Abstract
Tetramers of major histocompatability complex molecules (MHC) are now well-established reagents for the detection of antigen-specific T cells by flow cytometry. MHC tetramers are prepared by mixing enzymatically biotinylated MHC molecules with commercial preparations of streptavidin, usually conjugated to a fluorescent phycobiliprotein such as phycoerythrin (PE) or allophycocyanin (APC). While data obtained with MHC tetramers prepared with small molecule fluorophores has been reported, considerable lot-to-lot variation among conventional streptavidin conjugates to small molecules prevents routine preparation of such reagents. We now report robust preparation of MHC tetramers with small molecule fluorophores, using a recombinant mutant of streptavidin incorporating a carboxy-terminal cysteine in each of the four identical subunits that is conjugated to maleimide derivatives of any of several small molecule fluorophores. These reagents significantly expand the versatility of the MHC tetramer methodology.
Keywords: MHC tetramers, streptavidin, flow cytometry
Introduction
“Tetramers” of major histocompatability complex (MHC) molecules are now routinely used to identify antigen-specific T cells and have become an essential tool in nearly all areas of immunology, including vaccine development (Amara et al., 2001), tumor immunology (Yee et al., 1999), auto-immunity (Nepom et al., 2002), and basic studies of immune memory (Murali-Krishna et al., 1999). MHC tetramers are produced by mixing enzymatically biotinylated MHC fusions to a BirA substrate peptide (BSP) (Schatz, 1993) with fluorescent conjugates of streptavidin or avidin (Altman et al., 1996); the MHC molecules used for tetramer preparations are homogeneous with respect to the peptide ligand that occupies the peptide-binding groove. Although the interaction of monomeric MHC/peptide complexes with a specific T cell antigen receptor (TCR) has a short half-life (Davis et al., 1998), MHC tetramers have an enhanced avidity for specific T cells that allows them to be effective, specific staining reagents.
The most commonly used fluorophores for the preparation of MHC tetramers are the phycobiliproteins phycoerythrin (PE) and allophycocyanin (APC). Three factors account for the popularity of these fluorophores for tetramer reagents: (1) they are among the brightest of fluorophores available for most commercial flow cytometers; (2) reliable supplies of streptavidin-phycobiliprotein conjugates that routinely yield high quality tetramer reagents are available from a number of manufacturers; and (3) although they have occasionally been used (Kuroda et al., 2000; Plunkett et al., 2001; Champagne et al., 2001), commercial supplies of streptavidin conjugated to small molecule fluorophores give highly variable results, and many reagent lots completely fail to yield effective staining reagents. Despite the difficulty of preparing tetramers labeled with small molecule fluorophores, interest in them remains high for at least two reasons: (1) they will increase the flexibility of reagent panel construction, especially for new polychromatic flow cytometry applications (Perfetto et al., 2004); and (2) phycobiliproteins are not compatible with methods such as Flow-FISH which are used to measure telomere lengths in defined cell populations (Plunkett et al., 2001).
A possible explanation for the variability of tetramers prepared with small molecule fluorophores was provided by Cameron et al., who noted that many commercial preparations of streptavidin conjugated to fluorescein isothiocyanate (SA-FITC) exhibited substoichiometric biotin binding capacity and proposed that a fraction of the biotin binding sites were damaged during the process of modification of the streptavidin protein with FITC (Cameron et al., 2001). Examination of the crystal structure of streptavidin complexed with biotin (Freitag et al., 1997) demonstrates that the biotin carboxylate in the binding site of subunit D is within 7.25 □ of the ε -amino group of lysine 121 of the adjacent subunit A (Figure 1), suggesting that modification of this lysine side-chain with FITC is the most likely candidate for inhibiting biotin binding (and of course, there are three additional close encounter interactions in the rest of the streptavidin tetramer). Cameron et al. developed a protocol to protect the biotin-binding capacity of streptavidin during FITC modification by temporarily filling the biotin-binding site with 2-hydroxyazobenzene-4′-carboxylic acid (HABA) a mimic of biotin that binds to streptavidin with low-affinity and that is easily displaced by biotin in a subsequent reaction (Cameron et al., 2001). Though we have tried—and failed—to reproduce this procedure in our own laboratory, the original hypothesis led us to consider methods for targeted labeling of streptavidin at a site remote from the biotin-binding site. While wild-type streptavidin does not have a good candidate site such as a free cysteine thiol, the groups of Cantor and Sano have developed an E. coli expression system for recombinant streptavidin (Sano and Cantor, 1990), including a mutant with a single cysteine residue at the carboxyl-terminus of each of the four identical subunits that they refer to as StvC (Reznik et al., 2001). We now report the robust production of high-quality MHC tetramers using based upon StvC labeled with maleimide derivatives of a variety of fluorophores.
Figure 1. The carboxylate of biotin bound to subunit D (yellow) is 7.25 Å from the ε-amino group of lysine-121 (green) of the neighboring subunit A (otherwise magenta).
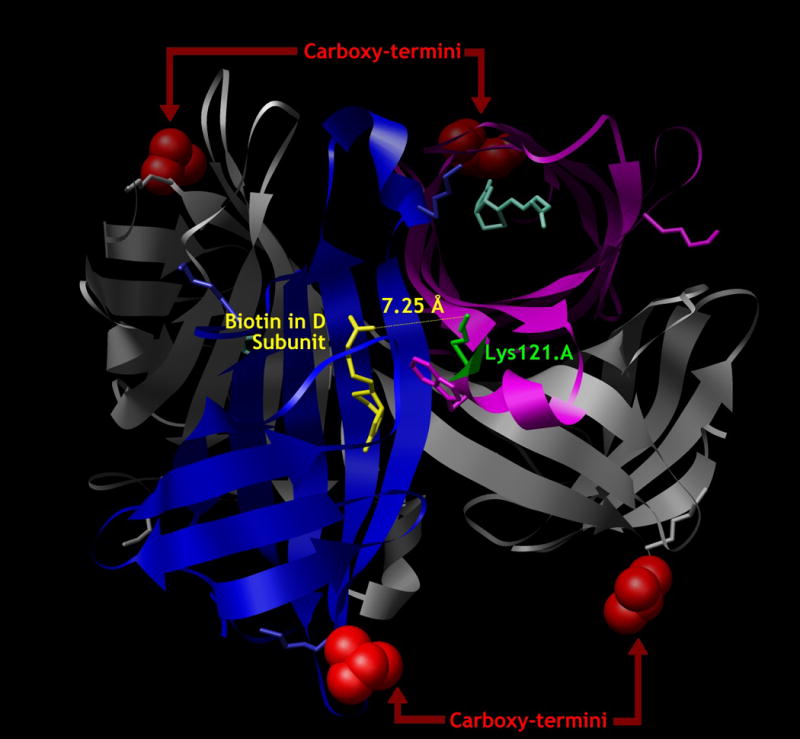
The carboxy-termini of all subunits, shown in red space-filling models, are very much further away from the biotin carboxylates.
Methods
Expression of StvC
Expression and purification of StvC was as described by Reznik et al (Reznik et al., 2001), with the following modifications. DTT was included throughout the entire purification procedure at a concentration of 5 mM. Following renaturation, precipitated protein was removed by centrifugation and filtration through 0.45 μm membranes, and ammonium sulfate was slowly added to a final concentration of 1.0 M. After filtration, again through a 0.45 μm membrane, the protein was loaded onto a Phenyl-Sepharose column (GE Healthcare), and was eluted with 50mM Tris pH8, 5mM DTT, without ammonium sulfate. A single peak was collected from the column and the purified StvC was stored at 2mg/ml in 50mM Tris buffer pH8 with 5 mM DTT prior to fluorescent labeling.
Fluorescent labeling of StvC
Just prior to labeling StvC with fluorophores, DTT was removed from the protein sample by desalting on PD-10 columns (GE Healthcare).
The following reagents were obtained from Molecular Probes: Pacific Blue-maleimide, Alexa-488-maleimide, fluorescein-maleimide, Alexa647-maleimide, and Alexa-680-maleimide.
Labeling reactions were carried out at a dye:protein molar ratio of 10:1 overnight at 4°C, according to the recommended protocol supplied by Molecular Probes. Unincorporated dye was removed by desalting on PD-10 columns or alternatively by dialysis. Labeled proteins are stored at 1.0 mg/ml in 0.1M sodium phosphate buffer pH7.2 containing 0.1M sodium chloride. Final F:P ratios are calculated based upon the extinction coefficients provided by Molecular Probes, correcting for absorbance of the dye at 280 nm.
Streptavidin labeled with NHS derivatives of the same dyes which were used to compare the fluorescence spectra, were obtained from Molecular Probes.
Fluorescence spectroscopy
Fluorescence spectra were recorded on a Fluoromax-2 scanning fluorimeter (ISA). Excitation and emission properties of the fluorophores used are as shown in Table 1. All the spectra were recorded with an integration time of 0.1 s with a slit width of 2 nm. Spectra of Streptavidin labeled with NHS derivatives of the same dyes were analyzed in parallel with the maleimide-modified streptavidin and the resulting spectra were normalized.
Table 1.
FACS suitable maleimide derivatives conjugated to StvC.
| Fluorophore | λmax(Excitation) | λmax(Emission) | Laser |
|---|---|---|---|
| Fluorescein | 494 | 518 | 488 |
| Alexa488 | 495 | 519 | 488 |
| Alexa647 | 650 | 668 | 633 |
| Alexa680 | 679 | 702 | 633 |
| Pacific Blue | 416 | 451 | 405 |
Tetramerization
Biotinylated MHC molecules prepared as described (Altman et al., 1996). All tests were done with HLA-A*0201 complexes with the well characterized immunodominant epitope from HCMV pp65, NLVPMVATV. Tetramers prepared by slow addition of labeled streptavidin to the biotinylated MHC, as described (Altman et al., 2003). Appropriate staining titers of tetramers were determined by testing serial dilutions of the tetramer stocks in staining experiments.
Size exclusion chromatography
HLA-A*0201 tetramers prepared with fluorophore-modified StvC were analyzed by size exclusion chromatography on TSKgel G3000swxl (TOSOH Bioscience LLC) column with 0.2 M sodium phosphate buffer pH7 as the mobile phase at a flow rate of 0.5 ml/min. Standard proteins for calibration of the column include ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), and chymotrypsinogen (25 kDa); all were obtained from GE Healthcare.
Flow cytometry
Whole blood samples from HLA-A*0201+ CMV-seropostive donors previously shown to have A2/NLV-tetramer+ cells were stained with anti-CD3, anti-CD8, and tetramers as described (Altman et al., 2003). The fluorophores on CD3 and CD8 antibodies were varied to accommodate the labels on the tetramers. Flow cytometry data files were collected on an LSR-II flow cytometer (Becton Dickinson), equipped with standard violet, blue, and red lasers and standard filter sets. Flow cytometry data were analyzed with FlowJo© software (TreeStar, Ashland, Oregon).
Results
Expression and purification
Following published protocols, we expressed StvC subunits as inclusion bodies in E. coli, washed them with a mild detergent, solubilized them in a denaturing buffer, and refolded them by dialysis (Reznik et al., 2001). We replaced the published purification on an iminobiotin affinity matrix with purification on a Phenyl-sepharose hydrophobic interaction column, permitting elution of the protein under milder conditions. Our approach was equally efficacious, as demonstrated by comparison of boiled and non-boiled samples of the purified protein on SDS-PAGE in Figure 2: the boiled sample migrated at the size expected for a monomer, while the non-boiled sample migrated with a size expected for a SDS-stable tetramer, with no detectable monomeric subunits.
Figure 2. Purified, recombinant StvC behaves like commercial preparations of stretpavidin on SDS-PAGE.
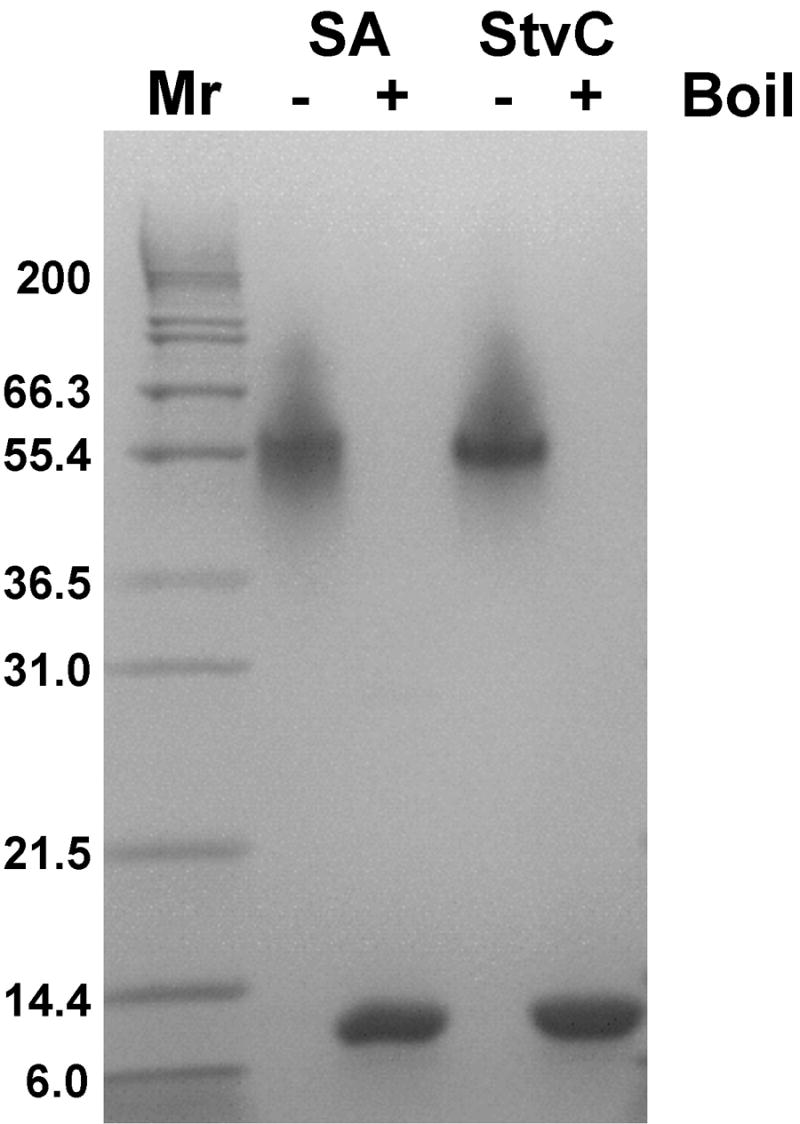
Non-boiled StvC and commercial streptavidin migrate as intact tetramers, while the boiled proteins migrate as monomeric subunits.
Labeling of StvC with fluorophores and comparison of emission spectra
For labeling StvC, we chose fluorophores that met the following two criteria: (1) they are available as thiol-reactive derivatives, (in practice, all are maleimides), and (2) they are excited by commonly available lasers on commercial flow cytometers. The fluorophores that we used and their characteristics are listed in Table 1. Following labeling and removal of the unincorporated dye, the dye:protein ratio for each conjugate was measured, and all fell in the range of 3–5, probably within experimental error of the expected value of 4.0, one for each of cysteine residue in each of the four identical subunits.
Since the reactive groups used to attach fluorophores to proteins can subtly influence the emission spectrum of the fluorophore—which can then lead to incorrect fluorescence compensation if the compensation settings are determined with one modified protein (e.g. an antibody labeled with an NHS derivative) and the experiment is performed with another (e.g. a maleimide modified streptavidin-based tetramer)—the emission spectra of the maleimide- and NHS-modified streptavidins were measured and compared. As seen in Figure 3, normalized spectra for NHS and maleimide modified streptavidins were nearly exact matches for Pacific Blue, and Alexa 64, and were very close for Alexa 488 and Alexa 680. The largest difference in emission spectra was seen when comparing the two fluorescein-modified streptavidins.
Figure 3. Normalized emission spectra of fluorophores conjugated to streptavidin.
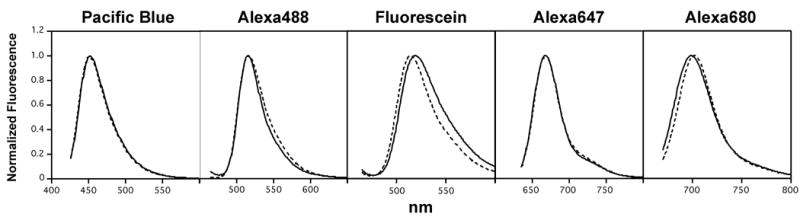
The emission spectra of the maleimide-modified StvC derivatives are shown as dotted lines, while the commercial preparations of NHS-modified streptavidins are shown as solid lines.
Determination of StvC/HLA-tetramer size by size exclusion chromatography
In order to obtain a rough characterization of the quaternary structure of the StvC tetramers, we analyzed them by size exclusion chromatography on TSKgel G3000swxl columns. The majority of the protein eluted with an interpolated molecular weight of 310 kDa (Figure 4), suggesting that the most predominant species is indeed a true tetramer (calculated molecular weight of 242 kDa), with no evidence of a significant concentration of higher-order aggregates.
Figure 4. StvC-based tetramers are predominantly tetramers, and do not contain high concentrations of higher-molecular weight aggregates.
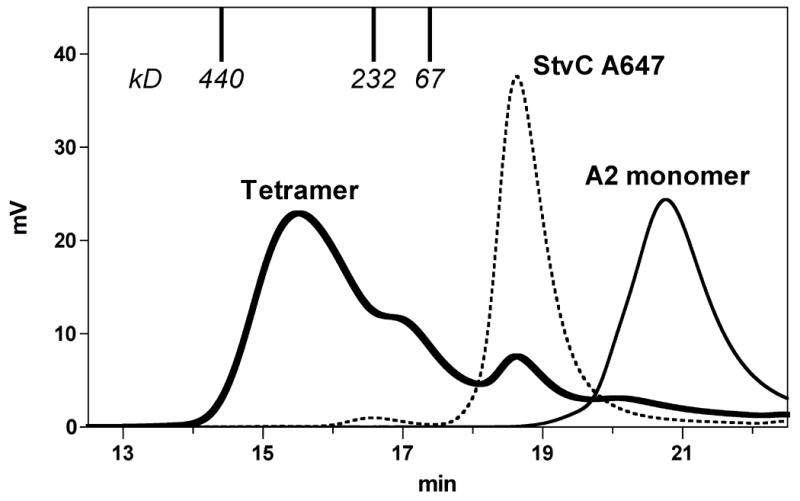
StvC modified with Alexa647-maleimide was mixed with a slight excess of HLA-A*0201 complexed with the HCMV.pp65 peptide NLVPMVATV an analyzed by size exclusion chromatography.
Flow cytometric analysis of tetramers labeled with fluorophores
Whole blood samples from HLA-A*0201+, CMV-seropostive donors were stained with anti-CD3 (APC or FITC labeled), anti-CD8 (PerCP labeled), and A2/CMV.pp65.NLVPMVATV tetramers prepared with each of the labeled StvC proteins. FACS data were acquired on an LSRII with or without compensation. As seen in Figure 5, each of the tetramers gave high quality stains with minimal background staining on an HLA-A2+ CMV-seronegative donor. Staining with a PE-labeled tetramer for comparison.
Figure 5. Flow cytometric analysis of tetramer staining from whole blood samples.
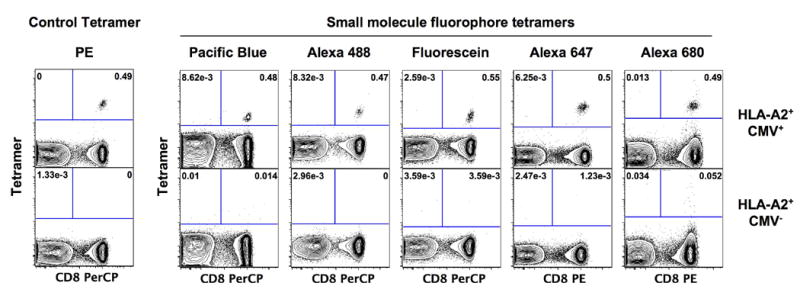
Whole blood samples from an HLA-A*0201+, HCMV+ donor were stained for 20 minutes at room temperature with A2/CMV.pp65.495–503 tetramers (peptide sequence NLVPMVATV) prepared with StvC labeled with fluorophores as indicated in the figure, together with anti-CD3 and anti-CD8 antibodies. Fluorophore labels on the anti-CD3 and anti-CD8 reagents vary from panel-to-panel and were chosen to minimize spillover from their fluorescence into the tetramer channel.
Discussion
Although we and others have occasionally prepared high-quality MHC tetramers labeled with small molecule fluorophores, our laboratory has long been frustrated by the difficulty that we have experienced in doing so reproducibly. We have screened a variety of labeled biotin-binding proteins—e.g. streptavidin, avidin, and various deglycosylated avidins—from a variety of suppliers, and none have consistently given us the excellent results that we uniformly achieve with phycoerythrin and allophycocyanin tetramers. We have entertained two possible, non-mutually exclusive, explanations to account for this mystery. First, our success with the phycobiliprotein tetramers might be attributable to their potentially greater propensity to aggregate relative to tetramers prepared with small molecule fluorophores, suggesting the possibility that the real “active ingredient” in a “MHC tetramer” prep is actually a higher order aggregate that has an even greater avidity for T cells than would a pure tetramer. Alternatively, we were intrigued by the observation of Cameron et al that commercially available FITC-labeled streptavidin had substoichiometric levels of biotin-binding activity, suggesting that the FITC modification interfered with one or more of the biotin binding sites, resulting in reagents that were mixtures of dimers and trimers, and that these lacked sufficient avidity to effectively stain T cells. Though Cameron’s proposal is certainly supported by the crystal structure of streptavidin, which reveals a potentially FITC-reactive lysine side chain that is only 7.25 Å from the carboxylate of a bound biotin ligand (Figure 1), we could not reproduce Cameron’s HABA-based “binding site protection protocol”, and we therefore sought alternative methods for introducing fluorescent labels at specific sites remote from the biotin-binding site. The most promising alternative was the StvC mutant produced by Reznik et al for the purpose of immobilization of streptavidin on a solid support, and which we successfully adapted to the production of fluorescent MHC tetramers. In our hands, tetramers prepared with fluorescently labeled StvC have so far given us the uniformly excellent results that we have come to expect from phycobiliprotein-based tetramers. The recombinant StvC has the further advantage in that it is based upon a truncated streptavidin core that is both more stable and better able to bind to biotinylated macromolecules than is native streptavidin (Sano et al., 1995). The fact that StvC-based tetramers appear to be “true tetramers” by size-exclusion chromatography Figure 4 suggests that higher-order aggregates are not required for efficacious staining.
In evaluating a MHC tetramer preparation, we ask the following questions: (1) does it specifically stain an identifiable population of T cells? (2) is that population well separated from the bulk population that is not stained with the tetramer, preferably with a clear baseline between the two populations? (3) how brightly stained is the tetramer+ population? and (4) are there a significant number of clearly “outlier” stained cells—for example those that are clearly CD8- when staining with a class I MHC tetramer, or those that fall outside a well defined cluster of staining intensity? Based upon the results in Figure 5, it appears as if the StvC tetramers labeled with the small molecule fluorophores are at least the equal of phycobiliprotein-based tetramers by three of these four criteria. The StvC tetramers clearly stain a defined population of T cells that is completely separated from the negative staining population (criteria 1 and 2), and essentially few or no outlier stained cells are observed (criteria 4). The only criteria upon which phycobiliproteins are clearly superior to the StvC tetramers is that the positive-staining cells are “brighter” and better separated from the negative population. This may be a decisive factor in some advanced multicolor flow cytometry experiments, where use of ancillary reagents labeled with fluorophores that significantly “spillover” into the detector for the tetramer fluorophore will reduce the ability to resolve the tetramer+ from the tetramer- populations (Perfetto et al., 2004). These effects require further testing, which is now ongoing in our laboratory. For most standard applications, the most important factor by far is whether or not the tetramer+ population is cleanly separated from the tetramer- population at all, and the question of “by how much” is very much a secondary issue. By this criterion, the StvC tetramers labeled with small molecule fluorophores are clearly better than adequate, and our experience so far is that they yield data with fewer “outlier” events than do phycobiliproteins-based tetramers (data not shown). Perhaps the only exception are the reagents prepared with Alexa680, but we do not yet have enough experience with these reagents to know if the slightly higher outlier background we see in Figure 5 (0.052% of CD8+ T cells) will be reproducible; even if that were the case, this level of outlier background is still low and is eminently usable.
For the tests shown in Figure 5, fluorescence compensation due to spillover of signals from the tetramer into the other fluorescence channels was not required. In situations where compensation is required, careful consideration must be given to which reagents are most appropriate for setting up the compensation controls. In general, we do not recommend use of the tetramers themselves, as the population stained by the tetramer is usually present at too low a frequency to be useful for determination of appropriate compensation settings. One alternative is to use an antibody specific for a lineage marker such as CD3 or CD8 that is labeled with the same fluorophore as the tetramer. While this will be adequate in many cases, it is not our preferred method, as there are subtle differences in the emission spectra of proteins modified with maleimide and NHS derivatives of the same fluorophore (see Figure 3B, 3C, and 3E) that may lead to incorrect compensation settings. Instead, we prefer a protocol in which compensation samples are prepared by staining a particle—such as a biotinylated bead or a cell stained with a biotinylated antibody—with the same maleimide-modified StvC protein as used in the tetramer. Of course, these samples must also contain appropriate internal negative controls.
Additional recombinant streptavidin variants may also prove to be useful tools in MHC tetramer technology. For example, it might be possible to improve the brightness of the stains through use of mutants that incorporate more than one cysteine, such as the Stv28 variant produced by Sano in which each subunit is tagged with a stretch of five cysteines, creating a streptavidin tetramer with 20 potential sites for labeling (Sano, 1999; Sano et al., 1993). We are currently setting out to test this mutant, but we are aware of the potential downsides to this approach, including possibly increased propensity of the multiply-labeled molecule to aggregate or for the fluorescent labels to homoquench. Streptavidin molecules with a mixture of wild type and cysteinylated subunits might also prove useful, such as in the production of streptavidins that are labeled with exactly one phycobiliprotein. Finally, it might also be possible to use the cysteine-mutant streptavidins to introduce appropriate FRET pairs at defined positions in the molecule.
In conclusion, using an established, efficient and inexpensive system for expression of mutant streptavidin molecules containing a single cysteine residue per subunit at a site remote from the biotin-binding site, we have successfully and reproducibly produced MHC tetramers incorporating small molecule fluorophores that prove to be excellent reagents for the identification of antigen-specific T cells. Together with newly emerging quantum dot technology, these reagents significantly expand the potential applications of MHC tetramer technology.
Acknowledgments
We thank Takeshi Sano (Harvard Medical School) for the StvC expression plasmid and advice on StvC refolding and purification. This work was supported by the NIAID MHC Tetramer Core Contract (N01-AI-25456). The NIAID Tetramer Core Facility (http://www.niaid.nih.gov/reposit/tetramer/index.html) will now prepare reagents labeled with the fluorophores described in this paper.
This paper is dedicated to the memory of Vitalii Grigoriev.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman JD, Davis MM, Shevach EM. MHC-Peptide Tetramers to Visualize Antigen-Specific T Cells. In: Coligan JE, Bierer BE, Margulies DH, editors. Current Protocols in Immunology. John Wiley & Sons, Inc; 2003. pp. 17.3.1–17.3.33. [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–6. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Amara RR, Villinger F, Altman JD, Lydy SL, O'Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma HL, Grimm BD, Hulsey ML, Miller J, McClure HM, McNicholl JM, Moss B, Robinson HL. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- Cameron TO, Cochran JR, Yassine-Diab B, Sékaly RP, Stern LJ. Cutting edge: detection of antigen-specific CD4+ T cells by HLA-DR1 oligomers is dependent on the T cell activation state. J Immunol. 2001;166:741–5. doi: 10.4049/jimmunol.166.2.741. [DOI] [PubMed] [Google Scholar]
- Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, Förster R, Rowland-Jones S, Sékaly RP, McMichael AJ, Pantaleo G. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106–11. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–44. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Freitag S, Le Trong I, Klumb L, Stayton PS, Stenkamp RE. Structural studies of the streptavidin binding loop. Protein Sci. 1997;6:1157–66. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Seth A, Veazey RS, Nickerson CE, Lifton MA, Dailey PJ, Forman MA, Racz P, Tenner-Racz K, Letvin NL. Simian immunodeficiency virus-specific cytotoxic T lymphocytes and cell-associated viral RNA levels in distinct lymphoid compartments of SIVmac-infected rhesus monkeys. Blood. 2000;96:1474–9. [PubMed] [Google Scholar]
- Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 1999;286:1377–81. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- Nepom GT, Buckner JH, Novak EJ, Reichstetter S, Reijonen H, Gebe J, Wang R, Swanson E, Kwok WW. HLA class II tetramers: tools for direct analysis of antigen-specific CD4+ T cells. Arthritis Rheum. 2002;46:5–12. doi: 10.1002/1529-0131(200201)46:1<5::AID-ART10063>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- Plunkett FJ, Soares MV, Annels N, Hislop A, Ivory K, Lowdell M, Salmon M, Rickinson A, Akbar AN. The flow cytometric analysis of telomere length in antigen-specific CD8+ T cells during acute Epstein-Barr virus infection. Blood. 2001;97:700–7. doi: 10.1182/blood.v97.3.700. [DOI] [PubMed] [Google Scholar]
- Reznik GO, Vajda S, Cantor CR, Sano T. A streptavidin mutant useful for directed immobilization on solid surfaces. Bioconjug Chem. 2001;12:1000–4. doi: 10.1021/bc015507t. [DOI] [PubMed] [Google Scholar]
- Sano T. Boron-enriched streptavidin potentially useful as a component of boron carriers for neutron capture therapy of cancer. Bioconjug Chem. 1999;10:905–11. doi: 10.1021/bc990029w. [DOI] [PubMed] [Google Scholar]
- Sano T, Cantor CR. Expression of a cloned streptavidin gene in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:142–6. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Pandori MW, Chen X, Smith CL, Cantor CR. Recombinant core streptavidins. A minimum-sized core streptavidin has enhanced structural stability and higher accessibility to biotinylated macromolecules. J Biol Chem. 1995;270:28204–9. doi: 10.1074/jbc.270.47.28204. [DOI] [PubMed] [Google Scholar]
- Sano T, Smith CL, Cantor CR. A streptavidin mutant containing a cysteine stretch that facilitates production of a variety of specific streptavidin conjugates. Biotechnology (N Y) 1993;11:201–6. doi: 10.1038/nbt0293-201. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–43. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Isolation of high avidity melanoma-reactive CTL from heterogeneous populations using peptide-MHC tetramers. J Immunol. 1999;162:2227–34. [PubMed] [Google Scholar]


