Abstract
The M2 mRNA of human respiratory syncytial virus (RSV) contains two overlapping ORFs, encoding the transcription antitermination protein (M2–1) and the 90-aa M2–2 protein of unknown function. Viable recombinant RSV was recovered in which expression of M2–2 was ablated, identifying it as an accessory factor dispensable for growth in vitro. Virus lacking M2–2 grew less efficiently than did the wild-type parent in vitro, with titers that were reduced 1,000-fold during the initial 2–5 days and 10-fold by days 7–8. Compared with wild-type virus, the intracellular accumulation of RNA by M2–2 knockout virus was reduced 3- to 4-fold or more for genomic RNA and increased 2- to 4-fold or more for mRNA. Synthesis of the F and G glycoproteins, the major RSV neutralization and protective antigens, was increased in proportion with that of mRNA. In cells infected with wild-type RSV, mRNA accumulation increased dramatically up to approximately 12–15 hr after infection and then leveled off, whereas accumulation continued to increase in cells infected with the M2–2 knockout viruses. These findings suggest that M2–2 mediates a regulatory “switch” from transcription to RNA replication, one that provides an initial high level of mRNA synthesis followed by a shift in the RNA synthetic program in favor of genomic RNA for virion assembly. With regard to vaccine development, the M2–2 knockout has a highly desirable phenotype in which virus growth is attenuated while gene expression is concomitantly increased.
Keywords: paramyxovirus, reverse genetics, RNA regulation, vaccine development
Human respiratory syncytial virus (RSV), the most important viral agent of serious respiratory tract disease in infants and children worldwide, is a nonsegmented negative-strand RNA virus of the paramyxovirus family (1). Its genome is 15,222 nt in length (strain A2) and is transcribed into 10 subgenomic mRNAs by a sequential start–stop mechanism. RNA replication involves synthesis of a genome-length positive-sense RNA intermediate (antigenome) that is the template for producing progeny genomes. The nucleocapsid protein N, the phosphoprotein P, and the large polymerase protein subunit L are associated with the negative-sense RNA genome in the nucleocapsid. There are three transmembrane surface proteins, namely the fusion glycoprotein F, involved in viral penetration and syncytium formation, the glycoprotein G, which is a major attachment protein, and the small hydrophobic protein SH, whose function is unknown and which is dispensable for growth in vitro and in vivo (2, 3). The internal virion matrix protein M plays a major role in virion assembly (4). There are two nonstructural proteins, NS1 and NS2, which can be deleted individually or together from recombinant RSV (rRSV), with decreases in growth in vitro and in vivo (3, 5) (M. N. Teng and P.L.C., unpublished data). NS1 was shown to inhibit viral RNA replication and transcription in a model minigenome system (6) and thus might be a negative regulatory protein.
The M2 mRNA contains two overlapping translational ORFs, which each express a protein (Fig. 1A) (7). The upstream translational ORF (ORF1) encodes the 194-aa protein encoded ORF1 (M2–1), which is a structural component of the virion (8) and is an antitermination factor that promotes transcriptional chain elongation and also increases the frequency of readthrough at gene junctions (9, 10). The second translational reading frame (ORF2) has three potential start sites at codons 1, 3, and 7, all of which overlap with ORF1 (Fig. 1A) and would give an M2–2 protein of 83–90 aa. M2 ORF2 is present in all pneumoviruses examined to date (7, 11, 12). Translation of M2 mRNA in a cell-free system yielded the M2–1 protein and a second 11-kDa protein that was of the appropriate size to be the M2–2 protein (7). Coexpression of M2–2 in a model minireplicon system was found to have a very potent down-regulatory effect on RNA synthesis (9, 10). More recently, the RSV M2–2 protein was detected as a minor species in RSV-infected cells (28). This was presumptive evidence that M2 ORF2 is an eleventh RSV gene, which was confirmed in the present study.
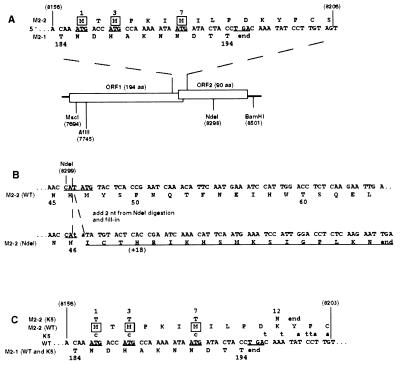
Figure 1.
Construction of the NdeI and K5 mutations, which interrupt M2 ORF2. Nucleotide sequences are in positive sense and blocked in triplets according to amino acid coding in ORF2. Nucleotide positions relative to the complete 15,223-nt recombinant antigenome are in parentheses; the other numbers refer to amino acid positions in the 194-aa M2–1 protein or 90-aa M2–2 protein. (A) Diagram of the two overlapping M2 ORFs. In the sequence at the top, the three potential translational start sites for M2–2 are underlined, and their encoded methionine residues are boxed. The termination codon for ORF1 is also underlined. In the diagram, restriction sites used for mutagenesis and cloning are indicated. (B) Construction of the NdeI mutation. The NdeI site at position 8299 in the middle of the M2–2 ORF was opened, filled in, and religated, which added 2 nt (lower case) to codon 47 of M2–2. This shifted the register to another reading frame, which was open for 18 additional codons encoding non-M2–2 amino acids (underlined). (C) Construction of the K5 mutation. The sequence shows the junction between ORF1 and ORF2, as in A. Potential ORF2 initiation codons in the wt parent are underlined, as is the ORF1 termination codon. Nucleotide changes in K5 are indicated above their wt counterparts. The three potential initiation codons for ORF2, codons 1, 3, and 7, were changed to ACG, which had no effect on amino acid coding in ORF1. The next potential methionyl start site in ORF2 is at codon 30. In addition, stop codons were introduced into all three frames immediately downstream of the M2–1 termination codon. In combination, these mutations had the effect of changing M2–2 amino acid 12 from K to N and terminating at codon 13.
In the present study, we have recovered rRSV in which M2 ORF2 was disrupted by a frame shift or by ablation of translational start sites and the introduction of stop codons. These viruses were viable but displayed altered growth kinetics, reduced RNA replication, and increased transcription, thus providing evidence of regulation between transcription and RNA replication that is mediated by the M2–2 protein.
MATERIALS AND METHODS
M2–2 Mutant Plasmid Constructions.
All rRSV and cDNA clones were based on RSV strain A2 of antigenic subgroup A. An 805-bp MscI–BamHI fragment (nt 7696–8501 in the complete rRSV antigenomic sequence) containing most of M2 ORF1 and all of ORF2 was subcloned and subjected to mutagenesis. A unique NdeI site in ORF2 was opened, filled in, and religated, creating a frame-shift mutation hereafter called the NdeI mutation (Fig. 1B). To create a second M2–2 mutant, hereafter called the K5 mutation, PCR mutagenesis (13) was carried out on the subcloned MscI–BamHI fragment in which the three potential ATG initiation codons of ORF2 were changed to ACG, these changes being silent in ORF1 (Fig. 1C). In addition, a stop codon was inserted into each register immediately after the termination codon of ORF1, which terminated ORF2 at codon 13. The mutagenic oligonucleotides, which were 5′-phosphorylated, were as follows:
forward primer 5′-TAATTAATTAAGTATAACTTCCATACTAATAACAAG-3′ (nt 8195–8231)
reverse primer 5′-TCAGGTAGTATCGTTATTTTTGGCGTGGTCGTTTGT-3′ (nt 8156–8191).
The presence of the NdeI or K5 mutations in their respective cDNAs was confirmed by sequencing the entire fragment, and each was subcloned into the AflII/BamHI sites of the support plasmid pTM-M2, which encodes both M2 ORFs and was used previously to supply both M2–1 and M2–2 proteins in a model minigenome system (9). The same fragment was also cloned into the AflII/BamHI sites of pUC118-FM2, which contained the F and M2 genes, and the StuI/BamHI fragment of this plasmid was subsequently transferred to the full-length antigenomic cDNA to create NdeI and K5 antigenomic cDNAs.
RESULTS
Mutagenesis of M2 ORF2.
Two separate mutations were made to ablate expression of a complete M2–2 protein. First, a unique NdeI restriction enzyme site (antigenome position 8299) within ORF2 was filled and religated to create a frame-shift mutation (2 nts added) at codon 47 of the 90-codon ORF (Fig. 1B). This frame shift fused the upstream 46 codons of ORF2 to 18 heterologous codons, which happened to be present in the other reading frame. PCR mutagenesis was utilized to construct a second mutation in which expression of ORF2 would be completely ablated. Specifically, each of the three potential initiation codons for ORF2 (Fig. 1 A and C) was mutated to ACG codons. Furthermore, to preclude the possibility of reversion or non-AUG initiation (14, 15), a stop codon was added in each register after the ORF1 termination codon, terminating M2 ORF2 at codon 13 (Fig. 1C). The M2–1 amino acid sequence was not affected in either mutant.
Effect of M2–2 Mutations on Minireplicon Transcription and Replication.
The function(s) of the M2–2 protein was not known, but it had previously been shown to strongly inhibit minigenome RNA synthesis (9, 10, 16). Therefore this highly sensitive assay was used to verify that the NdeI and K5 mutations ablated this inhibitory effect. RSV transcription and RNA replication were studied by using a negative-sense RSV-chloramphenicol acetyltransferase (CAT) minigenome C2, which contains the CAT ORF under the control of RSV transcription initiation and termination signals flanked by the 3′-leader and 5′-trailer regions of the RSV genome (9, 16). Intracellular synthesis of the C2 minigenome was driven from the transfected plasmid by T7 RNA polymerase supplied from the recombinant vaccinia virus vTF7–3 (17), and RSV proteins were expressed from cotransfected support plasmids.
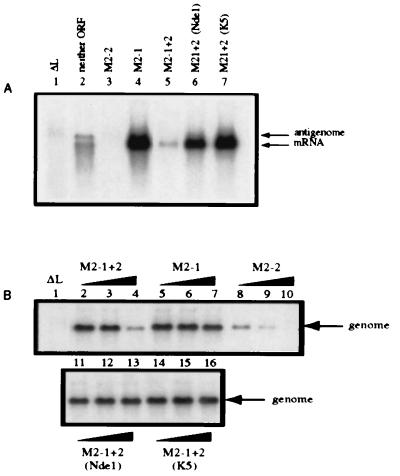
When minigenome C2 was complemented by N, P, and L, it directed the synthesis of antigenome and CAT mRNA (Fig. 2A, lane 2), with the latter being mostly prematurely terminated as observed previously (9, 16). When plasmid expressing M2–1 was added in addition, CAT mRNA was synthesized as full-length molecules (Fig. 2A, lane 4). Coexpression of M2–2 instead of M2–1 strongly inhibited the synthesis of antigenome and mRNA (Fig. 2A, compare lanes 2 and 3). When the M2 plasmid contained both ORF1 and ORF2 in their native configuration, M2–1 + 2, there was a significant reduction in transcription and replication products compared with that seen with M2–1, showing that the inhibitory activity of M2–2 predominated at this particular plasmid concentration (compare lanes 4 and 5). In contrast, M2–1 + 2 containing the NdeI or the K5 mutation behaved similarly to M2–1 alone, indicating that the inhibitory activity of M2–2 had been ablated without affecting M2–1 (compare lanes 6 and 7 with lane 4).
Figure 2.
The NdeI and K5 mutations each ablate the inhibitory function of M2–2 in a reconstituted minireplicon system. (A) HEp-2 cells were simultaneously infected with vTF7–3 (five plaque-forming units per cell) and transfected with plasmid encoding the negative-sense C2 minigenome cDNA (200 ng) and support plasmids (N, 400 ng; P, 200 ng; L, 100 ng per well of a six-well dish) and supplemented with pTM constructs (80 ng) expressing neither M2 ORF (lane 2), M2–2 (lane 3), M2–1 (lane 4), M2–1 + 2 (lane 5), or the M2–1 + 2 containing the NdeI (lane 6) or K5 (lane 7) mutations. Lane 1 contains RNA from a reaction that lacked L and is a negative control. Cells were exposed to 2 μg actinomycin D per milliliter from 24–26 hr after infection (21). At 48 hr after infection, total intracellular RNA was isolated and electrophoresed on formaldehyde gels for Northern blot analysis (16). Blots were hybridized to a negative-sense CAT-specific riboprobe to detect both mRNA and miniantigenome. (B) HEp-2 cells were transfected as described above with plasmid encoding positive-sense C4 miniantigenome complemented by the N, P, and L plasmids as in A. The transfection mixtures were supplemented with increasing amounts (0.008, 0.04, and 0.2 times the relative molar ratio of transfected pTM-N) of pTM constructs expressing M2–1 + 2 (lanes 2, 3, and 4), M2–1 (lanes 5, 6, and 7), M2–2 (lanes 8, 9, and 10) or M2–1 + 2 containing the NdeI (lanes 11, 12, and 13) or K5 (lanes 14, 15, and 16) mutation. Total intracellular RNA was analyzed by Northern blots hybridized with a positive-sense CAT-specific riboprobe to detect genomic RNA.
Comparable results were obtained when the C2 plasmid was replaced by the C4 plasmid, which expresses a positive-sense RNA representing the antigenome of the C2 minigenome (9, 16). In this case, Northern blots were analyzed with a positive-sense riboprobe to detect the synthesis of minigenome. Cotransfection of increasing amounts of M2–1 plasmid had no effect on the synthesis of minigenome (Fig. 2B, lanes 5, 6, and 7) consistent with previous findings that it does not affect RNA replication (9). However, increasing amounts of M2–2 alone or M2–1 + 2 led to a progressive reduction in the amounts of minigenome (Fig. 2B, lanes 8, 9, and 10 and lanes 2, 3, and 4), reflecting the inhibitory activity of M2–2. In contrast, increasing amounts of M2–1 + 2 containing either the NdeI or the K5 mutation did not detectably inhibit minigenome synthesis (Fig. 2B, lanes 11, 12, and 13 for the NdeI mutant, and lanes 14, 15, and 16 for the K5 mutant), indicating that neither mutant expressed an M2–2 protein active in this assay.
Recovery and Growth in Vitro of rRSV Containing the M2–2 Mutations.
The NdeI and K5 mutations were individually incorporated into a full-length antigenomic cDNA, and recovery of infectious rRSV was performed as described previously (18). Viruses containing either the NdeI or K5 mutations (rA2-NdeI and rA2-K5 respectively) were recovered with an efficiency comparable to that of wild type (wt), and the presence of the mutations was confirmed by sequencing reverse transcription–PCR products generated from infected cell RNA (data not shown). Thus, M2–2 is an accessory protein that is not required for RSV growth in cell culture.
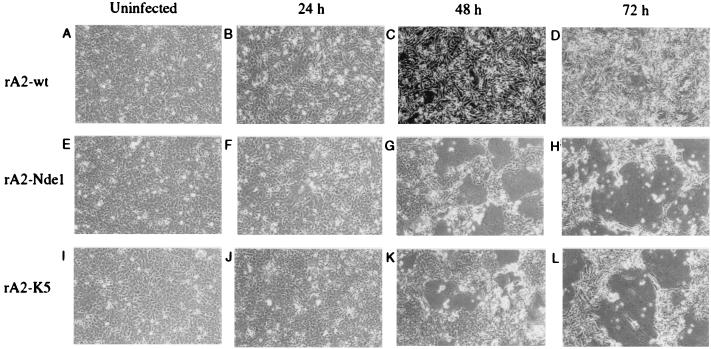
The rA2-NdeI and rA2-K5 viruses displayed a large plaque phenotype and accelerated syncytium formation. Specifically, when HEp-2 cells were infected at a multiplicity of infection (moi) of 1, syncytium formation was evident by 24 hr and was extensive by 48 hr after infection (Fig. 3), resembling those formed by the wt virus at day 4 after infection (not shown). These phenotypic changes suggested that the mutant viruses were either more fusogenic or had altered kinetics of growth or gene expression, or any combination thereof, compared with the parental virus.
Figure 3.
Cytopathogenicity of the rA2-NdeI and rA2-K5 viruses compared with rA2-wt. HEp-2 cells were mock infected or infected at an moi of 1 with the indicated virus, incubated for the indicated time, and photographed (×10). The 48-hr micrographs are darker because of a difference in exposure. Large syncytia are obvious in the two mutant viruses at 48 and 72 hr, and smaller ones are evident at 24 hr and in rA2-wt infected cells at the same three time points.
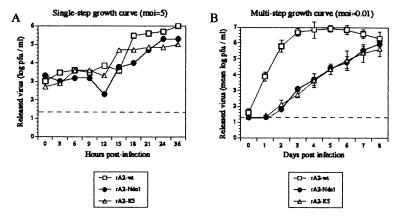
To examine single-cycle growth kinetics of the mutant viruses, HEp-2 cells were infected with rA2-wt, rA2-NdeI, or rA2-K5 at an moi of 5 (Fig. 4A). Both the rA2-NdeI and rA2-K5 recombinant viruses displayed slightly reduced growth kinetics compared with wt, with the final virus titers being approximately 10-fold less. To accentuate any potential differences, multistep growth cycles were evaluated in HEp-2 cells infected in triplicate at an moi of 0.01 (Fig. 4B). In cells infected with rA2-wt virus, peak virus titer was reached at 4–5 days after infection, whereupon it leveled and began to decline. This decline might be due to increased cytopathogenicity, which became evident after day 6 and was more pronounced than that observed for the mutants. Both of the mutant viruses had markedly delayed and reduced growth kinetics during the first few days of incubation (1,000-fold less mutant virus released compared with wt at days 2, 3, and 4 after infection), but by day 8 after infection, the titers approached those of the parental strain. However, the maximum titers of the two mutant viruses were found to be consistently 10-fold lower than that of wt. These results showed that the large plaque morphology exhibited by the mutant viruses was not associated with increased virus release when compared with the parental strain.
Figure 4.
Kinetics of growth of rA2-wt, rA2-NdeI, and rA2-K5 in cell culture. The dashed horizontal lines indicate the lower limit of detectability. (A) Single-step growth kinetics. HEp-2 cells were infected with rA2-wt, rA2-NdeI, or rA2-K5 at an moi of 5, and the entire medium overlay was harvested at the indicated times after infection and flash frozen. Viral titers were determined by plaque assay. (B) Multicycle growth kinetics. HEp-2 cells were infected in triplicate at an moi of 0.01 plaque-forming units per cell with the above viruses. At the indicated times after infection, the entire medium overlay was removed and flash frozen. Cells were washed twice in PBS and incubated with fresh medium. Mean virus titers determined by plaque assay (with error bars) are shown.
Northern Blot Analysis of Viral RNAs.
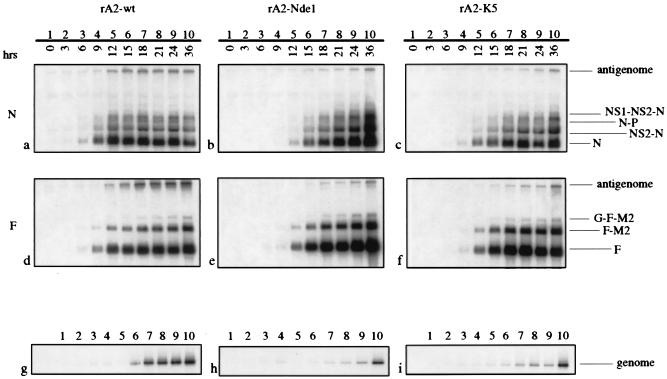
RNA replication and transcription by the M2–2 mutant viruses were examined. In the single-step growth experiment described above, cell monolayers from replicate plates were harvested at 3-hr intervals, and total intracellular RNA was analyzed by Northern blot hybridization. The accumulation of antigenome and mRNAs was monitored by hybridization with negative-sense riboprobes against the N gene (Fig. 5 a–c) or the F gene (d–f). The pattern of monocistronic, dicistronic, and tricistronic mRNAs produced by the mutant viruses was qualitatively similar to that of the wt strain. This suggested that ablation of the expression of M2–2 did not grossly alter the transcription antitermination effect of M2–1.
Figure 5.
Northern blot analysis of RNA replication and transcription. HEp-2 cells infected with rA2-wt (a, d, and g), rA2-NdeI (b, e, and h) and rA2-K5 (c, f, and i) were harvested at 3-hr intervals (lanes 1–10) from the single-cycle growth curve experiment described in Fig. 4A, and total intracellular RNA was isolated and subjected to Northern blot analysis. Blots were hybridized with a negative-sense N-specific riboprobe (a–c), a negative-sense F-specific riboprobe (d–f) or a positive-sense F specific riboprobe (g–i). Monocistronic mRNA, polycistronic readthrough mRNAs and antigenome and genome RNAs are indicated.
The accumulation of mRNA and antigenome in the cells infected with the rA2-wt virus was first detected at 6 hr after infection (Fig. 5 a and d, lane 3) and increased rapidly until approximately 12–15 hr after infection, and thereafter increased more slowly or leveled off (Fig. 5 a and d, lanes 5–10). In contrast, both the rA2-NdeI and rA2-K5 viruses displayed a marked delay in the synthesis of mRNA and antigenome, such that these RNAs became detectable at 9–12 hr after infection (b and e, lanes 4 and 5, for rA2-NdeI, c and f, lanes 4 and 5, for rA2-K5). The mRNA levels then increased to levels surpassing those of wt. In contrast, the accumulation of antigenome was considerably reduced compared with wt (b and e, lanes 7–10, for rA2-NdeI, and c and f, lanes 7–10, for rA2-K5). For example, PhosphorImager (Molecular Dynamics) analysis of the blots probed with the negative-sense F riboprobe (Fig. 5 d–f) revealed a 1.3- to 2-fold increase in accumulated mRNA at 24 hr after infection, with a simultaneous 3- to 4-fold reduction in the accumulation of antigenomic RNA compared with wt. Some experimental variability was evident. For example, the increase in N mRNA in c with rA2-K5 appeared less than that of rA2-NdeI in b. However, repeat experiments confirmed that on average the two mutants expressed similar amounts of mRNA, and that the amount of mRNA expressed by the mutants at later time points, such as 18–36 hr after infection, exceeded that of wt. PhosphorImager analysis of two independent experiments showed that the amount of N mRNA that accumulated in response to either mutant was 1.3- to 1.8-fold greater (K5 and NdeI, respectively) than wt at 24 hr and 4.1- to 4.4-fold greater (NdeI and K5, respectively) at 36 hr.
RNA replication was further examined by hybridization of replicate blots with a positive-sense F riboprobe, which detected the accumulation of genomic RNA (Fig. 5 g–i). PhosphorImager analysis indicated that the accumulation of genome by the mutant viruses was 15–18% that of the parental virus at 24 hr. Thus, the increase in mRNA accumulation described above occurred even though its genomic RNA template was less abundant. Overall, the molar ratio of mRNA to genome was approximately 7- to 18-fold greater in the mutant viruses. This provides evidence for a regulatory balance between transcription and RNA replication, one which swings in favor of transcription when expression of the M2–2 protein is ablated.
Increased Expression of Major Viral Antigens.
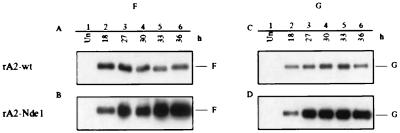
The increase in the accumulation of mRNA in cells infected with the rA2-NdeI and rA2-K5 viruses was mirrored by an increase in the accumulation of viral protein. Fig. 6 shows Western blot analysis of the F (A and B) and G (C and D) proteins synthesized in cells infected with rA2-wt virus or rA2-NdeI virus. The amount of F and G proteins present in the harvested cells at 36 hr after infection (A–D, lane 6) was 3-fold greater or more for rA2-NdeI than for rA2-wt.
Figure 6.
Western blot analysis of the accumulation of F and G glycoproteins in HEp-2 cells infected at an moi of 5 with rA2-wt (A and C) or rA2-NdeI (B and D). Cells were harvested at the indicated time, and total cellular protein was subjected to polyacrylamide gel electrophoresis under denaturing and reducing conditions, transferred to nitrocellulose (5), and reacted with rabbit antipeptide serum against the cytoplasmic domain of the F (A and B) or G (C and D) protein. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit IgG and visualized by enhanced chemiluminescence (Amersham Pharmacia).
DISCUSSION
M2 ORF2 can be interrupted in rRSV without loss of viability in cell culture. However, there were significant alterations in the viral RNA synthetic program, cell culture pathogenicity, and growth characteristics. This shows unambiguously that M2 ORF2 is an eleventh RSV gene, a finding that is somewhat unexpected because the ORF is located in the downstream half of the M2 mRNA, is preceded by 11 methionyl codons, and thus would not be expected to be efficiently translated. Because ORF2 is expressed as a separate protein (7), one possibility is that it is accessed by a ribosomal stop-restart mechanism, such as described for the second ORF of the M gene of influenza virus B (19). The activity described for M2–2 identifies an RNA regulatory protein in a negative-strand RNA virus. The NdeI and K5 mutations were indistinguishable phenotypically in this study, implying that the truncated M2–2 protein encoded by the former mutation could not substitute for the complete protein.
The absence of M2–2 was associated with a reduction in the accumulation of genomic and antigenomic RNA, the products of RNA replication, and an increase in the accumulation of mRNA, the product of transcription. This predicts two activities for M2–2. The first is to increase RNA replication. Whether the direct effect is at the level of antigenome synthesis, genome synthesis, or both, is not yet known. In addition, there is no information on the details of the mechanism of action of M2–2, such as whether it acts at the level of initiation or elongation. The second activity is to regulate transcription. In cells infected with rA2-wt, the accumulation of mRNA increased rapidly up to approximately 12 hr and more slowly thereafter, suggesting that transcription is down-regulated after that time. One of the effects of ablating M2–2 expression was a delay in the appearance of mRNA, which probably was caused by the delayed reduced synthesis of its genomic RNA template. Although the accumulation of mRNA by the mutant viruses was delayed, it reached wt levels by approximately 12–15 hr and continued to increase thereafter. This suggests that M2–2 mediates negative regulation of transcription late in infection, which is alleviated in its absence. Because the proteins of nonsegmented negative-strand RNA viruses typically increase in abundance during the course of infection, this negative regulatory effect likely is concentration dependent. Thus, RSV transcription is subject to negative autoregulation, and RNA replication is subject to positive regulation.
It is generally thought that there is a reversible “switch” between transcription and RNA replication by nonsegmented negative-strand RNA viruses. The synthesis of mRNA and antigenome ostensibly involves the same promoter and genomic nucleocapsid template and, for most viruses, the same protein components, N, P, and L. RSV is an exception in having the additional transcription-specific factor M2–1. One widely accepted model is that RNA synthesis switches from transcription to RNA replication when sufficient N protein accumulates to mediate cosynthetic encapsidation of the nascent RNA. This somehow switches the polymerase to read through gene junctions and synthesize a complete antigenome (20). However, in earlier work we were unable to reconstitute this “switch” in a model minireplicon system by overexpression of N protein (21). Unexpectedly, the present study implicates the M2–2 protein in this switch. One caveat is that we do not have direct evidence that the observed effects on transcription and replication are linked rather than independent events. In terms of a single-step growth cycle, the present results suggest that the M2–2 protein functions at around 12–15 hr after infection to reduce transcription (after which the already-made mRNAs continue to drive protein synthesis) and increase RNA replication, shifting the RNA synthetic program in favor of genomic RNA for virion assembly.
The other phenotypic differences observed for the M2–2 knockout viruses can also be explained by the changes in the RNA synthetic program described above. For example, the delay and reduction in virus production likely are a consequence of the delay and reduction in the synthesis of progeny genome and the initial delay in the accumulation of mRNA. Accelerated plaque formation likely is a consequence of increased synthesis of surface glycoproteins and accelerated cell-to-cell fusion. Nonetheless, this does not preclude other activities for M2–2, and it will be of particular interest to determine the phenotype of this knockout virus in experimental animals as well as to examine progeny virions biochemically and by electron microscopy.
Previous studies showed that the M2–2 protein inhibited RNA replication and transcription by RSV model minireplicons (9, 10). Inhibition of minigenome transcription would be fully consistent with the present report. However, the previously observed M2–2-mediated inhibition of minireplicon RNA replication is diametrically opposed to the present findings, where the absence, rather than presence, of M2–2 was associated with reduced RNA replication by rRSV. We presently do not have an explanation for the difference. It is presumed that the results with the knockout viruses represent the authentic situation. The results with the minireplicon system might be incomplete rather than artifactual. This could be a consequence of the differences between the minireplicon system and an authentic virus infection, for example: (i) the supply of proteins would be greatly affected by regulation in an authentic infection, but not in a reconstituted minireplicon system where proteins are supplied by transfected plasmids; (ii) the effects of M2–2 observed to date have been in minireplicon systems in which only a subset of viral proteins was supplied; and (iii) the relative level of M2–2 expressed in an authentic infection has not been determined but seems to be very low, and the minireplicon studies to date might have used levels that were too high.
The finding that M2–2 is not essential for growth in vitro defines this species as an accessory protein. Other paramyxovirus accessory proteins include the RSV SH, NS1, and NS2 proteins, the V and C proteins of Sendai virus, measles virus, and parainfluenza virus type 3 (PIV3), and the D protein of PIV3 (2, 3, 5, 22–27, 29) (M. N. Teng and P.L.C., unpublished data). Among these, the Sendai virus V and C proteins have been studied the most extensively. Ablation of the expression of the V protein in recombinant Sendai virus was associated with increases in transcription, RNA replication, and virus growth in vitro (24), although these differences were not obvious in a separate study (22). Growth of V-minus virus in vivo was attenuated, suggesting that the V protein might mitigate host immunity (22, 23). The Sendai virus C protein is expressed as four proteins, namely C′, C, Y1, and Y2, which arise from translational initiation at the first through fourth translational start sites in the C ORF, respectively. Deletion of these proteins from recombinant virus individually and in groups had complex effects that have not been completely defined. Deletion of both C′ and C delayed the appearance of genomic RNA and mRNA, after which these species were overproduced and greatly reduced the production of infectious virus (24, 25). Elimination of all four C-related proteins resulted in a virus that was extremely debilitated for RNA synthesis and growth in vitro (24). The functions of the V and various C proteins probably cannot be explained solely in terms of regulation of RNA synthesis and remain to be defined.
Ablation of expression of the M2–2 gene attenuated virus growth in cell culture by at least 1,000-fold during the initial days of a multicycle growth curve in vitro, with the final yield of infectious virus being reduced approximately 10-fold. This level of attenuation would be highly desirable for vaccine purposes, and the similarity in final yield between the wt and knockout viruses in cell culture suggests that this mutation should not interfere significantly with the production of vaccine virus. Although virus growth was attenuated, gene expression was enhanced. Typically, RSV gene expression is roughly proportional to virus growth, and attenuating mutations that reduce growth reduce antigen production. Thus, one of the problems in RSV vaccine development has been to find a level of attenuation that minimizes disease yet retains sufficient immunogenicity. The M2–2 knockout mutation offers the unique opportunity to significantly attenuate the virus with a concomitant increase, rather than decrease, in antigen expression. Although the mutant viruses exhibited increased fusion and cytopathogenicity during a high-multiplicity infection (Fig. 3), infection of the surface of the respiratory tract is more likely to resemble an infection at a low moi, such as in Fig. 4B. There, although the cytopathic effect of the M2–2 knockout virus was more rapid for individual cells and their immediate neighbors, the strong attenuation of virus production resulted in a slower dissemination of virus and much slower cytopathogenicity overall. This will be evaluated in vivo.
Acknowledgments
We thank Drs. Robert Chanock, Brian Murphy, Rachel Fearns, and Michael Teng for critical reading of the manuscript and Dr. Stephen Whitehead for supplying the pUC118-FM2 plasmid.
ABBREVIATIONS
- RSV
respiratory syncytial virus
- rRSV
recombinant RSV
- ORF1 and 2
translational ORF1 and 2
- wt
wild-type
- moi
multiplicity of infection
- CAT
chloramphenicol acetyltransferase
References
- 1.Collins P L, McIntosh K, Chanock R M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 1313–1352. [Google Scholar]
- 2.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead S S, Bukreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teng M N, Collins P L. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng M N, Collins P L. J Virol. 1999;73:466–473. doi: 10.1128/jvi.73.1.466-473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atreya P L, Peeples M E, Collins P L. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins P L, Hill M G, Johnson P R. J Gen Virol. 1990;71:3015–3020. doi: 10.1099/0022-1317-71-12-3015. [DOI] [PubMed] [Google Scholar]
- 8.Peeples M, Levine S. Virology. 1979;95:137–145. doi: 10.1016/0042-6822(79)90408-2. [DOI] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Cristina J, Grosfeld H. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy R W, Wertz G W. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling R, Easton A J, Pringle C R. J Gen Virol. 1992;73:1709–1715. doi: 10.1099/0022-1317-73-7-1709. [DOI] [PubMed] [Google Scholar]
- 12.Zamora M, Samal S K. J Gen Virol. 1992;73:737–741. doi: 10.1099/0022-1317-73-3-737. [DOI] [PubMed] [Google Scholar]
- 13.Byrappa S, Gavin D K, Gupta K C. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 14.Curran J, Kolakofsky D. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latorre P, Kolakofsky D, Curran J. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosfeld H, Hill M G, Collins P L. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath C M, Williams M A, Lamb R A. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb R A, Kolakofsky D. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Vol. 1. Philadelphia: Lippincott; 1996. pp. 1177–1204. [Google Scholar]
- 21.Fearns R, Peeples M E, Collins P L. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 22.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 23.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 25.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. J Virol. 1998;72:5984–5993. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radecke F, Billeter M A. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 27.Schneider H, Kaelin K, Billeter M A. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 28.Ahmadian G, Chambers P, Easton A J. J Gen Virol. 1999;80:2011–2016. doi: 10.1099/0022-1317-80-8-2011. [DOI] [PubMed] [Google Scholar]
- 29.Durbin, A., McAuliffe, J. M., Collins, P. L. & Murphy, B. R. (1999) Virology, in press. [DOI] [PubMed]