF2 or EGFR. Whereas PTEN loss was seen in only a minority of GBMs lacking IGF2 or EGFR OE, this alteration was frequent in both IGF2-OE and EGFR-OE GBMs. Differences in the frequency of PTEN loss across the three groups with EGFR OE, IGF2 OE, or lacking OE of either element were highly statistically significant (P < 0.0005; Fisher's exact test). A strong trend was seen for differences in frequency of p16 loss (P = 0.066; Fisher's exact test), with IGF2-OE cases showing the lowest frequency (Table 1).
Table 1.
CGH summary
| Case | PDGFR amplification, % | EGFR amplification, % | PTEN loss, % | PIK3R3 gain, % | p16 loss, % | RB loss, % | CDK4 amplification, % | MDM2 amplification, % |
|---|---|---|---|---|---|---|---|---|
| IGF2-OE (n = 11) | 0 | 0 | 73* | 9 | 27 | 27 | 9 | 9 |
| EGFR-OE (n = 25) | 0 | 84 | 80* | 0 | 64 | 21 | 12 | 4 |
| Neither (n = 52) | 10 | 2 | 35* | 10 | 42 | 12 | 12 | 0 |
Genomic copy number alterations for eight genes are presented as a percentage of total cases that overexpress EGFR (EGFROE), IGF2 (IGF2-OE), or neither element.
*, P < 0.0005; Fisher's exact test across all three groups.
Histological Analyses Confirm the Nonoverlap of IGF2 OE and EGFR OE and Reveal the Highly Proliferative Nature of IGF2-OE GBMs.
To validate our finding of IGF2 OE in independent GBM sample sets, we queried tissue blocks from GBM cases (Fig. 2). In situ hybridization (ISH) for IGF2 and immunohistochemistry (IHC) for EGFR in tissue microarray cores from 88 GBMs revealed that 6% (5/88) of all GBM cases were positive for IGF2 and 48% (41/88) showed strong IHC signal for EGFR. No cases were seen that were positive for both IGF2 OE and EGFR OE. In addition, 14 of 74 (19%) sections from an additional set of conventional GBM blocks examined by ISH showed an IGF2 signal in at least one region that was notably more intense than that seen in normal brain. Five (7%) of these cases were rated as intensely positive for IGF2 mRNA, none of which were positive for EGFR by IHC. As clinical histories and details of tissue processing were lacking for the tissues used for ISH, we chose not to perform further IHC analyses in this sample set.
Fig. 2.
Histological confirmation of nonoverlap between IGF2-OE and EGFR-OE GBMs. (A–D) IGF2 mRNA detection by ISH. (A and C) PhosphorImager scans of tissue microarrays hybridized for IGF2 mRNA using IGF2-antisense (A) or sense strand control probe (C). (B and D) Dark-field microphotographs of ISH of the tissue core indicated by red boxes in A and C. (Scale bar: 1 mm.) (E–J) Tissue sections from an EGFR-positive case (E–G) and an IGF2-positive case (H–J) showing IHC for EGFR, Ki-67, and p-akt. (Scale bar: 100 μm.)
Of the 120 primary GBM samples included in our expression profiling analysis, staining for Ki-67 and p-akt was available for 29 and 62 cases, respectively. Mean MIB1 labeling indices (± SEM) were significantly higher for IGF2-OE cases (36.0 ± 9.2, n = 5), than for either EGFR-OE cases (16.1 ± 2.3, n = 9) or GBMs that do not overexpress either growth factor element (20.1 ± 2.8, n = 15; SI Table 2; P < 0.05; Student's t test for both comparisons). Most IGF2-OE cases showed strong staining for p-akt (Fig. 2). Mean ratings of p-akt IHC for IGF2-OE GBMs (1.9 ± 0.1) were comparable to those of EGFR-OE samples (1.7 ± 0.15) and marginally elevated above cases that did not express either growth factor element (1.4 ± 0.1, P = 0.056, Student's t test; SI Table 2).
IGF2 Can Substitute for EGF in Promoting the Growth of Glioma-Derived Neurospheres.
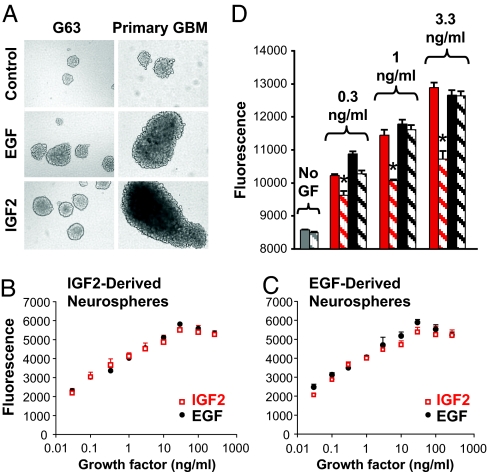
We next sought to determine whether IGF2 is capable of acting like EGF to support glioma tumor cell growth in vitro. Using serum-free cultures of neurospheres from GBM cell lines, we found that IGF2 induced growth responses that are indistinguishable from those shown in response to EGF (Fig. 3A Left). For each of two cell lines examined (G63 and G96), neurospheres from CD133+ sorted cells failed to grow appreciably in the absence of growth factors, but expanded rapidly in the presence of either EGF or IGF2. Maximal responses to both growth factors were seen at doses of ≈20 ng/ml. Growth of neurospheres from acutely dissociated primary GBM tissue (Fig. 3A Right) was also supported by similar concentrations of EGF or IGF2.
Fig. 3.
IGF2 can substitute EGF to support tumor-derived neurosphere growth. (A) (Left) G63 cell line was grown under basal control neurosphere conditions (Top), in the presence of 20 ng/ml EGF (Middle), or with 20 ng/ml IGF2 (Bottom). (Right) Similar results were obtained with neurospheres derived from primary GBM tissue. (B and C) Proliferation assay of cells from neurospheres formed in the presence of IGF2 (B) or EGF (C) shows equivalent growth effects for the two factors. (D) IGF1R blocking antibody (α-IR3, 10 μg/ml) partially inhibits IGF2-induced cell proliferation (∗, P < 0.03) but not the EGF-induced cell growth. Red bars, IGF2; black bars, EGF; solid bars, growth factor alone; striped bars, growth factor plus α-IR3. In B–D representative results of three experiments per cell line are shown. Data are presented as mean ± SD of triplicate samples.
To more directly compare the actions of EGF and IGF2, we investigated the ability of each factor to support growth of neurospheres that had been formed and expanded in the presence of either EGF or IGF2. When IGF2-dependent cell line-derived neurospheres were dissociated, both EGF and IGF2 induced rapid growth of newly formed spheres and dose–response curves for growth with each of the two factors were superimposable (Fig. 3B). Conversely, dissociated EGF-dependent spheres also showed virtually identical growth rates after treatment with either EGF or IGF2 (Fig. 3C). These results support the idea that IGF2 and EGF act on similar cell populations and demonstrate that both factors are equally effective in promoting growth of GBM-derived neurospheres. Growth induced by IGF2 but not EGF was significantly blocked by the presence of 10 μg/ml of the insulin-like growth factor receptor 1 (IGF1R)- blocking antibody IR3, proving specificity of the IGF2-induced cell growth (Fig. 3D).
IGF2 and Phosphoinositide 3-Kinase Regulatory Subunit 3 (PIK3R3) Are Overexpressed in Proliferative GBMs.
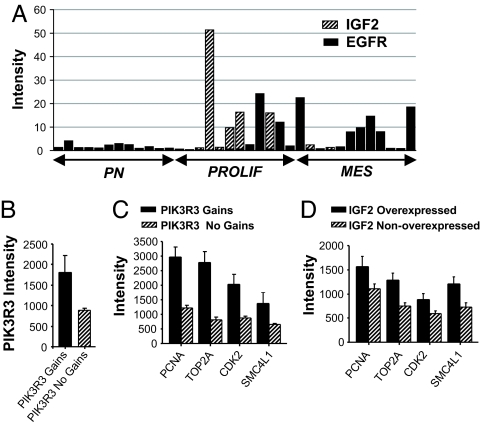
Recent studies conducted in our laboratories identified three molecularly defined prognostic subclasses of high-grade gliomas termed proneural, proliferative, and mesenchymal (5). When we examined a set of samples that included 12 prototypical cases of each of the three subgroups, we observed that IGF2-OE cases were limited to the proliferative subclass, whereas EGFR-OE tumors were present in both proliferative and mesenchymal subclasses (Fig. 4A). Taken together with our MIB1 results, these data support the hypothesis that IGF2-initiated signaling may be involved in the development of highly proliferative GBMs.
Fig. 4.
IGF2 and PIK3R3 are overexpressed in proliferative GBMs. (A) Stacked bar graph shows intensity values for IGF2 and EGFR in 36 samples representing three molecular subtypes of high-grade glioma: proneural (PN), proliferative (PROLIF), and mesenchymal (MES). Values plotted represent intensity values of EGFR or IGF2 for each tumor normalized to the mean intensity of the corresponding gene across all cases. (B and C) Affymetrix intensity values for both expression of PIK3R3 (B) and the proliferative markers PCNA, TOP2A, CDK2, and SMC4L1 (C) are all significantly elevated in cases with PIK3R3 genomic gains compared with those with no gain (n = 6 and 82, respectively; P < 0.001, t test; all comparisons.) (D) Expression of proliferative markers is elevated in IGF2-OE cases (n = 17) compared with IGF2-nonoverexpressing samples (n = 148; P < 0.05, t test; all comparisons).
Previous results revealed that a subset of proliferative subclass tumors show relative copy number gains for PIK3R3, a regulatory subunit of PI3K (5). Herein, we find that tumors with copy number gains at the PIK3R3 locus show significant elevations in PIK3R3 mRNA expression (Fig. 4B). GBMs with PIK3R3 genomic gains showed significantly higher expression of markers of proliferation than did GBMS without PIK3R3 gains (Fig. 4C; P < 0.001, all comparisons). The same markers showed increased expression levels in GBMs that overexpressed IGF2, compared with those with no IGF2 OE (Fig. 4D; P < 0.05, all comparisons). Thus, both IGF2-OE and PIK3R3-overexpressing GBMs appear to manifest a highly proliferative phenotype. Of six cases showing gains at the PIK3R3 locus, five lacked OE of either IGF2 or EGFR and one was an IGF2-OE GBM (Table 1).
PIK3R3 Is a Key Mediator of IGF2-Induced Intracellular Signaling in Human Gliomas.
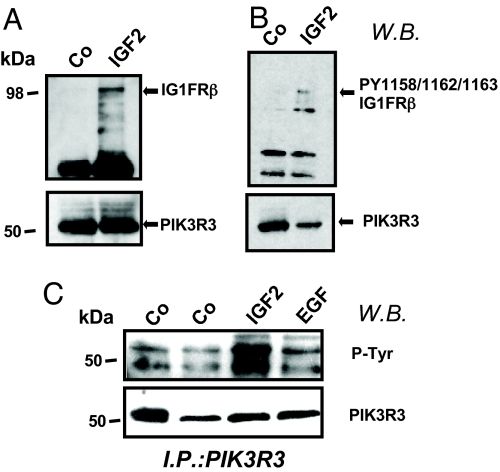
Previous OE studies have demonstrated the association between IGF1R and the PIK3R3 subunit upon growth factor stimulation (12). Given our findings showing that both IGF2 OE and PIK3R3 OE are associated with a proliferative phenotype in GBMs, we sought to investigate whether PIK3R3 mediates cellular effects of IGF2 in gliomas. G63 glioma cells, grown as neurospheres, were dissociated and plated in serum-free media for 48 h, followed by stimulation with IGF2 (20 ng/ml) for 15 min. Immunoprecipitation using an anti-PIK3R3 antibody showed that endogenous PIK3R3 associates with IGF1R (Fig. 5A). Furthermore, phosphorylated IGF1R was also pulled down by PIK3R3 in cells stimulated with IGF2 (Fig. 5B). Stimulation of G63 cells with IGF2 (20 ng/ml), EGF (10 ng/ml), or insulin (data not shown) resulted in formation of an intracellular tyrosine-phosphorylated complex that includes PIK3R3 (Fig. 5C).
Fig. 5.
IGF2 treatment induces association between phopho-IGF1R and PIK3R3 in glioma cells. Western blot show PIK3R3 immunoprecipitates of G96 cells after IGF2 stimulation. Cells were stimulated with IGF2 (20 ng/ml, 30 min) or EGF (10 ng/ml, 30 min) or were unstimulated (labeled Co). Blots were probed with antibodies as follows: antibody to IGF1R kinase domain (A Upper); antibody to PIK3R3 (A Lower); antibody to phospho-IGF1R (PY1158/Y1162/Y1163) (B Upper); antibody to PIK3R3 (B Lower); antibody to phospho-tyrosine (C Upper); antibody to PIK3R3 (C Lower).
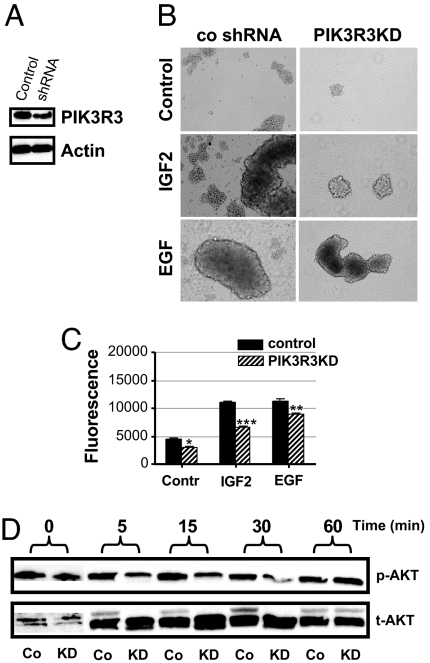
We next investigated the effects of PIK3R3 knockdown (KD) on IGF2- and EGF-induced neurosphere growth. Stable expression of PIK3R3 short hairpin RNA (shRNA) resulted in PIK3R3 mRNA reduction of 81% (G96) and 85% (G63) (data not shown) and KD of the protein detected by Western blot analyses of G96 (Fig. 6A) and G63 (SI Fig. 8A) cells. Pooled clones from G63 or G96 cells were sorted for CD-133 expression, and positively selected cells were used for in vitro neurosphere assays. Stable PIK3R3-KD resulted in inhibition of neurosphere growth in both cell lines (Fig. 6 B and C and SI Fig. 8 B and C). Although effects of PIK3R3-KD on neurosphere growth were seen either in the presence or absence of growth factors, the growth inhibition induced by PIK3R3-KD was most robust when cells were growth-stimulated with IGF2 (Fig. 6C and SI Fig. 8C). Of note, our basal neurosphere media contains insulin, thus growth inhibition with PIK3R3-KD seen in the absence of IGF2 may still be accounted for by reduced signaling through IGFR1. Because Akt is a major downstream target of PI3K, we analyzed the effects of PIK3R3-KD on phosphorylation of Akt upon IGF2 stimulation. G96 cells (PIK3R3-KD and control shRNA) were grown in neurosphere conditions, followed by growth factor withdrawal for 48 h and stimulation with IGF2 (20 ng/ml) for various periods of time. As shown in Fig. 6D, Akt phosphorylation at 5, 15, and 30 min is decreased in PIK3R3-KD cells compared with control. MAPK levels were unchanged (data not shown). Similar results were obtained with G63–derived neurospheres (data not shown).
Fig. 6.
PIK3R3 KD inhibits IGF2-induced growth of glioma-derived neurospheres. (A) Western blot reveals that stable PIK3R3 KD in G96 cells decreases protein levels compared with control shRNA-treated cells. (B) Representative neurospheres of PIK3R3KD and control cells demonstrate that PIK3R3KD inhibits sphere formation and/or growth induced by either EGF or IGF2 in G96 cells. (C) Viability assays of neurosphere growth show a decrease in the number of PIK3R3KD cells that is most profound in IGF2-stimulated cultures (∗, P < 0.05; ∗∗, P < 0.005; ∗∗∗, P < 0.0005). (D) In the absence of growth factor stimulation, p-akt levels are equivalent in control (Co) and KD cells. Under IGF2 (20 ng/ml) stimulation, p-Akt levels in G96PIK3R3KD cells are decreased compared with G96 control cells.
Discussion
Aberrant signaling initiated by growth factors and their receptors cooperates with loss of tumor suppressors to initiate and sustain glioma development. EGFR amplification represents a hallmark for a subclass of human GBMs (4), and mouse models have demonstrated that activating mutations of EGFR acting in concert with p16 loss can promote gliomagenesis (6). The current study identifies IGF2 OE as a novel molecular marker of a subgroup of high-grade gliomas that do not exhibit EGFR amplification. IGF2 has been previously implicated in several types of neoplastic growth. In humans, IGF2 has been implicated in the development of malignancies of the lung, prostate, and adrenal gland (13–16). Loss of IGF2 imprinting is linked to increased risk for developing colorectal cancer and Wilms' tumors (13, 17). In mouse models, OE of IGF2 can lead to the development of lung tumors (18, 19), loss of IGF2 imprinting promotes development of intestinal tumors (20), and IGF2 has been shown to play an essential role in the induction of medulloblastomas (21). Although previous findings reported loss of imprinting and OE of IGF2 in human meningiomas (22, 23), reports on IGF2 expression in glioma have not yielded a consistent picture (24, 25).
In the current study, we observed robust OE of IGF2 in a subset of GBMs that lack EGFR amplification or OE. CGH analysis confirmed the presence of EGFR amplification in one of four of the GBM cases we investigated, but did not reveal any evidence of genomic gains flanking the IGF2 locus. Regardless of the mechanism responsible for robust IGF2 OE, both the higher incidence of this event in grade IV vs. grade III astrocytomas and the association with a highly proliferative phenotype suggest that IGF2 plays a role in promoting development and growth of some GBMs.
Our data show that IGF2–OE GBMs are highly proliferative, a hallmark of aggressive disease, and that IGF2 supports the growth of GBM-derived neurospheres. Interestingly, another recent report (26) shows that IGF2-positive medulloblastoma cells in situ are restricted to a subpopulation that displays intense Ki-67 staining and that cultured medulloblastoma-derived cells and cerebellar neuronal precursors are growth-stimulated by IGF2. Thus, IGF2 may serve as an effective mitogen to promote growth of both medulloblastomas and GBMs, two forms of central nervous system malignancies both hypothesized to arise from neural stem and/or precursor cells.
Recent identification of brain tumor stem-like cells has provided new insights into parallels that exist between gliomagenesis and normal brain development. Two recent studies have used embryonic brain-derived neurospheres to demonstrate that loss of PTEN (11) or p53 (27) enhances renewal and expansion of neural stem cells and promotes their escape from homeostasis, mechanisms also believed to underlie tumor initiation and progression. Both neural stem cells and stem-like cells from brain tumors maintained as neurospheres under the influence of EGF are self-renewing and maintain the potential to differentiate along either neuronal or glial lineages (2, 3, 9, 28). Although no data exist to document the effects of IGF2 on adult neural stem cells, IGF2 has been shown to induce proliferation of cerebellar neuron precursors (29) and IGF2 is implicated as a key factor in supporting expansion of hematopoetic stem cells ex vivo (30). In the current study, we show that IGF2 can support the growth of neurospheres derived from GBMs to the same extent as EGF and that this IGF2-induced effect is mediated, at least in part, through IGF1R. Using multiple methods in independent sample sets, we find that EGFR OE and IGF2 OE are mutually exclusive in GBMs, suggesting that either alteration is capable of supporting tumor growth. Our study of 27 primary and recurrent case pairs is especially intriguing. Primary tumors arising without OE of either IGF2 or EGFR invariably gave rise to recurrent lesions also lacking OE of both elements, suggesting that some lesions arise and are sustained by mechanisms independent of either EGFR or IGF1R signaling. Primary tumors with EGFR or IGF2 OE most frequently were associated with recurrences showing OE of mRNA for the same element. One case, however, was striking in its complete switch from an initial EGFR-OE primary tumor to an IGF2–OE recurrence. These findings suggest that tumors arising under the influence of either EGFR or IGF2 require continued growth factor signaling to sustain tumor growth and that IGF2-induced signaling may substitute for EGFR signaling in driving tumor growth. Taken together, these data support the notion that IGF2 OE may represent an alternate pathway to EGFR amplification in the development and growth of GBMs.
The PI3K-Akt pathway plays a crucial role in supporting growth of several malignancies (31). PTEN, a negative regulator of this pathway, has been called both a “master regulator” of neural precursor development (32), as well as a potent tumor suppressor for gliomas. Several studies have shown that loss of PTEN is an important negative prognostic factor for GBM patients (see ref. 5). Genetic alterations in various catalytic subunits of the PI3K (PIK3CA and PIK3CD) have been recently described in human glioblastomas (33, 34), supporting the role of the PI3K pathway as an integrator of multiple signals essential for tumorigenesis (31). In the current work, we show that PTEN loss (assessed by CGH) and activation of the PI3K-Akt axis (assessed by p-akt IHC) are frequent occurrences in both EGFR-OE and IGF2-OE tumors. Furthermore, we present evidence that implicates a specific PI3K regulatory subunit in mediating IGF2 signaling in gliomas.
The regulatory subunit PIK3R3, also known as p55PIK(p55 γ), was originally isolated by expression library screening for proteins interacting with phosphorylated IRS-1 (35) and has been found to interact with the IGF1R in a yeast two-hybrid screening approach (12). During development, PIK3R3 is highly expressed in the cerebellum, where IGF1R and PIK3R3 were found colocalized in Purkinje cells (36). We identified a subgroup of proliferative GBMs that exhibit genomic gains for PIK3R3 and observed that these gains were associated with increased expression of mRNA of this molecule. Given that both GBMs with IGF2 OE and those with gains of PIK3R3 manifest a proliferative phenotype, we hypothesized that PIK3R3 may mediate some growth-promoting effects of IGF2 on GBM cells. We present evidence that IGF2 stimulation induces endogenous PIK3R3 to associate with phosphorylated IGF1R and incorporate into a tyrosine-phosphorylated intracellular complex. In addition, using glioma-derived neurospheres, we show that induction of both Akt phosphorylation and growth stimulation by IGF2 (and, to a lesser extent, EGF) is inhibited by stable KD of PIK3R3. Importantly, our KD experiments were performed in two cell lines and with different shRNA constructs, arguing in favor of the specificity of the observed effect. These results are unexpected as mRNA for other regulatory subunits of PI3K (i.e., p85α and p85β) are present in both cell lines examined (data not shown) and might have been anticipated to substitute for the action of PIK3R3.
In summary, our results reveal that robust expression of IGF2 is a marker for a subset of high-grade GBMs lacking EGFR amplification and provide evidence that IGF2 signaling via engagement of PIK3R3 can support growth of GBM cells in vitro. These findings suggest that IGF2 OE may serve as an alternate mechanism to EGFR amplification for driving the formation and growth of GBMs.
Experimental Procedures
Expression Arrays, CGH Arrays, and Taqman.
Microarray, CGH, and Taqman were performed as described (5). Both published data and eight new cases from juvenile patients are included in this study (SI Table 2). EGFR OE was defined as >5-fold increase over the median value of all tumors. This cutoff represents the major inflection point in a plot of log expression across all cases. For IGF2, expression in normal brain and most tumors was near the lower detection limit, hence a more conservative standard of OE was defined as >50-fold increase over the median value for all tumors. For scoring relative gains and losses in CGH data, a previously described method was used (37). For more details see SI Methods.
ISH and IHC.
ISH and IHC were performed as described (38). In addition to samples listed in SI Table 2, tissues from commercial sources were used for IGF2 ISH and EGFR IHC. For ISH of IGF2, probe represented nucleotides 468-1341 of GenBank accession no. NM_000612. For more details see SI Methods.
In Vitro Neurosphere and Cell Proliferation Assays.
For neurosphere cultures, cells isolated from GBM cell lines (39) with a CD133 cell isolation kit (Miltenyi Biotech, Auburn, CA) or unsorted cells from primary GBM were maintained in culture as described (40). For growth assays, neurosphere cultures were mechanically triturated into single-cell dissociates and monitored for growth by Alamar blue after 3 days in culture with daily addition of EGF or IGF2 (Fig. 3) or 14 days for shRNA experiments (Fig. 6). Where indicated, IR3 antibody to IGF1R was added to cultures 1 h before growth factor stimulation. For more details see SI Methods.
Immunoprecipitation and Western Blot.
Glioma cell lines grown under neurosphere conditions were dissociated and grown an additional 48 h in the absence of growth factors. For growth factor stimulation experiments, recombinant human IGF2 (R & D Systems, Minneapolis, MN) was added to cultures (20 ng/ml) after which cells were harvested and subjected to immunoprecipitation and Western blotting. For more details see SI Methods.
shRNA Experiments.
Two retroviral shRNA constructs purchased from Open Biosystems (Huntsville, AL) were used in generating stable cell lines: RHS1764–9494180 for G96 and RHS1764–9208343 for G63. Neurospheres from CD133-sorted cells of both G63 and G96 were tested for growth responses in three replicate experiments yielding similar results. For more details see SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. David Eberhard for confirmation of histopathology of some tissues, Sheila Bhedda for help with IHC, Drs. Manfred Westphal and Katrin Lamszus (University of Hamburg, Hamburg, Germany) for glioma cell lines and glioma neurosphere culture, Genentech sequencing and oligo synthesis laboratories, Ian Kasman for assistance with photomicroscopy, Mike Ward for assistance with Bioinformatics software, and Allison Bruce for help with graphics.
Abbreviations
- CGH
comparative genomic hybridization
- EGFR
epidermal growth factor receptor
- EGFR-OE
EGFR overexpressing
- GBM
glioblastoma
- IGF1R
insulin-like growth factor receptor 1
- IGF2
insulin-like growth factor 2
- IGF2-OE
IGF2 overexpressing
- IHC
immunohistochemistry
- ISH
in situ hybridization
- KD
knockdown
- OE
overexpression
- PI3K
phosphoinositide 3-kinase
- PIK3R3
PI3K regulatory subunit 3
- shRNA
short hairpin RNA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611271104/DC1.
References
- 1.Holland EC. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 2.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N, Alvarez-Buylla A, Berger MS. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 4.Nutt C, Louis DN. Cancer of the Nervous System. 2nd Ed. New York: McGraw–Hill; 2005. pp. 837–847. [Google Scholar]
- 5.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JA, Colman H, Soroceanu L, et al. Cancer Cell. 2006;9:157–163. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Holland EC, Hively WP, DePinho RA, Varmus HE. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell MA, He X, Wilkie N, Pollack S, Marshall G, Wafford KA, Svendsen CN. Nat Biotechnol. 2001;19:475–479. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- 9.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 10.Sung JY, Lee SY, Min DS, Eom TY, Ahn YS, Choi MU, Kwon YK, Chung KC. J Neurochem. 2001;78:1044–1053. doi: 10.1046/j.1471-4159.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- 11.Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H. Proc Natl Acad Sci USA. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey BR, Furlanetto RW, Nissley SP. Gene. 1998;209:175–183. doi: 10.1016/s0378-1119(98)00045-6. [DOI] [PubMed] [Google Scholar]
- 13.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 14.Giordano TJ, Thomas DG, Kuick R, Lizyness M, Misek DE, Smith AL, Sanders D, Aljundi RT, Gauger PG, Thompson NW, et al. Am J Pathol. 2003;162:521–531. doi: 10.1016/S0002-9440(10)63846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SL, Goko H, Xu ZD, Kimura G, Sun Y, Kawachi MH, Wilson TG, Wilczynski S, Fujita-Yamaguchi Y. Cell Tissue Res. 1998;291:469–479. doi: 10.1007/s004410051016. [DOI] [PubMed] [Google Scholar]
- 16.Pollak M, Beamer W, Zhang JC. Cancer Metastasis Rev. 1998;17:383–390. doi: 10.1023/a:1006154108619. [DOI] [PubMed] [Google Scholar]
- 17.Vu TH, Chuyen NV, Li T, Hoffman AR. Cancer Res. 2003;63:1900–1905. [PubMed] [Google Scholar]
- 18.Fults DW. Neurosurg Focus. 2005;19:1–8. doi: 10.3171/foc.2005.19.5.8. [DOI] [PubMed] [Google Scholar]
- 19.Moorehead RA, Sanchez OH, Baldwin RM, Khokha R. Oncogene. 2003;22:853–857. doi: 10.1038/sj.onc.1206188. [DOI] [PubMed] [Google Scholar]
- 20.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 21.Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D, Zimmer A, Muller U, Samson E, Quintanilla-Martinez L, Zimmer A. J Biol Chem. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 22.Hultberg BM, Haselbacher G, Nielsen FC, Wulff BS, Gammeltoft S. Cancer. 1993;72:3282–3286. doi: 10.1002/1097-0142(19931201)72:11<3282::aid-cncr2820721125>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Muller S, Zirkel D, Westphal M, Zumkeller W. Eur J Cancer. 2000;36:651–655. doi: 10.1016/s0959-8049(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 24.Sandberg AC, Engberg C, Lake M, von Holst H, Sara VR. Neurosci Lett. 1988;93:114–119. doi: 10.1016/0304-3940(88)90022-5. [DOI] [PubMed] [Google Scholar]
- 25.Uyeno S, Aoki Y, Nata M, Sagisaka K, Kayama T, Yoshimoto T, Ono T. Cancer Res. 1996;56:5356–5359. [PubMed] [Google Scholar]
- 26.Hartmann W, Koch A, Brune H, Waha A, Schuller U, Dani I, Denkhaus D, Langmann W, Bode U, Wiestler OD, et al. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J. Development (Cambridge, UK) 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 28.Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 29.Brooker GJ, Kalloniatis M, Russo VC, Murphy M, Werther GA, Bartlett PF. J Neurosci Res. 2000;59:332–341. doi: 10.1002/(sici)1097-4547(20000201)59:3<332::aid-jnr6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhang CC, Lodish LH. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- 31.Cully M, You H, Levine AJ, Mak TW. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 32.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 33.Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 34.Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pons S, Asano T, Glasheen E, Miralpeix M, Zhang Y, Fisher TL, Myers MG, Jr, Sun XJ, White MF. Mol Cell Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trejo JL, Pons S. J Neurobiol. 2001;47:39–50. doi: 10.1002/neu.1014. [DOI] [PubMed] [Google Scholar]
- 37.Misra A, Pellarin M, Nigro J, Smirnov I, Moore D, Lamborn KR, Pinkel D, Albertson DG, Feuerstein BG. Clin Cancer Res. 2005;11:2907–2918. doi: 10.1158/1078-0432.CCR-04-0708. [DOI] [PubMed] [Google Scholar]
- 38.Phillips HS, Hains JM, Laramee GR, Rosenthal A, Winslow JW. Science. 1990;250:290–294. doi: 10.1126/science.1688328. [DOI] [PubMed] [Google Scholar]
- 39.Hartmann C, Kluwe L, Lucke M, Westphal M. Int J Oncol. 1999;15:975–982. doi: 10.3892/ijo.15.5.975. [DOI] [PubMed] [Google Scholar]
- 40.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.