Abstract
Circadian rhythms of cell and organismal physiology are controlled by an autoregulatory transcription-translation feedback loop that regulates the expression of rhythmic genes in a tissue-specific manner. Recent studies have suggested that components of the circadian pacemaker, such as the Clock and Per2 gene products, regulate a wide variety of processes, including obesity, sensitization to cocaine, cancer susceptibility, and morbidity to chemotherapeutic agents. To identify a more complete cohort of genes that are transcriptionally regulated by CLOCK and/or circadian rhythms, we used a DNA array interrogating the mouse protein-encoding transcriptome to measure gene expression in liver and skeletal muscle from WT and Clock mutant mice. In WT tissue, we found that a large percentage of expressed genes were transcription factors that were rhythmic in either muscle or liver, but not in both, suggesting that tissue-specific output of the pacemaker is regulated in part by a transcriptional cascade. In comparing tissues from WT and Clock mutant mice, we found that the Clock mutation affects the expression of many genes that are rhythmic in WT tissue, but also profoundly affects many nonrhythmic genes. In both liver and skeletal muscle, a significant number of CLOCK-regulated genes were associated with the cell cycle and cell proliferation. To determine whether the observed patterns in cell-cycle gene expression in Clock mutants resulted in functional dysregulation, we compared proliferation rates of fibroblasts derived from WT or Clock mutant embryos and found that the Clock mutation significantly inhibits cell growth and proliferation.
Keywords: cell cycle, circadian rhythms, Clock mutation, gene expression, protein-encoding transcriptome
Many organisms have ≈24-h rhythms in metabolism, physiology, and behavior that are driven by cell autonomous circadian pacemakers (1). These circadian rhythms allow organisms to coordinate a myriad of physiological processes with the changing environment. In mammals, the circadian pacemaker is composed of interlocked transcription-translation feedback loops: the primary loop is composed of the basic helix–loop–helix transcription factors CLOCK and BMAL1, which drive transcription of the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes (1, 2). PER and CRY proteins form the negative limb of the feedback loop by inhibiting their own CLOCK:BMAL1-induced transcription; turnover of PER and CRY allows the cycle to begin anew. The interlocked loop consists of REV-ERB-α and RORα, which repress and activate the Bmal1 gene, thereby modulating its function (3, 4). Mutation or deletion of Clock (5), Bmal1 (6), Per1/2 genes (7, 8), or Cry1/2 (9, 10) genes results in behavioral arrhythmicity and disruption of the autoregulatory loop, whereas disruption of components of the secondary loop results in short period-length phenotypes (3, 4).
The molecular components of the circadian clock are present in the majority of neurons in the suprachiasmatic nucleus (SCN), a bilateral body in the anterior hypothalamus (1, 2, 11). Circadian transcription and neuronal activity in the SCN persist in the prolonged absence of environmental input, and SCN-lesioned animals exhibit behavioral locomotor arrhythmicity. Therefore, these nuclei act coordinately as the primary timekeeper in mammals. It was long thought that SCN neurons were the only cells in the body with an intrinsic clock, but more recent studies have shown that neurons in different brain regions, glia, and diverse peripheral tissues such as liver, kidney, and lung express functional molecular clocks (12–16). These peripheral tissue clocks can sustain rhythmicity even in the absence of the SCN, although the phases of the different tissues are coordinated by signals originating from the SCN (17).
Microarray studies performed on brain, liver, and heart tissue have shown that up to ≈10% of the transcriptome is under circadian regulation (18–21). Interestingly, studies comparing two or more tissues suggested that, although the genetic components of the pacemaker are similar across tissues, the genes exhibiting rhythmic expression are tissue-specific: Panda et al. (19) found only 28 genes, of >7,000 known genes, that were rhythmically expressed in both SCN and liver, while Storch et al. (20) identified a similarly low percentage of genes that were rhythmic in both heart and liver. These experiments raise the question of how circadian oscillators, expressed ubiquitously throughout the body, are able to regulate the expression of diverse genes in a tissue-dependent manner. It has been proposed that the majority of circadian output is achieved via a transcriptional cascade, with the CLOCK–BMAL1 complex directly inducing the expression of other transcription factors that are subsequently responsible for the majority of rhythmic, tissue-specific expression (22). This hypothesis has been supported by the observation that many transcription factors are rhythmic and are direct targets of the CLOCK–BMAL1 complex (23, 24). However, those studies were performed with microarrays that interrogate only approximately one-fourth of the protein-encoding mouse transcriptome, and therefore did not fully represent the genome.
Studies of mice harboring the Clock mutation have suggested a role for CLOCK in a wide variety of physiological processes. Recent studies have implicated circadian genes in functions as diverse as obesity and glucose tolerance (25, 26), reproduction (27), tumor susceptibility (28), and response to chemotherapeutic agents (29). One of the most interesting roles identified for peripheral circadian rhythms is in regulation of the cell cycle: Matsuo et al. (30) found that, after partial hepatectomy, livers from mice lacking both Cry genes regenerated more slowly than livers from WT mice. In the same study, the expression of several genes key to cell-cycle progression, including Wee1 and Cdc2, was found to be under transcriptional regulation by the CLOCK–BMAL1 complex. Other groups have identified circadian regulation of transcription of cyclin D1 and c-myc and protein levels of BCL2 and BAX, all key factors in cell-cycle regulation (28, 31). Furthermore, it has been shown that individual cells continue to rhythmically express clock proteins during cell division, and that the timing of cell division is gated by the circadian clock (14). In addition to regulating circadian gene transcription, the Clock mutation has been shown to affect many nonrhythmic genes, suggesting that core circadian components, including CLOCK, play important roles in noncircadian functions (19).
To address the role that the circadian clock plays in whole-genome gene regulation and in cell growth and proliferation, we analyzed RNA expression from WT liver and skeletal muscle tissue by using a microarray chip (GNFIM) designed to interrogate 36,182 nonredundant transcripts derived from protein-encoding genes (32). We also examined the same tissues from Clock mutant mice to investigate both circadian and noncircadian roles for CLOCK. We then used targets generated from analysis of microarray data to identify pathways disrupted in Clock mutants, including cell growth and proliferation, and tested whether changes in gene expression resulted in functional abnormalities of cell-cycle progression in Clock mutant fibroblasts.
Results and Discussion
Rhythmic Gene Expression in Liver and Skeletal Muscle from WT Mice.
To identify circadian-regulated transcripts, WT mice were entrained to a light/dark (LD) 12:12 cycle and then released into constant darkness (DD). Starting at 30 h in DD, corresponding to circadian time (CT) 18 (CT18), liver and skeletal muscle samples were collected from mice every 4 h for 48 h. For each time point, RNA was extracted from each tissue and pooled into two separate samples (each representing five mice), providing two independent biological replicates per time point. The samples were then hybridized to duplicate sets of custom-made genome arrays composed of 36,182 probe sets designed against the mouse protein-encoding transcriptome. Duplicate intensity values for two full circadian cycles (2 days) for each probe set were evaluated for circadian rhythmicity by COSOPT, as described (19). If significance of fit to a cosine wave was determined, the COSOPT algorithm provided a multiple measures correct β (MMCβ) value indicating goodness of fit to a cosine wave form over a ≈24-h period. MMCβ values for genes with a known circadian expression pattern were then used to set the MMCβ significance cutoff, in this case 0.2; any probe sets with MMCβ ≤ 0.2 were considered rhythmic [see supporting information (SI) Fig. 5 and Table 1]. To assess the validity of this cutoff further, four genes (Timp3, Herpud1, Dscr1, and Gilz) that were not previously known to be rhythmic but that had an MMCβ < 0.3 in both tissues were chosen for follow-up analysis. WT tissues were collected every 4 h for 24 h, and mRNA expression levels were measured by real-time PCR. In all cases where RT-PCR was performed on separate tissue samples, microarray results were confirmed, suggesting that the combination of separate replicates for each genotype and time point with the GCRMA algorithm produces replicable results (33).
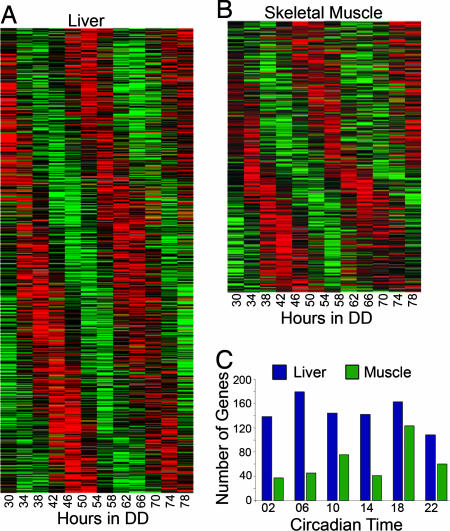
COSOPT analysis identified 854 probe sets in liver and 383 probe sets in muscle that had an MMCβ ≤ 0.2 (Fig. 1 A and B and SI Tables 1 and 2). These data were then subjected to an intensity cutoff value of 500 as the optimal discriminant between expressed and nonexpressed genes. This analysis resulted in the identification of 716 rhythmic genes of 6,818 genes expressed in the liver (10.5%) and 267 rhythmic genes of 7,824 genes expressed in skeletal muscle (3.4%). The liver and skeletal muscle values are consistent with past reports (19, 20) regarding the percentage of rhythmically expressed genes in these tissues. As in previous studies, our results indicate that only a small number of genes are rhythmically expressed in more than one tissue, even when no intensity value cutoff is imposed. Only 57 genes had an MMCβ value ≤ 0.2 in both tissues sampled, many of which have previously been shown to be circadian (SI Table 3). These genes include core pacemaker components (Per2 and Cry2), their regulators (Nr1d1 = Rev-erbα and Stra13 = Dec1), and their immediate downstream targets (Dbp and Tef).
Fig. 1.
Rhythmic expressed genes in WT liver and skeletal muscle. (A and B) Liver and gastrocnemius muscle tissues were collected from WT mice over a 48-h period (30–74 h in DD), and mRNA expression was determined by using a custom Affymetrix whole mouse genome microarray. Expression data were subjected to COSOPT cosine analysis to identify transcripts that were expressed with an ≈24-h period. Rhythmic (MMCβ < 0.2) genes were plotted by peak phase for liver (A, 854 genes) and skeletal muscle (B, 383 genes). (C) These genes were then separated into phase clusters derived from COSOPT analysis. Rhythmic liver genes exhibited large-phase clusters at CT06 and CT18, whereas the largest skeletal muscle-phase cluster occurred at CT18.
Using information derived from COSOPT analysis, we determined the phase distribution of rhythmic genes (Fig. 1C). In liver, we found the largest clusters of rhythmic genes at CT06 and CT18, 6 h after the onset of subjective day and subjective night, respectively. These data are in agreement with previous studies on the phase distribution of gene expression in the liver and likely represent physiological demands on metabolism and catabolism (19). In contrast, in muscle we identified a single large-phase cluster at CT18. As CT18 represents the midpoint of the active phase, ≈6 h after the onset of activity, this peak may be activity-induced. It has previously been shown that resistance exercise can directly affect expression levels of key clock components and downstream targets in muscle (34); our data suggest that locomotor activity may act to phase-coordinate expression of rhythmic genes in skeletal muscle.
We used GeneSpring software to sort highly significant rhythmically expressed genes (MMCβ < 0.1) into functional categories and hand-annotated the list for accuracy. In both tissues, genes involved in biosynthesis and metabolism represented the largest Gene Ontology cluster, as might be expected given the physiological roles of the tissues analyzed (35% in liver and 18% in muscle; SI Fig. 6). However, we also found that proteins involved in the regulation of gene transcription were abundant, representing 10% of highly rhythmic genes in liver and 17% of rhythmic genes in muscle. Many of these transcription factors were rhythmic in one tissue but not the other, suggesting a mechanism for tissue-specific regulation of rhythmic processes. It has been shown that circadian regulation of the transcriptome occurs via both induction (mediated by CLOCK–BMAL1) and repression (mediated by PER–CRY) of gene expression (4, 35). In the transcriptional cascade model, core clock proteins directly regulate the transcription of a limited set of genes that are themselves transcription factors. Spatial and/or temporal restrictions on the expression and dimerization of the transcription factors in the second or third step of the cascade could then determine the tissue-specific cohort of rhythmically expressed genes.
The functional categorization also suggested that up to 16% of rhythmic genes in WT tissues were associated with the cell cycle. We confirmed the rhythmicity of known cell-cycle genes such as Wee1 and p21 (Cdkna1), with real-time PCR (SI Fig. 7). We also found that several Gadd45 isoforms and apoptosis-associated genes such as Bclaf were under circadian control in at least one tissue. Finally, we identified rhythmic expression of genes involved in inducing growth and proliferation, such as Inhbc, Igfbp5, Kit ligand, the PDGF receptor, and Vegf. These data support an association between the circadian clock and cell-cycle regulation, as has recently been suggested by work in mammalian cells (14, 30, 36, 37) and Neurospora (38).
Effect of the Clock Mutation on Rhythmically Expressed Genes.
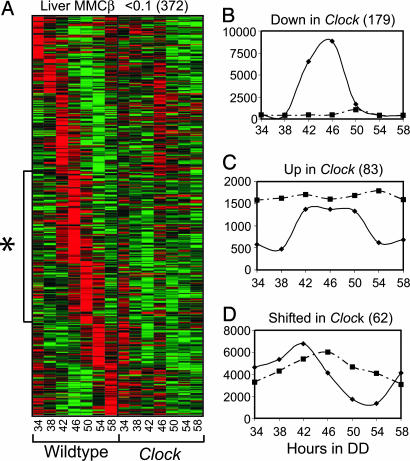
To determine the effect of the Clock mutation on circadian gene expression, we collected liver and skeletal muscle from Clock mutant mice for seven time points every 4 h starting 34 h after the onset of DD (CT22—CT22). We then repeated the genome array analyses and evaluated any differences in gene expression in Clock tissues by using two-way ANOVA comparisons with corresponding WT data (SI Tables 4 and 5). In general, expression of genes with MMCβ < 0.1 in WT tissue exhibited a significant effect of time, genotype, or time and genotype, when compared with expression in tissue from Clock mutant mice (P < 0.05). The majority (71% in liver and 78% in muscle) of rhythmic genes in WT tissue had significantly different expression intensities in Clock tissue (P < 0.05 with respect to genotype; Fig. 2A). Of these genes, the majority (68% and 70%) were down-regulated (<1-fold change; Fig. 2B), including known CLOCK–BMAL1 targets such as Per2, Dbp, Cry2, and Stra13. Circadian genes that were phase-coordinated with known CLOCK–BMAL1 targets in WT tissue (such as Per2 and Dbp) also tended to be down-regulated in Clock mutants, suggesting that many of these genes may also be direct CLOCK–BMAL1 targets. Alternatively, it has recently been proposed that CLOCK–BMAL1 may confer circadian regulation of gene expression not only by activating transcription, but also via specific repression of downstream targets (39). Therefore, circadian genes that normally are repressed by CLOCK–BMAL1 at specific times would likely exhibit up-regulation in Clock tissue. Overall, only a minority (32% and 30% in liver and skeletal muscle, respectively) of all WT circadian genes that were affected by the Clock mutation were up-regulated (Fig. 2C), including Oat, which was previously shown to be both rhythmic in WT mice and up-regulated in Stra13−/− mice (13). Many genes, both up- and down-regulated, exhibited a significant reduction in amplitude in Clock tissues compared with WT tissues, but generally retained a significant variation with time in Clock mutant tissue. It is important to note that this analysis cannot discriminate between genes that are directly regulated by CLOCK and genes that are regulated by other aspects of the molecular pacemaker. Future experiments that compare tissue-specific gene expression among several different circadian mutant or knockout strains will be required to tease apart the effects of single versus multiple components of the pacemaker.
Fig. 2.
The effect of the Clock mutation on WT circadian genes in mouse liver. (A) Pseudocolored graph depicting highly significant (MMCβ < 0.1) WT circadian genes (y axis) versus hours after DD (x axis) for WT or Clock mice. The graph is based on Z-score comparisons of each gene's individual time point intensity in relation to its average intensity for all times and genotypes, where red indicates higher expression and green indicates lower expression. Genes were ordered based on peak time in WT samples. The asterisk indicates genes that peak at CT10–14, which are in-phase with known CLOCK-BMAL1 target genes, such as Per2, Dbp, and Cry2. (B–D) Shown are graphs of fluorescence intensity (y axis) versus hours in DD (x axis) for examples of genes that are overall down-regulated (Dbp; B), up-regulated (Tmlhe; C), or peak-shifted (Casp6; D) in Clock (dashed line) versus WT (solid line) liver. Numbers in parentheses indicate how many genes fit the criteria for each grouping.
We also identified a population of genes in Clock peripheral tissues (16% and 11% in liver and muscle, respectively) that showed a significant shift in the peak time of gene expression compared with WT, but did not exhibit a change in average expression levels (Fig. 2D). Most of these phase-shifted genes were delayed by 4–8 h relative to WT expression. Behaviorally, the Clock mutation results in a 4-h lengthening of the activity/rest cycle period in DD (5). It is possible, therefore, that these peak-shifted genes control, or are controlled by, physiological rhythms (such as feeding and metabolism) in Clock mutants.
CLOCK Regulation of the Noncircadian Transcriptome.
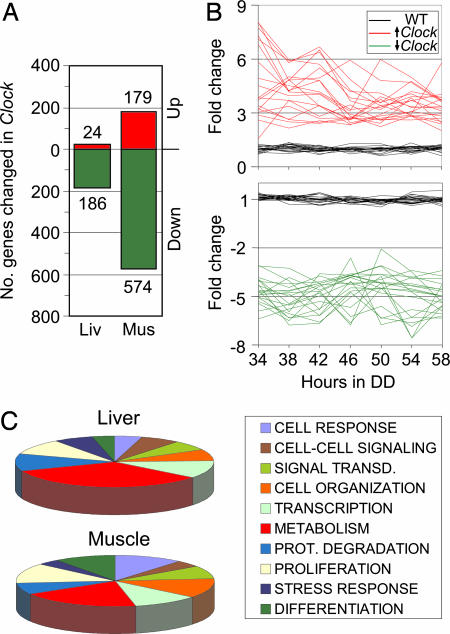
In addition to investigating the effect of the Clock mutation on rhythmic gene expression, we searched for nonrhythmic CLOCK-regulated targets in liver and skeletal muscle. For each probe set, the intensity value for all time points between 34 and 58 h in DD was considered for WT and Clock mutants separately. A t test was performed to compare the WT versus Clock mutant average intensity for each probe set, and the Clock mutant average intensity was divided by the WT average intensity to determine fold change. This method allowed us to analyze average expression level differences regardless of circadian regulation. Genes that had a t test P < 0.01 and a fold change of ≥2.0 or ≤0.5 were considered to be differently expressed between WT and Clock mutants (SI Tables 6 and 7). Using no intensity cut-off, we found that ≈200 genes in the liver and 750 genes in the muscle differed in expression level between WT and Clock mice (Fig. 3 A and B). In both liver and skeletal muscle, the Clock mutation resulted in the down-regulation of gene expression (86% and 76% of significantly changed genes in liver and muscle, respectively) more frequently than up-regulation. In sum, these results suggest that many genes regulated by CLOCK are nonrhythmic, and, furthermore, that CLOCK is directly or indirectly involved in both transcriptional repression and activation in the periphery.
Fig. 3.
Effect of the Clock mutation on gene expression. Gene expression in liver and gastrocnemius muscle from Clock mice was compared with gene expression in WT mice. (A) Bar graphs represent the number of genes whose average expression over a 24-h period showed significant, >2-fold change in expression in Clock tissue. In both liver and muscle, the majority of CLOCK-controlled genes were down-regulated compared with WT expression. (B) Each graph shows 20 examples of genes whose average expression was up-regulated (Upper) or down-regulated (Lower) in Clock muscle. (C) Pie charts depict the most highly represented functional categories of genes changed in Clock mouse liver or muscle.
We used GeneSpring to categorize CLOCK-regulated genes by function and found that genes involved in cell growth and proliferation represented one of the largest functional categories in both liver and skeletal muscle (Fig. 3C). In the liver, 10% of CLOCK-regulated genes were involved in cell growth and proliferation, and in muscle 13% of CLOCK-regulated genes fell into the growth and proliferation category. Included in the cell proliferation-associated genes whose expression was affected by the Clock mutation were p21 (Cdkn1a), a key inhibitor of cell-cycle progression that is up-regulated in peripheral Clock mutant tissue, ki-67, a marker of cell proliferation, and hspca (SI Table 8). Coupled with the above observations, these results suggest that the circadian oscillator and the CLOCK protein may play regulatory roles in cell-cycle control.
CLOCK Regulation of Cell Growth and Proliferation.
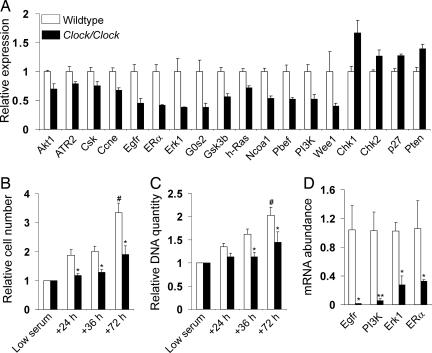
Disrupting circadian rhythms has been shown to have an impact on cell cycle in vivo. In Cry double knockout mice, liver regeneration proceeds more slowly than normal, likely because of constitutive up-regulation of Wee1, a checkpoint kinase (30). Per2 knockout mice show elevated levels of c-myc (a proliferative gene) and reduced p53 (antiproliferative) expression concomitant with increased γ-irradiation-induced rates of cell proliferation and lymphoma (28). In contrast, overexpression of Per1 sensitizes cells and inhibition of Per1 abrogates the response to DNA damage-induced apoptosis (37). In Clock mutant mice, we found that cell-cycle inhibitory genes were generally up-regulated (e.g., p21, p27, Chk1, Chk2, and Atr1) or nonrhythmic (Wee1), whereas proproliferative genes such as Jak2, ERα, Pbef, Akt1, Cdk2, cyclins D3 and E1, and the TGFβ and EGF receptors were down-regulated (Fig. 4A and SI Table 8).
Fig. 4.
DNA synthesis and cell proliferation is diminished in Clock primary cells. (A) The average expression intensities from 24-h microarray data are shown for many genes that are involved in cell proliferation and exhibit significantly (P < 0.05; see SI Table 8) changed expression in Clock liver. (B and C) MEFs from WT and Clock embryos were cultured in 2% BCS for 48 h and then switched to media containing 10% FBS. The number of live cells (B) and quantity of DNA (C) were measured during low-serum and high-serum (+24 h, +36 h, +72 h) conditions. At least three wells per culture were measured at each time point, and all data were normalized to the average low-serum values for each culture. Asterisks represent the significant difference observed for both the relative cell number and relative DNA quantity of WT MEF cells versus Clock MEF cells at +36 h and +72 h, and the relative cell number of WT MEFs versus Clock MEFs at +24 h (∗, effect of genotype; two-way ANOVA). In addition, WT MEFs exhibited significantly higher relative cell number and DNA quantitation at +72h, compared with other time points (#, effect of time; two-way ANOVA). Error bars represent standard deviation from the means of normalized data from 12 independent WT and 10 independent Clock MEF cultures. (D) mRNA expression from MEFs grown in 10% FBS was determined by RT-PCR. Expression of the proproliferative genes Erk1, ERα, Egfr, and PI3K was significantly reduced in Clock MEFs (∗, P < 0.05; ∗∗,P < 0.01; Student's t test).
We hypothesized that the effect of the Clock mutation on cell-cycle gene expression might result in altered proliferation of Clock mutant cells. To test this, we measured DNA synthesis and cell growth in 12 WT and 10 Clock/Clock independently derived mouse embryonic fibroblast (MEF) lines. Cells were plated in 96-well plates at 1–2 × 104 cells per well (three to five replicates per MEF line). To synchronize cell growth and circadian rhythms, the cells were subjected to 48 h of low serum (2% BCS) conditions, then reintroduced to 10% FBS. The amount of DNA and the number of cells were determined after 3 and 48 h in low-serum conditions and at 24, 36, and 72 h after introduction of high serum. As neither genotype showed a change in DNA quantity or cell number during low serum, the 3- and 48-h low-serum values for each genotype were averaged and used to normalize high-serum values by dividing each replicate's high-serum reading by its average low-serum reading. We observed that Clock MEFs exhibit reduced DNA synthesis and cell proliferation compared with WT MEFs (Fig. 4 B and C). Seventy-two hours after introducing 10% FBS, DNA quantity and cell number had increased significantly in WT cells; in contrast, there was no significant increase in Clock MEF DNA quantity or cell number at any time point examined. For both DNA quantity and cell number, there were main effects of genotype, with WT values significantly elevated over Clock values, and time, with 72 h significantly different from all other time points (two-way ANOVA with repeated measures; P < 0.0001). These effects of the Clock mutation depended on cell plating density and were seen at low, but not high, cell densities.
The deficiency in cell proliferation and lack of DNA synthesis suggests that Clock cells fail to respond to mitogenic signals and/or experience a block at the G1/S transition. S-phase entry is dually regulated by the Src-Shc-Ras-Raf-MEK pathway and the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (40, 41), both of which are activated by growth factors, including EGF and estrogen, present in the fetal serum used in cell culture media. Therefore, we measured the expression of components involved in stimulating the G1/S transition, including epidermal growth factor receptor (Egfr), estradiol receptor α (ERα), and the kinases ERK1 and PI3K, and found that all four proproliferative genes were significantly down-regulated in Clock MEFs, in agreement with the microarray results described above (P < 0.05 for all; t test) (Fig. 4D). Thus, Clock cells appear to combine overexpression of factors that inhibit cellular proliferation, such as p27 and p21, with a reduced ability to respond to mitogenic signals.
Despite the failure of Clock MEFs to proliferate in vitro, Clock mutant mice show no apparent developmental abnormalities (5). One possibility, as suggested by DeBruyne et al. (42), is that other genes, such as Npas2, can compensate for the lack of functional CLOCK. It would be of interest to examine whether the Npas2 knockout mice also exhibit deficits in MEF proliferation (43). Another possibility is that Clock mutant mice may not have been carefully examined for developmental defects, and that these mice may have subtle deficits. Adult Clock mutants have been shown to be more susceptible than WT mice to side effects of the chemotherapeutic drug cyclophosphamide, exhibiting reduced B cell survival and recovery after treatment (29). Although the genes responsible for reduced B cell proliferation after chemotherapy in Clock mutants were not identified, our present results suggest that CLOCK-mediated up-regulation of antiproliferative signals, particularly those sensitive to DNA damage that would be activated by cyclophosphamide (44), combined with a reduced response to proliferative signals, could underlie the observed sensitivity. As in the liver regeneration model in Cry knockout mice, deficits in cell cycle and development may become apparent only upon perturbation of the system (30). It is also possible that during development maternal factors override the inhibitory effects of the Clock mutation on growth in fetal cells; after birth, proliferation occurs at a much lower level, and therefore defects may not be apparent unless the system is challenged. Given that studies in humans have shown an association between circadian disruption induced by shift work and the development of breast cancer (45), and that several clock gene mutations have been identified in humans (46, 47), the association between circadian rhythms and development, cell proliferation, and physiological responses to environmental insults presents a fertile field for future research.
Materials and Methods
Animals.
A total of 190 WT male C57BL/6J mice age 7–10 weeks were purchased from The Jackson Laboratory (Bar Harbor, ME) and entrained to a L/D 12:12 cycle for 2 weeks. Mice were then placed in light-tight boxes on a L/D 12:12 cycle for 4 weeks, then released into DD. Starting 30 h after entry into DD (CT18), tissues from 5 (skeletal muscle, gastrocnemius) or 10 (liver) WT mice were collected every 4 h for 48 h, for a total of 12 time points. At time points 34–58 h in DD, tissues from age-matched male C57BL/6J Clock homozygous mutant mice that had been treated with the same light entrainment protocol as the WT mice were collected. Tissues were collected from 5 Clock mutants at each time point except for 34 and 46 h after the onset of DD, when tissues from 10 Clock mice were collected and run as independent replicates. Mice were euthanized by cervical dislocation, and the liver was removed, divided into several 2-mm cubed sections, and snap-frozen on dry ice. The gastrocnemius muscle was removed from each hind leg and frozen in liquid nitrogen.
For verification of microarray data by real-time PCR (TaqMan), 60 male WT mice, aged 12–15 weeks, and 27 age-matched Clock male mice were subjected to the same light entrainment protocol described above. Beginning 34 h after entry into DD (CT22), liver and muscle were collected from eight WT mice and four Clock mutant mice per time point every 4 h for 24 h.
RNA Extraction and Probe Hybridization.
RNA was extracted from ≈100 mg of frozen tissue by using TRIzol (Invitrogen, Carlsbad, CA). Tissues destined for microarray analysis were further processed with a RNAeasy miniprep kit (Qiagen, Chatsworth, CA), and pooled into two independent groups representing five mice each per time point. These groups were kept separate throughout the microarray process. Five micrograms of total RNA per group was used as a template to synthesize cDNA and biotinylated cRNA (Enzo kit; Affymetrix, Santa Clara, CA) using standard Affymetrix protocols. The microarray used was a custom design by Affymetrix and the Genomics Institute of the Novartis Research Foundation (GNF) and is described in Su et al. (32). After hybridization, the arrays were washed with a custom GNF fluidics machine, scanned with an Affymetrix GCS3000 scanner, and analyzed with the GCRMA algorithm (33).
Real-Time PCR.
TaqMan RT-PCR technology (Applied Biosystems, Foster City, CA) was used to validate microarray results and measure expression of genes in adult tissue and MEF cells. Probe and primer sets were designed with Primer Express software (Applied Biosystems) to span exon-exon junctions of target cDNA sequences (see SI Table 9 for sequence information). One hundred nanograms of each RNA sample was reverse-transcribed, and the resulting cDNA was amplified by using the TaqMan EZ RT-PCR kit (Applied Biosystems). For adult tissues, Gapdh was used as an internal reference control, and probe and primer concentrations were optimized so that the target and control reactions could be performed in the same tube. Expression of the target gene was determined by using the comparative Ct (cycle of threshold detection) method to normalize target expression relative to Gapdh expression. For MEF RNA samples, RNA concentration was verified by spectrophotometry, and target gene Ct values were normalized to average WT values, as Gapdh was significantly lowered in Clock MEF cells (data not shown).
MEF Cell Culture.
Embryos were removed from mice on days 12–14 of pregnancy. The uterine and placental tissues were dissected away, and the embryos were washed in cold PBS. The head and peritoneal organs were removed and used to genotype embryos, and the remaining tissue was minced and agitated at 37°C for 30 min in PBS containing 1× trypsin/EDTA with 100 μg/ml DNaseI, followed by two more agitations with equal volumes of PBS containing 1× trypsin/EDTA for 30 min each. Enzymatic dissociation of the cells was stopped with DMEM plus 10% FBS, and cells were centrifuged and resuspended in DMEM plus 10% FBS. Cells were plated on 10-cm dishes at a density of 3 × 106 cells per dish. Passage 2 and 3 cells were plated in 96-well plates at 1–2 × 103 cells per well in DMEM plus 2% BCS (low-serum condition). Twelve WT and 10 Clock/Clock MEF cell lines were used, and each line was represented by three to five replicates. Three and 48 h after plating, cells were collected to quantitate the number of live cells and amount of DNA. After 48 h, cells were placed in media with 10% FBS. The number of live cells and the amount of DNA were quantitated at 0, 24, 48, and 72 h after placing cells in 10% serum. The number of cells per well was measured with an MTT-conversion assay (VyBrant, Molecular Probes, Eugene, OR), and the amount of DNA per well was measured by using a fluorescence-based assay (CyQuant; Molecular Probes) per the manufacturer's instructions. To measure the expression of genes involved in the regulation of cell proliferation, WT and Clock MEFs were grown to confluency in 10% FBS. RNA from the cells was extracted by using TRIzol, diluted to 100 ng/μl, and relative abundance of gene expression was measured by RT-PCR, as described above.
Supplementary Material
Acknowledgments
We thank Ethan Buhr and Martha Vitaterna for expert assistance with tissue sampling. This work was supported by the Novartis Research Foundation (J.B.H., J.R.W., and J.Z.), a predoctoral National Research Service Award from the National Institute of Neurological Disorders and Stroke (to B.H.M.), National Institutes of Health Grants AR050717 (to K.A.E.) and U01 MH61915 (to J.S.T.), The Whitehall Foundation (S.P.), and Silvio O. Conte Center National Institutes of Health Grant P50 MH074924 (to J.S.T.). J.S.T. is an Investigator and E.L.M. is a Research Associate in the Howard Hughes Medical Institute.
Abbreviations
- CT
circadian time
- DD
constant darkness
- LD
light/dark
- MEF
mouse embryonic fibroblast
- MMCβ
multiple measures correct β
- SCN
suprachiasmatic nucleus.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE3751).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611724104/DC1.
References
- 1.Lowrey PL, Takahashi JS. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 4.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitaterna MH, King DP, Chang A, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 8.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 9.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M., de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 10.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 13.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prolo LM, Takahashi JS, Herzog ED. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. Proc Natl Acad Sci. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Grant TW, Hastings MH, Kyriacou CP. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 19.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 20.Storch KF, Lipna OIL, Viswanathan N, Davis F, Wong W, Weitz C. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 21.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 22.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 23.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 24.Grechez-Cassiau A, Panda S, Lacoche S, Teboul M, Azmil S, Laudet V, Hogenesch JB, Taneja R, Delaunay F. J Biol Chem. 2004;279:1141–1150. doi: 10.1074/jbc.M305369200. [DOI] [PubMed] [Google Scholar]
- 25.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L, Pelicano H, Liu J, Huang P, Lee C. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 29.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Proc Natl Acad Sci USA. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 31.Granda TG, Liu XH, Smaaland R, Cermakian N, Filipski E, Sassone-Corsi P, Levi F. FASEB J. 2005;19:304–306. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- 32.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker JR, Hogenesch JB. Methods Enzymol. 2005;393:366–376. doi: 10.1016/S0076-6879(05)93016-4. [DOI] [PubMed] [Google Scholar]
- 34.Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, Takahashi JS, Schambelan M, Conklin BR. Genome Biol. 2003;4:R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 36.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 38.Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- 39.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. FASEB J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 40.Bartek J, Lukas J. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 41.Chang L, Karin M. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 42.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 43.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 44.Antoch MP, Kondratov RV, Takahashi JS. Cell Cycle. 2005;4:901–907. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Colditz GA. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 46.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.