Abstract
Efficient incorporation of novel DNA sequences into a specific site in the genome of living human cells remains a challenge despite its potential utility to genetic medicine, biotechnology, and basic research. We find that a precisely placed double-strand break induced by engineered zinc finger nucleases (ZFNs) can stimulate integration of long DNA stretches into a predetermined genomic location, resulting in high-efficiency site-specific gene addition. Using an extrachromosomal DNA donor carrying a 12-bp tag, a 900-bp ORF, or a 1.5-kb promoter-transcription unit flanked by locus-specific homology arms, we find targeted integration frequencies of 15%, 6%, and 5%, respectively, within 72 h of treatment, and with no selection for the desired event. Importantly, we find that the integration event occurs in a homology-directed manner and leads to the accurate reconstruction of the donor-specified genotype at the endogenous chromosomal locus, and hence presumably results from synthesis-dependent strand annealing repair of the break using the donor DNA as a template. This site-specific gene addition occurs with no measurable increase in the rate of random integration. Remarkably, we also find that ZFNs can drive the addition of an 8-kb sequence carrying three distinct promoter-transcription units into an endogenous locus at a frequency of 6%, also in the absence of any selection. These data reveal the surprising versatility of the specialized polymerase machinery involved in double-strand break repair, illuminate a powerful approach to mammalian cell engineering, and open the possibility of ZFN-driven gene addition therapy for human genetic disease.
Keywords: gene therapy, protein production, somatic cell genetics
The C2H2 zinc finger (1), the most abundant DNA recognition motif in eukarya (2, 3), is highly amenable to engineering for the recognition of virtually any DNA sequence (4–6). These properties have been successfully exploited to enable the modulation of gene expression via their application as designed transcription factors (ZFP-TFs) (7), as well as direct modification of the DNA itself via engineered zinc finger nucleases (ZFNs) for human gene correction (8). The latter process, based on work from several laboratories including our own (9–16), overcomes the exceedingly low frequency of spontaneous homologous recombination in mammalian cells, which until recently has made the targeted modification of human genome sequence in vivo impractical (17, 18). Although this limitation has been addressed in settings where drug-based selection schemes can be applied (19, 20), it is restricted to particular cell types, e.g., fibroblasts and mouse embryonic stem cells. Such traditional “gene targeting” requires the construction of elaborate vectors, a 6- to 8-week regimen of treatment with two distinct selective agents, and the isolation of individual cell clones by limiting dilution, only a subset of which carries the desired targeting event (18).
ZFN-mediated gene correction (8), in contrast, occurs at high frequency without selection, is applicable to a broad range of primary and transformed cells, and does not require cell cloning because it invokes a natural process of genetic information transfer via a double-strand break (DSB). A DSB evoked by a stalled DNA replication fork or by an environmental insult is normally eliminated via end-joining (21) or homology-directed repair (HDR). The latter is a specialized form of homologous recombination that transfers genetic information to the broken chromosome from a DNA molecule of related sequence (22–25). Indeed, we have earlier shown that targeting a DSB to a specific site in the genome with engineered ZFNs (Fig. 1A) transfers single-base-pair changes from a donor plasmid into the chromosome with efficiencies that can exceed 20% (16).
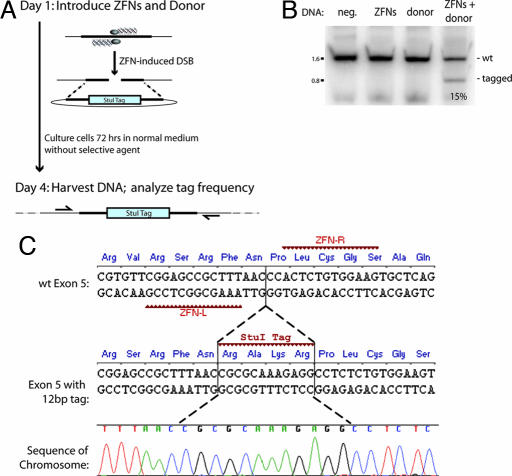
Fig. 1.
A ZFN-induced DSB leads to efficient, homology-based tag transfer into a native chromosomal locus. (A) Experimental outline and a schematic of the process whereby a ZFN-induced DSB is repaired by using an extrachromosomal donor as a template. (B) PCR-based measurements of ZFN-driven tag integration frequency into the IL2Rγ locus in K562 cells. Cells were left untransfected (first lane, “neg.” for negative control) or were transfected with an expression cassette for ZFNs that induce a DSB at exon 5 of IL2Rγ (16) (second lane), and donor plasmids carrying a 12-bp tag flanked by 750-bp homology arms, in the absence (third lane) and presence (fourth lane) of the IL2Rγ ZFNs. Genomic DNA was extracted 72 h later. The IL2Rγ locus was amplified by 20 cycles of PCR in the presence of radiolabeled dNTPs by using primers that hybridize to the chromosome outside of the donor homology arms, and the PCR products were digested with StuI, resolved by 10% PAGE, and autoradiographed. The percentage of StuI-sensitive DNA is indicated below the fourth lane. (C) Sequence analysis of ZFN-edited chromatids. The primary DNA sequence, and the amino acid sequence it encodes, of exon 5 of the human IL2Rγ gene, along with the target sites of the designed ZFNs, are indicated. The central portion of the donor sequence, along with the tag, is shown below. A representative chromatogram of the DNA sequence of one of the chromatids obtained from sample 4 (in B) is provided, showing the chromosomal sequence to be altered precisely in the manner specified by the donor, i.e., by copy-pasting of codons for four new amino acids in-frame with the endogenous ORF. Note that an additional silent SNP (Pro229 CCA→CCT), introduced for cloning purposes, is also transferred from the donor.
However rapid and efficient, gene correction is a localized event, and a single DSB, whether induced by a homing endonuclease (26) or by a ZFN (M.C.H., Y.-L.L., and F.D.U., unpublished data), can allow efficient correction of mutations only within an ≈200-bp window surrounding the break. The complex mutational spectrum underlying many human monogenic diseases would therefore require tailoring ZFNs to each cluster of mutations. This requirement has prompted us to investigate the feasibility of using ZFNs to drive site-specific “gene addition,” specifically, the integration of long DNA segments into a predetermined locus. Both medical (gene therapy) and industrial (e.g., engineering cell lines for protein production) gene addition is currently achieved via random integration of the transgene into the genome, a process that presents safety concerns from a clinical perspective (27) and is costly and time-consuming in industrial applications because of chromatin-based effects on expression of a randomly integrated transgene (28). A considerable effort notwithstanding (29, 30), only limited progress has been made so far in controlling the location of gene insertion, and extensive screening or selection for the desired event is almost invariably a prerequisite.
The present work shows that efficient, site-specific gene addition into a predetermined endogenous locus in human cells can occur in the absence of selection. We show that if a ZFN-cleaved locus is provided with an engineered template that consists of novel genetic information flanked by appropriate regions of target site homology, then break repair occurs via HDR in a manner most consistent with the synthesis-dependent strand annealing (SDSA) model of DSB repair (31, 32). Our data illuminate the surprising versatility of the currently unidentified DNA polymerase machinery involved in HDR: we observe accurate and highly efficient transfer of up to 8,000 bp of genetic information from an episomal donor to an endogenous locus in human cells in the absence of selection.
Results
Efficient ZFN-Induced Tag Integration into a Native Chromosomal Locus.
In earlier work (16) we showed that a ZFN-induced DSB can transfer a single-base-pair change into an endogenous locus from a plasmid donor in human cells at a frequency of ≈20% in the absence of selection. Such “gene correction” invokes a process formally known as “short-patch gene conversion” (24). Studies in budding yeast (33) and Drosophila, however, have indicated that considerably larger segments of genetic information can be transferred in a homology-dependent way from one DNA molecule to another in the wake of a DSB. For example, the best-studied such system, mating type switching in Saccharomyces cerevisiae, relocates 700 bp in this manner (34).
As originally proposed by Gloor and colleagues (31) based on their analysis of P-element excision in Drosophila, providing a novel DNA stretch within a donor molecule, such that it becomes invisible to the HDR machinery during the homology search, could allow a break-driven integration event to occur via a process termed SDSA (see Discussion for a detailed explanation) (32, 33). Evidence from budding yeast (35) and Drosophila (31, 36) supports the notion that SDSA is one of the two pathways of homology-based DNA break repair in mitotic cells (an alternative process, known as the “double Holliday junction,” appears to operate in some settings) (32).
From a somatic cell genetics perspective, SDSA-based resolution of a nuclease-induced break could, in principle, allow the transfer of extended stretches of genetic information to endogenous chromosomal locations from a plasmid donor. In our studies of ZFN-driven gene correction, however, we found that relatively minor sequence differences between the chromosomal locus being targeted and the homology arms contained on the donor molecule significantly lower genome editing frequency. This “donor–target” divergence penalty was imposed even when the mismatches were only a few base pairs from the position of the DSB (Y.-L.L., F.D.U., and M.C.H., unpublished data), suggesting a mechanistic distinction of this phenomenon from the well established requirement for donor–target isogeny in conventional gene targeting (18). Based on the assumption that SDSA is the major break repair pathway for endogenous loci in mammalian cells, we hypothesized that confining this divergence to the position corresponding to the DSB could lower this penalty (Fig. 1 A and C).
To investigate this issue, we built a donor DNA plasmid engineered to introduce a 4-aa tag in-frame with an endogenous locus (Fig. 1C). In this construct, 750-bp homology arms isogenic to the IL2Rγ locus are interrupted with a 12-bp stretch engineered to introduce four new amino acids, RAKR, i.e., a furin cleavage site, in-frame with the endogenous IL2Rγ ORF (for ease of subsequent detection, this tag was engineered to carry a StuI recognition site). Importantly (Fig. 1C), the tag was placed between Asn228 and Pro229, i.e., precisely at the position of the ZFN-induced break. We introduced this donor plasmid along with an expression plasmid encoding designed ZFNs engineered to introduce a DSB in exon 5 of IL2Rγ into K562 cells. After 72 h of culturing the cells in normal medium and in the absence of any selection, we harvested genomic DNA and measured the frequency of tag integration by a highly quantitative PCR-based assay (16) that measures the percentage of total chromatids that have acquired an StuI site. In agreement with expectation, no measurable tag integration into the chromosome was observed in the absence of ZFNs (Fig. 1B, third lane). In contrast, 15% of the chromatids were StuI-sensitive in cells exposed both to the donor plasmid and the ZFNs (Fig. 1B, fourth lane). We isolated, cloned, and sequenced the cognate stretch of the X chromosome from this cell sample; this analysis (Fig. 1C) demonstrated the precise “copy-pasting” of the four codons from the donor plasmid into the chromosome, i.e., the reconstruction of the donor-specified genotype at an endogenous locus in human cells.
The speed (72 h) and frequency (15%) of this genome editing event was identical to that we had earlier observed in transferring single-base-pair changes to the chromosome (16). These data suggest that a properly placed exogenous DNA sequence can be transferred with high efficiency and in the absence of selection from a plasmid donor into a native locus in mammalian cells by inducing a DSB at the target using designed ZFNs.
A ZFN-Invoked DSB Can Be Used to Target the Efficient Integration of an ORF and an Expression Cassette in the Absence of Selection.
The high efficiency of ZFN-driven tag transfer from a plasmid donor into an endogenous locus in the absence of selection provided encouraging evidence in support of SDSA as a major break resolution pathway for an endogenous locus in human cells. It remained unclear whether this method could be used to obtain practically useful frequencies of targeted integration of longer DNA stretches into the chromosome, such as ORFs and expression cassettes, namely, DNA fragments that would be useful in typical real-world applications of this technology.
We therefore first used a chromosomal reporter system (Fig. 2A) to investigate whether ZFNs can be used to integrate a 900-bp ORF into a defined locus in human cells. To generate a reporter cell line, we introduced an ORF-less promoter cassette followed by a ZFN recognition site (16) into the genome of HEK293 cells at single copy. In agreement with published estimates of the rate of homologous recombination in human cells (18), transfection of these cells with only a donor DNA plasmid construct containing a GFP ORF flanked by stretches of homology to the integrated construct yielded <0.06% GFP-positive cells [Fig. 2A Center and supporting information (SI) Fig. 5]. In striking contrast, 72 h after transfection of these reporter cells with the donor DNA construct together with an expression construct encoding the ZFNs, 1.4% of the cells were GFP-fluorescent (Fig. 2A Right and SI Fig. 5). Only background levels of GFP-positive cells were seen when the ZFN-encoding and donor DNA plasmids were introduced into cells that did not have the ZFN target site engineered into their genome (data not shown). Together these data showed that ZFNs can be used to drive transgene integration into a defined chromosomal location in human cells.
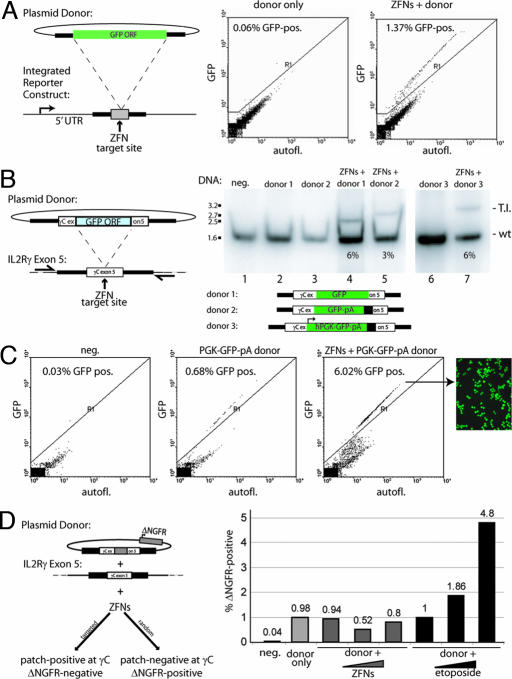
Fig. 2.
ZFN-driven targeted integration of a series of progressively larger DNA sequences into an endogenous locus. (A Left) A schematic of a chromosomal reporter construct in HEK293 cells that contains the recognition site for two ZFNs (gray box) and a donor molecule that carries the GFP ORF (green rectangle) flanked by homology arms. The percentage of GFP-positive HEK293 cells was measured by FACS (Center and Right) and is indicated in each panel. (B) PCR-based measurements of ZFN-driven integration frequency into the IL2Rγ locus in K562 cells. Cells were left untransfected (lane 1) or were transfected with an expression cassette for ZFNs that induce a DSB at exon 5 of IL2Rγ (16) (lane 2), and donor plasmids carrying the indicated inserts flanked by 750-bp homology arms, in the absence (lanes 3 and 6) and presence (lanes 4, 5, and 7) of the IL2Rγ ZFNs. The donor DNAs tested were as follows: a 900-bp GFP ORF (lane 4) or the same ORF followed by a polyA sequence (lane 5) and an autonomous expression cassette (human phosphoglycerokinase promoter–GFP–polyA; lane 7). Genomic DNA was amplified by using primers outside the donor homology arms, and the level of targeted integration was determined by PAGE and autoradiography (the integrant-carrying chromosome migrates above the wild-type one). The integration frequency is indicated for each panel. Note that the autoradiograph for lanes 6 and 7 was generated in an experiment distinct from that for lanes 1–5. (C) Functional measurement of targeted integration frequency. The percentage of GFP-positive cells was measured by FACS in K562 cells transfected with an IL2Rγ donor molecule carrying an autonomous GFP expression cassette (Center; see donor schematic at the bottom B), transfected with this donor and IL2Rγ-specific ZFNs (Right) or untreated control cells (Left), and is shown within each panel. All measurements were taken 3 weeks after transfection to permit decay of expression from the donor episome. A fluorescence micrograph of an aliquot of the GFP-positive cells from Right is also shown. (D) FACS-based measurement of the rate of plasmid DNA random integration. (Left) The plasmid donor construct (a tag-interrupted homology stretch flanked by an autonomous expression cassette for a cell surface marker, ΔNGFR). Cell phenotypes expected from a targeted (lower left) or random (lower right) integration event are shown. (Right) FACS data from an experiment in which K562 cells were treated with only the donor molecule, the donor molecule together with the ZFN expression cassette, or the donor molecule and an increasing concentration of etoposide. The percentage of cells positive for the ΔNGFR marker (as measured by FACS after sufficient cell passaging to allow for donor DNA decay) in each sample is indicated.
To measure the frequency of ZFN-driven targeted integration at an endogenous locus, rather than into a reporter gene, we prepared a series of donor DNA molecules carrying progressively larger insert sequences, each flanked by invariant short regions of DNA homologous to the IL2Rγ gene. We treated K562 cells with these donor DNA molecules and the IL2Rγ-specific ZFNs, expanded them for 72 h in the absence of selection, and analyzed integration frequency by a quantitative PCR assay. We found that a 900-bp GFP ORF, a 1.1-kb GFP ORF-poly(A) tail cassette, and a 1.5-kb promoter-transcription unit integrated into this stretch of the X-chromosome in a ZFN-dependent manner at a frequency of 6%, 3%, and 6%, respectively (Fig. 2B). In agreement with data obtained on the integration of short tags (Fig. 1B), direct sequencing of IL2Rγ-derived PCR products confirmed precise, homology-dependent integration of the donor-provided sequence at the ZFN target site (data not shown).
To demonstrate the functionality of the transgene when integrated into the target locus, we then took cells corresponding to lanes 1, 6, and 7 (Fig. 2B) and FACS-sorted them for GFP expression. As in all previous experiments, the cells were grown in the absence of any selection. After sufficient cell passaging to allow for the episomal donor to be eliminated from the cells, we observed 0.7% GFP-positive cells in the “donor alone” sample resulting from random integration (17) and 6% GFP-positive cells in the “ZFN + donor” DNA-treated population (Fig. 2C). This value agrees precisely with genotyping data on the ZFN-targeted locus obtained by a quantitative PCR assay (Fig. 2B) and indicates that targeted integration into exon 5 of IL2Rγ results in a functional level of transgene expression. Analysis of insert-positive clonal lines revealed, similar to our published results (16), an ≈2:1 ratio of cells heterozygous and homozygous for the insert (data not shown), indicating the feasibility of ZFN-driven biallelic targeted integration.
DNA introduced into human cells integrates randomly at a measurable frequency (17). To determine whether ZFN treatment would alter the rate of this spontaneously occurring random integration, we used a circular donor molecule in which a cell surface marker gene (ΔNGFR) was placed outside one of the homology arms (Fig. 2D) to specifically detect nonhomologous integration events. In three independent experiments (Fig. 2D and data not shown), we observed no change in the random integration of ΔNGFR in cells transfected with both the donor and ZFN-encoding plasmids relative to cells transfected with only the donor DNA plasmid (Fig. 2D, donor + ZFN samples). In contrast, when we exposed donor-transfected cells to etoposide, which is known to introduce random DSBs into the genome, we observed a marked increase in the rate of random integration events (Fig. 2D, donor + etoposide).
Rapid and High Efficiency of ZFN-Driven Targeted Integration of an 8-kb Expression Cassette in the Absence of Selection.
The utility of ZFN-driven targeted integration would be significantly expanded if a large “payload,” e.g., a long cDNA or multiple expression cassettes, could be introduced into the genome at high frequency by using a single donor DNA. We constructed a donor in which the 750-bp homology arms specific for IL2Rγ were interrupted with a 7,762-bp insert carrying three separate promoter-transcription units, two of which encode the heavy and light chains of a human IgG antibody molecule, and the third of which encodes a marker (Fig. 3A). Cells were exposed to the ZFNs and this donor molecule and cultured in nonselective medium. A qualitative PCR-based assay confirmed that ZFN-treated cells acquired transgene-specific sequences in the X chromosome (data not shown); remarkably, in three independent experiments, Southern blotting indicated that ≈5–8% of the chromatids had acquired the transgene (Fig. 3C Left and data not shown). The cells were then passaged in the absence of any selective pressure for 5 weeks to reduce episomal donor DNA levels to background. We observed measurable levels of human IgG in the medium harboring cells exposed to the ZFNs and the donor DNA construct, significantly above control samples (Fig. 3B).
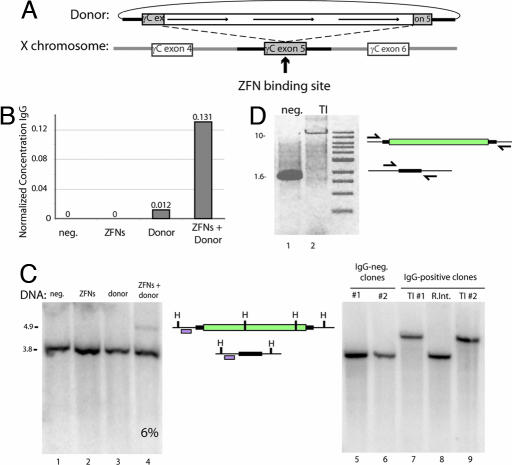
Fig. 3.
ZFN-driven integration of an ≈8-kb DNA sequence encoding multiple transgenes into an endogenous locus. (A) A schematic representation indicating the design of the donor DNA plasmid containing the 750-bp homologous flanking sequence of the IL2Rγ exon5 region and the three promoter-transcription units (line arrows). The site of cleavage of the ZFNs for IL2Rγ is indicated. (B) ELISA for IgG in culture medium. Medium was collected from K562 cells after the indicated treatments, and levels of secreted IgG were measured by performing an ELISA with an antibody for the heavy and light chains of IgG. IgG concentration is expressed in nanograms per milliliter per cell. (C) Southern blot-based measurement of targeted integration frequency. (Left) The same genomic DNA preparations as in B were digested with HindIII, and Southern blotting was performed with a fragment of the IL2Rγ locus that lies adjacent to the left donor homology arm. The locus maps in Center indicate the restriction map of a wild-type chromosome (bottom, 3.8 kb) and a chromosome carrying the integrated transgene (top, 4.9 kb), and the probe used is indicated with an open box. H, HindIII site. (Right) Southern blot analysis of cell clones (see SI Fig. 6C for their genotypes). Lanes 5 and 6, control clones that do not secrete IgG into the medium; lanes 7 and 9, cells that secrete IgG into the medium and appear to carry the insert at IL2Rγ as gauged by PCR (SI Fig. 6C); lane 8, cells that secrete IgG into the medium and do not appear to carry the insert at IL2Rγ as gauged by PCR (SI Fig. 6C). Genomic DNA from single-cell-derived clones was isolated and digested with HindIII and probed as in Left. (D) PCR across the integrated expression cassette. PCR was performed on genomic DNA isolated from clones either negative (lane 1) or positive (lane 2) for transgene integration after transfection with the ZFNs and the donor plasmid, using a high-fidelity polymerase and primers outside of the donor homology arms (arrows in locus map).
To investigate events at the single-cell level, we performed limiting dilution of the cells in the absence of selection. Eighty five single-cell-derived clones were studied, and 8 (9.4%) were found to secrete human IgG into the medium (SI Fig. 6C). We used a PCR-based assay (SI Fig. 6C) as well as Southern blotting (Fig. 3C Right) to genotype these along with control (i.e., IgG nonsecreting) clones at the ZFN-targeted locus. We found seven of eight clones carried the transgene at exon 5 of IL2Rγ, whereas the remaining clone represented a random integration event elsewhere in the genome (SI Fig. 6C), indicating an 8.2% targeted integration frequency, a value fully consistent with our Southern blotting data (Fig. 3C). Remarkably, long-range PCR (Fig. 3D) and sequencing across the insert (data not shown) indicated that the transgene had integrated in a precise, homology-dependent fashion.
In agreement with data obtained after integration of single-ORF constructs (Fig. 2D), extensive Southern blotting analysis revealed no randomly integrated donor DNA in clones carrying the transgene at IL2Rγ (data not shown). Interestingly, both PCR and Southern blotting indicated the absence of wild-type X chromosome-derived signal in transgene-carrying single-cell clones (Fig. 3C, lanes 7 and 9, and SI Fig. 6). To address the possibility that this resulted from the loss of X chromosomal sequence [i.e., a loss-of-heterozygosity event (25)], we performed FISH using a probe located at XqC3, a chromosomal position 3.5 Mb away from the IL2Rγ locus, and found that cells from these clonal lines maintained euploidy for the X chromosome (SI Fig. 6A). In subsequent experiments, we observed an overall 2:1 ratio of cells heterozygous and homozygous for the transgene (data not shown), demonstrating that, as expected, biallelic integration is not the exclusive outcome of ZFN-mediated gene addition. Taken together, these data indicate that designed ZFNs can be used to drive the targeted integration of inserts up to 7.6 kb in length at a frequency of >5% in the absence of any selection into a native locus in human cells. Importantly, as gauged by PCR, Southern blotting, and direct sequencing, cells carrying this large transgene have integrated it into the chromosome via a homology-driven process.
Discussion
The present work offers two conclusions. From a basic biology standpoint, our data reflect on the repair of DSBs by homology-based mechanisms, a process highly conserved between yeast (32, 33) and mammals (22, 23). In a recent study inspired by the classic Meselson–Stahl experiment (37), Haber and colleagues (35) used in vivo metabolic labeling to conclusively identify SDSA (Fig. 4A) as the primary homology-based mechanism for DSB repair in budding yeast. As predicted by studies in yeast and in Drosophila (32), the outcome of this process is the highly accurate reconstruction of a donor-specified genotype at the chromosomal locus. Although other interpretations are possible, our data are most simply explained if repair of a DSB at a native chromosomal locus in human cells also employs this SDSA mechanism.
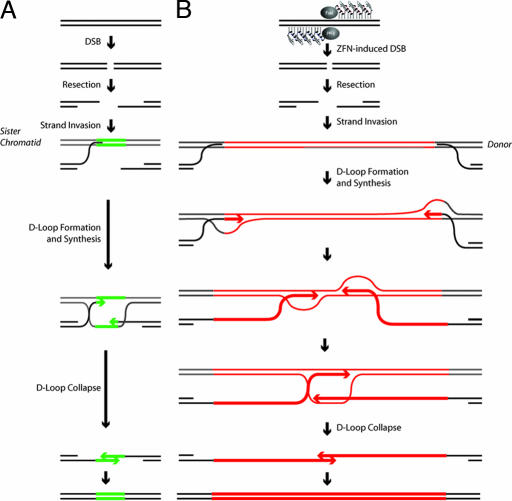
Fig. 4.
ZFN-driven repair of endogenous DSBs and of ZFN-induced breaks followed by homology-directed targeted integration. (A) SDSA-based HDR of an endogenous break (32). After a DSB a single-stranded chromosomal tail invades a sister chromatid, and after DNA synthesis of a short stretch the D loop collapses the newly synthesized DNA. (B) Homology-directed targeted integration after a ZFN-induced break and SDSA-based transfer of genetic information into the break, a model most consistent with the data presented in the current work and evidence in the literature. After a ZFN-induced break, the single-stranded chromosome end homes into the homology arm carried by the donor (this process is unimpeded by the presence of the insert because the latter is located precisely at the position corresponding to the break and hence remains “invisible” to the homology search mechanism). Synthesis then proceeds across at least 50% of the insert length, with the newly synthesized single-stranded DNA trailing the D loop. Irrespective of whether the two broken chromosomal ends use the same (as shown) or two different donor molecules as templates, once synthesis in each D loop has proceeded long enough for the two DNA stretches to overlap, the newly synthesized DNA molecules can leave the D loop, anneal to each other, and then use each other as templates to restore an intact chromatid, now carrying the donor-specified transgene at the chromosome.
Our discovery that transgenes as long as 8 kb can be transferred into a native chromosomal locus (presumably via SDSA) is surprising. The DNA polymerase responsible for the majority of HDR events in yeast and in mammalian cells remains unidentified (38–41). Our analysis shows this polymerase to be remarkably versatile and capable of synthesizing at least a 4,000-nt single strand before donor escape and annealing with a similar such single strand emerging from the other chromosome end (Fig. 4B). This versatility is unexpected, given that this enzyme has presumably evolved to perform short-patch gene conversion between sister chromatids, i.e., synthesize <100 nt before SDSA heals the broken chromosome (Fig. 4A).
Our results have significant implications for somatic cell genetics. We find that designed ZFNs can drive site-specific addition of long DNA stretches into a predetermined locus in the human genome, and observe a high frequency (5–15%) of integration of DNA segments between 12 bp and ≈8 kb in length. The largest cassette we have integrated to date (7.7 kb) carries three autonomous promoter-transcription units, which would be sufficient for a large number of the anticipated applications of this technology, although an important direction for future work is the investigation of the upper size limit on insert size in ZFN-driven gene addition. Importantly, whatever the transgene size, we find, in agreement with published work on the ZFN-driven transfer of small-scale changes (e.g., single base pair) into chromosomal loci (16), that cells carrying mono- or biallelic integration events can be generated in a single step, within 72 h, and in the absence of any selection for the desired event.
Our findings, therefore, considerably expand the existing toolbox of mammalian somatic cell genetics in two significant ways: (i) in contrast to canonical gene targeting protocols (19, 42), no drug-based selection or limiting dilution is required to obtain large numbers of cells carrying entire ORFs, or cassettes of multiple autonomous promoter-transcription units, at investigator-specified locations; and (ii) ZFN-driven gene correction, originally developed to repair point mutations (8), has now been expanded to gene addition, namely, a process that can repair a broad range of recessive genotypes in trans via the targeted integration of an entire ORF. We note that, in independent experiments employing distinct transgenes (Fig. 2D, SI Fig. 6C, and data not shown), we observe no measurable ZFN-driven change in donor plasmid random integration rates. This finding has important implications for the safety of this technology particularly when applied to human therapeutic settings, e.g., stem cell modification.
Over the past 10 years a large directory of designed zinc fingers has been generated that allow the construction of engineered nucleases capable of inducing a DSB at essentially any locus in the human genome (6, 43). This versatility, combined with the precision and efficiency of ZFN-driven integration, provides the foundation to rapidly and efficiently direct a broad range of inserts to essentially any location in the genome of living human cells. Indeed, we have observed equally rapid and efficient ZFN-driven gene addition at three distinct native human loci (R.C.D., E.A.M., J.M.R., F.D.U., M.C.H., unpublished data). Moreover, elaborating on our earlier finding on efficient gene correction in primary human T cells (16), we have also observed efficient gene addition across numerous distinct cell types, including both human primary as well as human stem cells (M.C.H., unpublished data). Several uses of this technology in basic science, industrial settings, and potentially even clinical applications can therefore be envisaged. Endogenous ORFs could be tagged with peptide motifs at their amino or carboxyl-termini for allowing their biochemical purification at endogenous expression levels to enable accurate functional characterization of the protein (and any associated peptides) in vitro and to facilitate the investigation of gene function in vivo under normal physiologic levels of the gene product. Furthermore, endogenous genes could be specifically modified (e.g., fused in-frame with a GFP marker gene) to allow for the creation of endogenous reporter genes that could serve as markers of cell fate or be used in the context of small-molecule screens. As shown here, constructs encoding a particular biologic could also be integrated into a specific genomic location known to be compatible with stable, high-level, long-term expression for protein production applications. Last but not least, our data on the ability to direct the transgene to a specific location in the genome provides evidence that ZFN-driven targeted gene addition may present an attractive future direction in developing therapy for monogenic disease.
Materials and Methods
ZFN and Donor DNA Constructs.
ZFNs targeting exon 5 of the human IL2Rγ gene have been previously described (16). A chromosomal reporter system to test ZFN-driven targeted integration was constructed as described (16) with modifications: the chromosomal transgene contained a 5′ UTR from the human β-globin gene, followed by a ZFN recognition site and a polyadenylation signal, whereas the donor DNA construct contained a promoterless GFP ORF flanked by homology arms to the transgene. For all experiments on targeted integration into IL2Rγ, we used a 1.5-kb donor DNA construct homologous to the chromosomal locus (16) and introduced heterologous stretches described in Figs. 1–3 immediately upstream of the codon corresponding to Pro229 using standard recombinant DNA techniques. An autonomous expression cassette for the ΔNGFR cell surface marker was introduced outside the left homology arm of the donor construct using a unique PmeI site. Full details are available from the authors upon request. HEK293 and K562 cells were cultured and transfected with DNA constructs as described (16).
Analysis of Targeted Integration Events.
FACS analysis was performed by using a bench-top miniFACS device (Guava Technologies, Hayward, CA), and the data were further analyzed by using WinMDI software. DNA-based analysis of targeted integration frequency was performed by a highly quantitative PCR assay as described (16), except the restriction enzyme digestion step was omitted (the sole exception being experiments with a donor that introduces a 12-bp patch that contains a StuI recognition site, in which the small size difference between a wild-type and integrant-carrying chromosome necessitated the use of restriction enzyme digestion). Southern blotting on genomic DNA digested with DpnI to eliminate excess donor DNA and the indicated restriction enzymes was performed as described (16). ELISA for human Ig in tissue culture medium was performed by using an EasyTiter IgG kit (Pierce, Rockford, IL). Genotyping of the X chromosome for long transgenes was performed by using primer pairs in which one primer annealed to the chromosomal locus and the second primer annealed to the transgene. Long-range PCR analysis used Accuprime HiFi DNA polymerase and a Topo-XL cloning kit (both from Invitrogen, Carlsbad, CA). FISH for the androgen receptor locus was performed by using a commercially available FITC-labeled probe for the androgen receptor locus as described (44).
Supplementary Material
Acknowledgments
We thank the University of California (Berkeley, CA) microscopy and FACS facilities for use of equipment; Aaron Klug, Sean Brennan, and the anonymous referees for comments on the manuscript; and Luigi Naldini for helpful discussions. This work was supported in part by Grant 70NANB4H3006 from the U.S. Department of Energy/National Institute of Standards and Technology Advanced Technology Program.
Abbreviations
- ZFN
zinc finger nuclease
- DSB
double-strand break
- HDR
homology-directed repair
- SDSA
synthesis-dependent strand annealing.
Footnotes
Conflict of interest statement: C.O.P. is chair of the Scientific Advisory Board for Sangamo BioSciences, Inc. E.A.M., J.M.R., Y.-L.L., Y.J., R.C.D., P.D.G., F.D.U., and M.C.H. are full-time employees of Sangamo BioSciences, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611478104/DC1.
References
- 1.Miller J, McLachlan AD, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klug A. Ann NY Acad Sci. 1995;758:143–160. doi: 10.1111/j.1749-6632.1995.tb24814.x. [DOI] [PubMed] [Google Scholar]
- 3.Tupler R, Perini G, Green MR. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 4.Pavletich NP, Pabo CO. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 5.Choo Y, Sanchez-Garcia I, Klug A. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 6.Pabo CO, Peisach E, Grant RA. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 7.Reik A, Gregory PD, Urnov FD. Curr Opin Genet Dev. 2002;12:233–242. doi: 10.1016/s0959-437x(02)00291-5. [DOI] [PubMed] [Google Scholar]
- 8.High KA. Nature. 2005;435:577–579. doi: 10.1038/435577a. [DOI] [PubMed] [Google Scholar]
- 9.Rouet P, Smih F, Jasin M. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouet P, Smih F, Jasin M. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YG, Cha J, Chandrasegaran S. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J, Berg JM, Chandrasegaran S. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova M, Beumer K, Trautman JK, Carroll D. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 15.Porteus MH, Baltimore D. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 16.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 17.Yanez RJ, Porter AC. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- 18.Sedivy JM, Joyner AL. Gene Targeting. Oxford: Oxford Univ Press; 1992. [Google Scholar]
- 19.Thomas KR, Folger KR, Capecchi MR. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 20.Brown JP, Wei W, Sedivy JM. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 21.Downs JA, Jackson SP. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 22.Jackson SP. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 23.West SC. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 24.Valerie K, Povirk LF. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 25.Hoeijmakers JH. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 26.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohn DB, Sadelain M, Glorioso JC. Nat Rev Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- 28.Kwaks TH, Otte AP. Trends Biotechnol. 2006;24:137–142. doi: 10.1016/j.tibtech.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Hendrie PC, Russell DW. Mol Ther. 2005;12:9–17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Ginsburg DS, Calos MP. Adv Genet. 2005;54:179–187. doi: 10.1016/S0065-2660(05)54008-2. [DOI] [PubMed] [Google Scholar]
- 31.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Symington LS. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paques F, Haber JE. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herskowitz I, Rine J, Strathern JN. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones EW, Pringle JR, Broach JR, editors. Vol 2. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1992. pp. 583–656. [Google Scholar]
- 35.Ira G, Satory D, Haber JE. Mol Cell Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preston CR, Flores CC, Engels WR. Genetics. 2006;172:1055–1068. doi: 10.1534/genetics.105.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meselson M, Stahl FW. Proc Natl Acad Sci USA. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE. Mol Cell Biol. 2004;24:6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Rattray AJ, Strathern JN. Mol Cell. 2005;20:658–659. doi: 10.1016/j.molcel.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klug A. Proc Jpn Acad. 2005;81:87–102. [Google Scholar]
- 44.Jouvenot Y, Poirier F, Jami J, Paldi A. Curr Biol. 1999;9:1199–1202. doi: 10.1016/S0960-9822(00)80026-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.