Abstract
RNA interference (RNAi) is implicated in maintaining tandem DNA arrays as constitutive heterochromatin. We used chromatin immunoprecipitation with antibodies to RNA polymerase II (RNAPol-ChIP) to test for transcription of the following repeat arrays in human cells: subtelomeric D4Z4, pericentromeric satellite 2, and centromeric satellite α. D4Z4 has a promoter-like sequence upstream of an ORF in its 3.3-kb repeat unit. A short D4Z4 array at 4q35 is linked to facioscapulohumeral muscular dystrophy (FSHD). By RNAPol-ChIP and RT-PCR, little or no transcription of D4Z4 was detected in FSHD and normal myoblasts; lymphoblasts from an FSHD patient, a control, and a patient with D4Z4 hypomethylation due to mutation of DNMT3B (ICF syndrome); and normal or cancer tissues. However, RNAPol-ChIP assays indicated transcription of D4Z4 in a chromosome 4-containing human-mouse somatic cell hybrid. ChIP and RT-PCR showed satellite DNA transcription in some cancers and lymphoblastoid cell lines, although only at a low level. Given the evidence for the involvement of RNAi in satellite DNA heterochromatinization, it is surprising that, at most, a very small fraction of satellite DNA was associated with RNA Pol II. In addition, our results do not support the previously hypothesized disease-linked differential transcription of D4Z4 sequences in short, FSHD-linked arrays.
Keywords: FSHD, transcription analysis, gene expression, chromatin immunoprecipitation, satellite DNA, DNA repeat arrays
1. Introduction
Constitutive heterochromatin containing long tandem arrays of DNA repeats, such as pericentromeric chromatin, has generally been considered transcriptionally inert. However, there are some exceptions, such as certain Drosophila pericentromeric genes [1]. Moreover, transcription of arrays of tandem DNA repeats has recently been implicated in maintaining constitutive heterochromatin in fission yeast, nematodes, plants, and vertebrates through the production of small interfering RNAs (siRNAs) from larger precursor RNAs by the RNA interference (RNAi) pathway [2]. For example, in a human-chick somatic cell hybrid (SCH), the importance of the RNAi machinery for normal structure and function of centromeres was demonstrated by conditionally knocking down the function of Dicer, the RNAi nuclease [3]. In normal murine tissues and embryonic stem cell cultures, low levels of transcripts from major and minor satellite DNAs adjacent to or within centromeres were detected [4–6]. The pericentromeric satellite III DNA at 9qh undergoes induction of RNA polymerase II-associated transcription in heat-shocked HeLa cells [7, 8]. Accumulation of centromeric satellite DNA-derived transcripts was seen in murine cells induced to differentiate in vitro or treated with the DNA methylation inhibitor 5-azacytidine or the apoptosis-inducer staurosporine [9].
In this study, we addressed the question of whether we could detect and quantitate transcription of two satellite DNAs and a subtelomeric array of tandem DNA repeats by chromatin immunoprecipitation using antibodies to RNA polymerase II (RNAPol-ChIP) [10–12; www.genpathway.com]. For each DNA region investigated, we monitored the amount of immunoprecipitation by real-time quantitative PCR (qPCR). Recently, RNAPol-ChIP has been used to quantitate occupancy of promoters [e.g., 13–15] and to quantify transcription itself [10, 12]. We analyzed the following repeat arrays: human satellite II DNA (Sat2), most of which is in the very long pericentromeric heterochromatin of chromosome 1 (1qh); centromeric satellite α DNA (Satα); and the subtelomeric D4Z4, which is associated with facioscapulohumeral muscular dystrophy (FSHD) [16, 17]. D4Z4 arrays were included in this study because they contain a putative gene within their 3.3-kb repeat unit [18, 19]. Furthermore, they might be subject to RNAi because it was proposed that all large arrays of tandem DNA repeats are templates for production of siRNA [6]. RNAPol-ChIP was chosen for the transcription analysis because it can directly measure transcription and is independent of posttranscriptional processing, differential RNA stability in vivo, and the size of the RNA product.
The analysis of transcription from D4Z4 arrays was of particular interest because of their involvement, by an unknown mechanism [20], in FSHD. These arrays are present at 4q35 and 10q26 and are highly polymorphic in size [21, 22]. FSHD, a dominant disorder, is tightly linked to the number of repeat units in the D4Z4 array. Patients almost always have <11 repeat units at one of their two allelic 4q35 arrays while unaffected individuals have 11–100 repeat units at both 4q35 alleles [23] (Fig. 1A). By RT-PCR, Northern blotting, or cDNA cloning, Ding et al. [24] found D4Z4-like transcripts related to DUX4, an open reading frame (ORF) in the D4Z4 repeat unit, in several normal human tissues and a rhabdomyosarcoma cell line. However, sequencing revealed that they did not arise from D4Z4, but rather from partially homologous sequences elsewhere. Because DUX4 could encode a double-homeodomain protein [18, 19] and a decreased size of murine transgene arrays can be associated with increased expression [25], it was hypothesized that a decrease in the number of D4Z4 repeat units at one allelic 4q35 position causes FSHD by upregulation of DUX4. We compared transcription at D4Z4 in disease and control cell populations, including myoblasts from FSHD patients, lymphoblasts from patients with the ICF syndrome (immunodeficiency, centromeric region instability, facial anomalies) that results in strong hypomethylation of D4Z4 [26] due to mutations in DNMT3B [27], and cancers with D4Z4 hypomethylation. The effect of hypomethylation of D4Z4 on its transcription is of special interest because FSHD cells exhibit moderate hypomethylation of their abnormally short, disease-linked D4Z4 array at 4q35 [28]. We also report the first analyses of transcription of Sat2 and Satα in control or transformed human cell populations displaying either the high levels of methylation that are normal for these sequences or hypomethylation [29, 30].
Fig. 1.
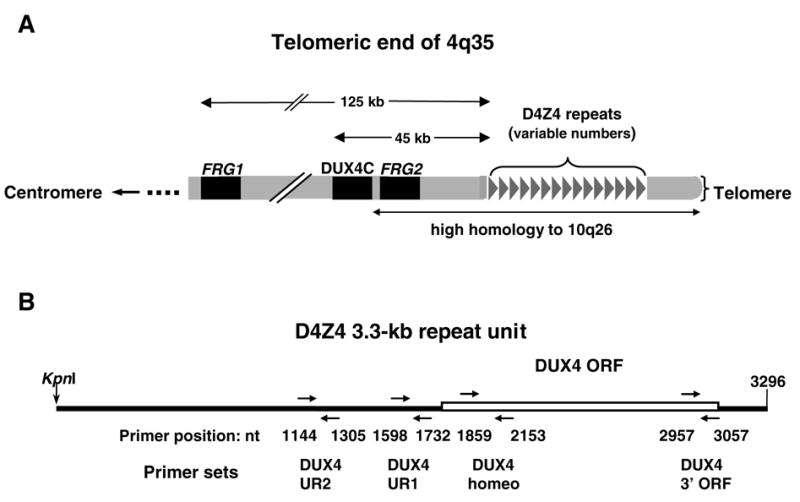
Schematic illustration of the distal region of 4q35 and the repeat unit of D4Z4. (A), positions of the size-polymorphic D4Z4 array; FRG1 and FRG2, the only known genes in the distal portion of 4q35; and DUX4C, a putative gene, are illustrated (not to scale). From one to about 100 repeat units can be present in the D4Z4 array. Small numbers of repeat units at 4q35 are linked to FSHD. The region of 4q35 that is highly homologous to the subtelomeric region of 10q, including the D4Z4 array, is indicated. DUX4C is highly homologous to, but in the opposite orientation from, DUX4 in the D4Z4 array; FRG2 is in the same direction as DUX4C. (B), The DUX4 ORF within the 3.3-kb D4Z4 repeat unit and the primers used to test for transcription from D4Z4 are shown with positions given relative to the first base of the KpnI site in the 3.3-kb D4Z4 repeat unit (from GenBank AF11753). The DUX4 UR1 primers are partially homologous to a subrepeat called hhspm3 (GenBank X06587).
2. Materials and methods
2.1. Cell culture and tissue samples
Myoblast cell strains (GM17899 from Coriell Institute and F1010 from our lab) were from biopsies of moderately affected deltoid skeletal muscle of FSHD patients with a confirmed short D4Z4 array at 4q35. A non-FSHD myoblast cell strain (GM17901, Coriell Institute) was established from a deltoid muscle biopsy of a patient with generalized muscle weakness and a diagnosis of dematomyositis. At the passage used for analysis, most of the cells in the myoblast cultures stained desmin-positive, indicating that they were myoblasts, and not contaminating fibroblast-like cells. Lymphoblastoid cell lines (LCLs) were B-cell lines from FSHD patients, GM17939 and GM17868 (Coriell Institute); normal controls, AG14836 and AG15022 (Coriell Institute) and three previously described controls [31]; and ICF patients, ICF C (GM08714, Coriell Institute), ICF B, ICF K, ICF G, ICF S [31] and ICF 1 and 3 [32]. The cancer samples were previously described [29, 30]. All human tissue samples were obtained with IRB approval for these studies from Tulane Medical Center and the University of Mississippi Medical Center at Jackson. Somatic cell hybrids were from the Coriell Institute and harbor only one human chromosome each, Chr4 (GM11687), Chr14 (GM10479), or Chr15 (GM11715).
2.2. Chromatin Immunoprecipitation
Genpathway’s TranscriptionPath method was carried out as described [12]. Cultured cells were fixed in 1% formaldehyde for 15 min according to standard procedures (http://www.genpathway.com/order.html). Fixation was stopped with 0.125 M glycine, and cells were washed with PBS containing 0.5% Igepal (Sigma) and collected. Frozen tissues for ChIP were minced in formaldehyde, fixed for 15 min, and processed similarly to the cells. Cells and tissues were homogenized (Dounce homogenizer and Tissue Tearor, BioSpec Products, respectively), and the lysates were sonicated (Misonix sonicator). The majority of DNA fragments were about 300–500 bp long. The RNA Pol II antibodies were directed against the carboxyterminal domain of the largest subunit of RNA Pol II (ab5095, Abcam) or the N-terminal 224 residues (H224, sc-9001, Santa Cruz Biotechnology). In several assays, we used antibodies against RNA Pol I and TBP (c-17913 and sc-273, respectively, Santa Cruz Biotechnology). Generally, 5–30 μg of DNA in the form of sonicated chromatin was immunoprecipitated. Then, protein A-agarose (Invitrogen) was used to isolate the immune complexes, followed by DNA purification. About 2–5% of the final immunoprecipitated fraction and 25 ng of the non-immunoprecipitated sonicated chromatin (input DNA) were subjected to qPCR (MyIQ Cycler, Bio-Rad) catalyzed by Taq polymerase (iQ SYBR Green Supermix, Bio-Rad) using an annealing temperature of 58°C and detecting the amplified DNA by SYBR Green fluorescence. For a given sample, the same amount of template was amplified with each of the primer-pairs (Table 1) except that, in the case of Sat2 primers, the input DNA was diluted 1:100 before PCR, with corresponding normalization. For each set of primers, the dissociation curve was checked to make sure that it was unimodal. To determine the level of transcription for each DNA region tested from the threshold cycle number, a standard curve of three to four 10-fold serial dilutions of input DNA from one of the chromatin samples (assigned values of 100–100,000) was included in each multiwell plate and amplified in parallel with a primer-pair for one of the reference genes. The slope of the standard curves were −3.3 ± 0.4 and the correlation coefficients were 0.95–s0.99. The average Ct values of triplicate amplifications from immunoprecipitated DNA or input DNA were compared to those from the above-mentioned standard curve and converted to the appropriate values. To normalize for differences in amplification efficiency for various primer-pairs using the same DNA input, values obtained from amplification of immunoprecipitated DNA were divided by the values obtained from amplification with input DNA. The resulting numbers were multiplied by 100 and called “normalized transcription units.” Positive and negative controls, using well-characterized primer-pairs for constitutively expressed genes and untranscribed genomic regions (Untr) were included in every ChIP experiment.
Table 1.
PCR primers for ChIP and RT-PCR
| Gene or untranscribed region | DNA accession number | Amplicon | Primer sequence | Product size (bp) |
|---|---|---|---|---|
| DUX4 UR2 | AF117653 | 5′end of DUX4 putative promoter in D4Z4c | GGGTCTCGCTCTGGTCTTCT
GAGGGGAGTGTGGAACTGAA |
162 |
| DUX4 UR1 | AF117653 | putative promoter region of DUX4 in D4Z4 | GGGCTCACCGCCATTCAT
TGCACCTCAGCCGGACTGT |
135 |
| DUX4 Homeo | AF117653 | DUX4 putative double homeodomain in D4Z4 | ACGGAGACTCGTTTGGAC
TGGAAAGCGATCCTTCTC |
295 |
| DUX4 3′ORF | AF117653 | 3′end of putative DUX4 gene in D4Z4 | GCGCAACCTCTCCTAGAAAC
AGCAGAGCCCGGTATTCTTC |
101 |
| DUX4C UR | AY500824 | 777–957; putative promoter of DUX4C through 5′ end of ORF | AACGCGGTGGAGGTGGTAGGTCTT
CCGCGGGGAGGGTGCTGTC |
181 |
| Chr4 Satα | M38467 | 162–301 | CTGCACTACCTGAAGAGGAC
GATGGTTCAACACTCTTACA |
139 |
| Chr1 Sat2 | X72623 | 838–997 | CATCGAATGGAAATGAAAGGAGTC
ACCATTGGATGATTGCAGTCAA |
160 |
| FRG1 (ChIP)c | AF146191 | Intron 2 | TGCTACTGATGCCTTTACTTCA
AAGCAGTCTAATGGTGCCTACT |
78 |
| FRG1 (RT-PCR) | AF146191 | Exon1 | TCTACAGAGACGTAGGCTGTCA
CTTGAGCACGAGCTTGGTAG |
178 |
| ACTB (ChIP) | NM_001101 | Intron 3 | CCTCATGGCCTTGTCACAC
GCCCTTTCTCACTGGTTCTCT |
200 |
| PPIB (ChIP) | NM_000942 | Intron 4 | TATGGCTCAGGAGGGCTAAG
GCCCAACCTGGGTCTTACTT |
155 |
| PPIB (RT-PCR) | NM_000942 | Exon 4 & 5 | GCACAGGAGGAAAGAGCATCTA
ACCACCTCCATGCCCTCTA |
202 |
| ADH5 (ChIP) | NM_000671 | Intron 2 | GCTCAGCCCAAGATCAAAAC
TCCAGCCTTACAATCACAACA |
89 |
| HPRT (ChIP) | NM_000194 | Intron 1 | CGAGCTTACCCTGCCAATAA
TCTATTTCCTGGTGGTCACAAA |
150 |
| HPRT (RT-PCR) | NM_000194 | Exon 3 & 4 | GGGAGGCCATCACATTGTAG
CCCTGTTGACTGGTCATTACA |
166 |
| rRNA 18S (ChIP) | HSU13369 | 18S rRNA | AGTTGGTGGAGCGATTTGTC
CGCTGAGCCAGTCAGTGTAG |
205 |
| Untr 4 | Chr4: 53,081,269-53,081,451 | Chromosome 4 | CTCCCTCCTGTGCTTCTCAG
AATGAACGTGTCTCCCAGAA |
183 |
| Untr 5 | Chr5: 3,000,608-3,000,815 | Chromosome 5 | CTGTACCTGGGGTTCATTCATT
CAGTAAGCCGTTCACTCTCACA |
208 |
| Murine ActB (ChIP) | NM_007393 | Intron 3 | TGATAAGTGGCCTTGGAGTG
CTCAGGGCAGGTGAAACTGT |
150 |
| Murine Ppib (ChIP) | NM_011149 | Intron 3 | GATGGATGGGAGTTCTGTGA
CATTCTCACCCTCAAGACACAA |
133 |
| Murine Ppib (RT-PCR) | NM_011149 | Exon 4 & 5 | CAAAGACACCAATGGCTCAC
CGGAGTCGACAATGATGACA |
176 |
| Murine Actb (prom; ChIP) | NM_007393 | Region -163 to -24 relative to TSS | CATGGTGTCCGTTCTGAGTG
ACAGCTTCTTTGCAGCTCCT |
140 |
| Murine Ppib (prom; ChIP) | NM_011149 | Region +115 to +328 relative to TSS | GCGCAATATGAAGGTGCTCT
GACGTCAGGTCGGAACTAC |
214 |
| Murine Untr 6 | Chr6: 120,740,575-120,740,790 | Murine chromosome 6 | TCAGGCATGAACCACCATAC
AACATCCACACGTCCAGTGA |
216 |
The primer-pairs were used for Q PCR in ChIP assays or RT-PCR, as indicated, or if not otherwise specified, they were used for both. The sequences are human if not specified as murine. TSS, transcription start site; prom, promoter primers used only for TBP ChIP assays. As described in the text, the D4Z4 primers also amplify partially homologous sequences not in tandem D4Z4 arrays. Also, the FRG1 primers from intron 1 are not 4q35-specific. Gene-internal primers for standards were used for RNAPol-ChIP to avoid 5′ gene regions at which RNA Pol II might accumulate in RNA Pol II ChIP assays. The positions of D4Z4 primers are given in Fig. 1B.
2.3. RT-PCR
For real-time RT-PCR, total RNA was extracted with Trizol (Invitrogen) and digested with DNaseI (either RQ1, Promega, 1 unit for 10 μg of RNA, 1 h at 37ºC, or Amplification Grade, Invitrogen, 3 units for 3 μg of RNA, 45 min at room temperature). After inactivation of the enzyme, the RNA was used for reverse transcription primed with random hexanucleotides (Superscript III, Invitrogen). The amount of the different transcripts from a given sample is expressed as arbitrary units of RNA and was determined from a standard curve, as described for ChIP.
For semi-quantitative (sq) RT-PCR (28 cycles), random hexanucleotides were used to generate the cDNA unless oligo(dT) priming is specified. The quality of each cDNA preparation was tested by PCR with primers for GAPD. For the test PCR primers, the corresponding minus-RT template was always shown to be negative upon concurrent amplification followed by gel electrophoresis and visualization of bands with ethidium bromide.
3. Results
3.1. Strategy for assaying transcription
We tested tandem arrays of DNA repeats present in subtelomeric, pericentromeric, or centromeric regions for the level of associated RNA Pol II by ChIP assays as an indicator of transcription. ChIP-deduced levels of transcription are expressed as “normalized transcription” (normalized for primer-pair efficiency of amplification as described in Section 2.2 above). Real-time RT-PCR assays were also done on random-primed cDNA to approximate steady-state levels of transcripts from the repeat arrays relative to those of constitutively expressed genes.
Because the 3.3-kb repeat unit of the D4Z4 array contains an ORF called DUX4 (Fig. 1B) and an upstream sequence that can function as a promoter in reporter assays [19], we chose four primer-pairs within or upstream of DUX4 (Fig. 1B, Table 1) for RNAPol-ChIP PCR and RT-PCR. These primers amplify part of DUX4 that could encode two homeodomain regions (Homeo); the 3′ end of DUX4 (3′ ORF); the proposed promoter upstream of the DUX4 ORF (upstream region 1, UR1); or a further upstream sequence (UR2; Fig. 1B). These sequences are shared between the D4Z4 arrays at 4q35 and 10q26 (Fig. 1A). They are also related to many closely homologous sequences elsewhere in the genome [18] as evidenced by testing a panel of human-rodent SCH DNAs for amplification with some of the primer-pairs used here [33]. This complication was addressed by the use of human-rodent SCHs, as described below. In addition, there is a solitary, highly homologous ORF proximal to the D4Z4 array, DUX4C, which is present on 4q35 (Fig. 1A). Homeo primers are completely homologous to sequences in DUX4C at 4q35 and DUX4 at 4q35 and 10q26 and so should amplify all three corresponding sequences. The other D4Z4 primer-pairs do not have sufficient homology to the DUX4C region to amplify it. We also used a primer-pair that amplifies the upstream region of DUX4C (DUX4C UR), but not that of DUX4 within the array.
For Sat2 and Satα, which are composed of variants of oligonucleotide repeats [34, 35], we used a primer-pair that was derived from chromosome 1 (Chr1) or chromosome 4 (Chr4), respectively (Table 1). The Sat2 primers amplified DNA from several human chromosomes in addition to Chr1, as determined by PCR testing of a panel of human-rodent SCH DNAs. However, most of the Sat2 sequences are at 1qh [34]. The Satα primer-pair amplified sequences only from Chr4 (data not shown). Specific primer-pairs were used for positive transcription standards and for negative controls of untranscribed regions in the human genome (Untr; Table 1). In addition, we examined transcription of FRG1, the 4q35-specific gene that is closest to the D4Z4 array (Fig. 1A).
3.2. Testing for transcription of D4Z4 arrays in disease and control human cell cultures
The size of the D4Z4 array at 4q35 is tightly linked to FSHD, a disease that seems to involve the in vivo-counterpart of myoblasts, i.e. satellite cells [36, 37]. Therefore, we first assayed for D4Z4 transcription in myoblast cell strains (two FSHD and one control). The normalized transcription signals for the D4Z4 sequences were no higher than the background level for the untranscribed control sequence in these cells (Fig. 2A and C, and data not shown). For the DUX4C upstream region (DUX4C UR, Fig. 1B), which is located proximal to the D4Z4 array in 4q35 (Fig. 1A), there was a slight increase in signal over that of the non-transcribed sequence (~30%; Fig. 2A and C). However, there was no signal above background with the Homeo primers, which amplify a nearby sequence in the DUX4C ORF and the DUX4 ORF (Fig. 1). Real-time RT-PCR on RNA from three myoblast cell strains indicated no reverse transcriptase-dependent signal for any of five tested D4Z4 or D4Z4-like sequences (Fig. 2B and D and data not shown).
Fig. 2.
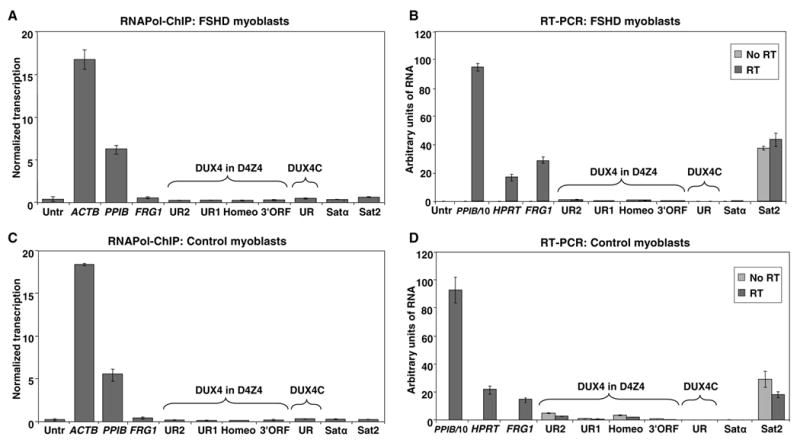
Transcriptional analysis of D4Z4 and satellite DNA sequences in FSHD and control myoblasts. (A) and (C), in FSHD myoblasts (FM1010) and control myoblasts (GM17901), ChIP was done with an antibody specific for the largest subunit of RNA Pol II (ab5095), and the immunoprecipitated DNA was analyzed by qPCR using the indicated primers (Table 1). Levels of transcription were determined as described in Materials and methods and normalized to 25 ng of chromatin per immunoprecipitation. Error bars indicate maximal and minimal values from triplicate amplifications. An untranscribed region on Chr5 was the negative control (Untr); ACTB and PPIB are the positive controls. Tested subregions of the putative DUX4 gene in D4Z4 are indicated by a bracket, as is DUX4C, the putative DUX4-related gene 42 kb proximal to the D4Z4 array (Fig. 1A). FRG1, like DUX4 and DUX4C, is located on 4q35 (Fig. 1A). The tested satellite DNA repeats were Satα and Sat2. (B) and (D), total RNA from control or FSHD myoblasts was reverse transcribed with random priming and analyzed by qPCR using the indicated primers (Table 1). Samples amplified without reverse transcription (no RT) served as controls. The qPCR values obtained for PPIB were divided by 10 to bring them into the range of the other tested genomic regions before plotting. The normalized transcription levels for Panel A for Untr, FRG1, UR1, UR2, Homeo, 3′ORF, UR, Satα, and Sat2 were as follows: 0.32, 0.44, 0.22, 0.20, 0.20, 0.26, 0.40, 0.30, and 0.52, respectively. The respective values for Panel C were 0.26, 0.46, 0.16, 0.22, 0.16, 0.20, 0.34, 0.28 and 0.28. VASSILI, IN PANEL C, THE BAR FOR UR1 IS LOWER THAN THAT FOR UR2 BUT THE NUMERICAL VALUE ABOVE FOR UR1 IS HIGHER THAN FOR UR2. PLEASE CHECK THIS AND THE HOMEO, 3′ORF, AND SAT2 NUMBERS AND BARS, PLEASE.
In contrast, transcription of the 4q35 FRG1 gene was evident by RNAPol-ChIP in all myoblast cell strains (Fig. 2A and C), although at a low level (signals of 1.4–1.7 times the untranscribed control and 3–5% the level of the PPIB positive control). Transcription was confirmed by RT-PCR (Fig. 2B and D). It has been reported that a shortened D4Z4 array at 4q35 results in massive overexpression of FRG1 in myoblasts and muscle as determined by semi-quantitative RT-PCR [38, 39], although this is controversial [33, 37]. By real-time RT-PCR, we observed a difference of only about two-fold in FRG1 RNA levels relative to the reference HPRT or PPIB RNA levels in a comparison of FSHD and control myoblasts (Fig. 2B and D) using FRG1 exon 1 primers [40]. Furthermore, by ChIP assay using FRG1 intron 2 primers (Table 1), only a modest difference between the FSHD and control samples was seen (Fig. 2A and C).
We also looked for transcription of D4Z4 in B-cell LCLs, which are fast-growing cell lines established by transformation with Epstein-Barr virus. The LCLs were from controls, FSHD patients, or patients with ICF, a rare recessive DNA methyltransferase 3B-deficiency disease involving strong hypomethylation of certain tandem DNA repeats, including D4Z4 [26, 41]. Because DNA hypomethylation is often associated with increased transcription, ICF cells were especially good candidates for D4Z4 transcription. By real-time RT-PCR, only one D4Z4 sequence showed consistent, albeit very low levels, of transcripts, namely DUX4 UR2 (Fig. 3B, D, and F) with the following ratios of RNA-dependent qRT-PCR product from DUX4 UR2 relative to those from the HPRT standard: 0.03, 0.08, and 0.01, for the ICF, FSHD, and control LCLs, respectively. However, the lack of signal from nearby DUX4 ORF and UR1 sequences casts doubt on whether the very low DUX4 UR2 signal is indicative of transcription from D4Z4. By RNAPol-ChIP, neither transcription at DUX4 UR2 nor at the other D4Z4 sequences was evident in the ICF, FSHD, or control LCLs (Fig. 3A, C, and E).
Fig. 3.
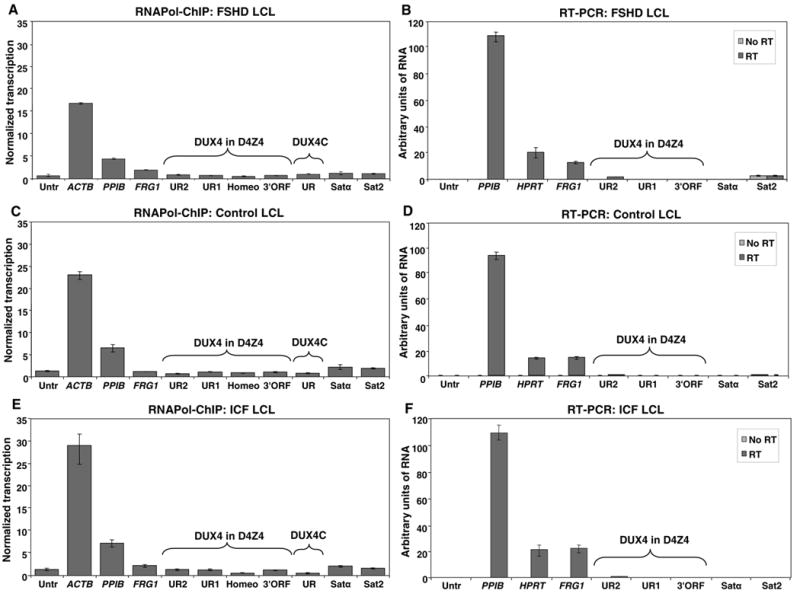
Transcriptional analysis of D4Z4 and satellite DNA sequences in FSHD, ICF, and control lymphoblastoid cell lines. (A), (C), and (E), an FSHD LCL (GM17939), a control LCL (AG14836), and an ICF LCL (GM08714), respectively, were used for ChIP with an antibody specific for RNA Pol II (H-224). Immunoprecipitated DNA was analyzed by qPCR with the indicated primers, as for Fig. 2. (B), (D), and (F), RNA from the same ICF and control LCLs and a different FSHD LCL (GM17868) were analyzed by qPCR with or without reverse transcription. The normalized transcription levels for Panel A for Untr, FRG1, UR1, UR2, Homeo, 3′ORF, UR, Satα, and Sat2 were as follows: 0.7, 1.97, 0.74, 0.91, 0.62, 0.74, 1.05, 1.27, 1.13. For panel B the respective values were: 1.35, 1.19, 1.12, 0.71, 0.92, 1.03, 0.8, 2.1, 1.94. For panel C the respective values were: 1.41, 2.23, 1.27, 1.38, 0.54, 1.19, 0.48, 2.03, 1.53. VASSILI, THE BAR FOR UR2 LOOKS QUITE A BIT HIGHER THAN FOR UNTR BUT UR2’S VALUE IS ONLY ~3% HIGHER THAN UR1’S. ALSO PLEASE CHECK THE REST OF THE BARS VS. VALUES FOR HIS FIG.
We also used all three LCLs for ChIP with an antibody against the 135-kDa subunit of RNA Pol I to determine if this enzyme was transcribing D4Z4. As expected, a strong signal was obtained with primers corresponding to genomic sequences for 18S rRNA, but the primer-pairs for D4Z4 and related sequences gave no more signal than obtained for the untranscribed control sequence (data not shown). In summary, there was no consistent evidence for appreciable transcription of D4Z4 in disease or control myoblasts and LCLs.
3.3 Testing for transcription of satellite DNA in human cell cultures
Because it has been postulated that D4Z4 repeat units reside in heterochromatin [21, 39, 42, 43], we tested transcription of Sat2, located in pericentromeric heterochromatin, and Satα, which is found in chromatin with a partially heterochromatic nature [44], in the above-described cell cultures. From the RNAPol-ChIP analyses of these cultures, only one of the myoblast cell strains exhibited a considerable transcription signal for Sat2 over background. The respective ratios of normalized transcription of Sat2 and Satα relative to the background from the untranscribed control sequence were 1.6 and 0.9 for this FSHD myoblast cell strain (Fig. 2A), but only 0.7 and 0.6 for the other FSHD myoblast cell strain (data not shown), and 1.1 and 1.1 for the normal myoblast cell strain (Fig. 2C). In RT-PCR assays for RNA from these satellite DNAs, there was slightly more PCR signal after RT than without RT, but only for the FSHD myoblasts (Fig. 2B and D and data not shown). However, the background signal in the minus-RT reactions for Sat2 makes interpretation of these results difficult and reflects the problem of completely removing all highly repeated DNA in RNA preparations without degrading the RNA during incubation with “RNase-free” DNase. For some preparations, more of the contaminating DNA was removed (Fig. 3B, D, and F) than others (Fig. 2B and D). The stronger background with primers for Sat2 than Satα can be ascribed to the Sat2 primers recognizing pericentromeric DNA in many of the chromosomes, including in the very long 1qh region, while the primers for Satα recognize only centromeric DNA from Chr4.
With the three LCLs, ChIP assays indicated small amounts of RNA Pol II engagement at Sat2 or Satα relative to the untranscribed control sequence. However, there was no more evidence of transcription in the patient LCLs than in the control LCL (Fig. 3 A, C, and E). When tested with the RNA Pol I antibody, no signal over background was seen for the satellite DNAs (data not shown). When the same LCLs were examined by real-time RT-PCR for Sat2 or Satα transcripts, the only positive result was from the ICF LCL with the Sat2 primers. However, the ratio of RNA-dependent qRT-PCR product from Sat2 vs. that from the HPRT standard was only 0.008 (Fig. 3F; the signal was too low to be visible in this panel). Given the strong hypomethylation of Sat2 in ICF LCLs, we also tested ICF and control LCLs by sqRT-PCR (28 cycles) with Sat2 primers with or without priming by oligo(dT). Four of the seven ICF LCLs and one of the control LCLs gave reverse transcription-dependent bands of the expected size (Fig. 4 and data not shown). Therefore, the low levels of engagement of RNA Pol II at Sat2 observed in ChIP assays were confirmed for Sat2 in ICF LCLs by Q or sqRT-PCR for some of the disease and one of the control LCLs.
Fig. 4.
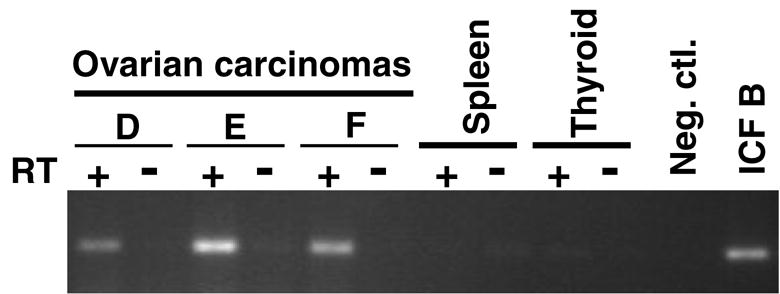
Semi-quantitative RT-PCR analysis for Sat2 RNA transcripts. The indicated cancer samples, the LCL from ICF patient B, and normal spleen and thyroid were analyzed by RT-PCR for 28 cycles with Sat2 primers either with (+ RT) or without (- RT) reverse transcription. Priming of cDNA synthesis was done with oligo(dT).
Satellite III transcripts from the pericentromeric heterochromatin of Chr9 were reported to be induced by heat shock [7, 8]. Therefore, we tried to stimulate transcription of Sat2 sequences in a control LCL by incubation at 44ºC for 30 min followed by a 2-h recovery at 37ºC. We saw no induction of Sat2 transcription by sqRT-PCR (28 cycles) after this heat-shock treatment. Similarly, Jolly et al. [8] did not observe Sat2 or Satα transcripts in heat-shocked HeLa cells although induction of satellite III transcripts was evident. We did however see a modest induction of Sat2 transcripts upon 24-h treatment of the same control LCL with 2 μM trichostatin A (TCA), a histone deacetylase inhibitor (data not shown).
3.4. Testing for transcription of D4Z4 arrays in human-rodent somatic cell hybrids
We tested for D4Z4 transcription in a human-mouse SCH containing human Chr4 as the only human chromosome and no detectable murine D4Z4-like sequences [18]. Use of this SCH also minimized the possible dilution of the ChIP-derived normalized transcription values for D4Z4 by numerous D4Z4-like sequences in the acrocentric chromosomes. Upon ChIP assay, stronger signals were observed for the D4Z4 sequences (DUX4 UR2, UR1, and 3′ORF) than for the untranscribed control sequences with two RNA Pol II antibodies in three immunoprecipitates (ab5095 and H224, Fig. 5A–C). High specificity of the assay was confirmed using both murine and human untranscribed control sequences, non-immune serum, and a Chr1 SCH (Fig. 5A–C). Upon qRT-PCR, the Chr4 SCH showed only a small increase in the RT-dependent signal for DUX4 UR2 but not for the other D4Z4 sequences (data not shown).
Fig. 5.
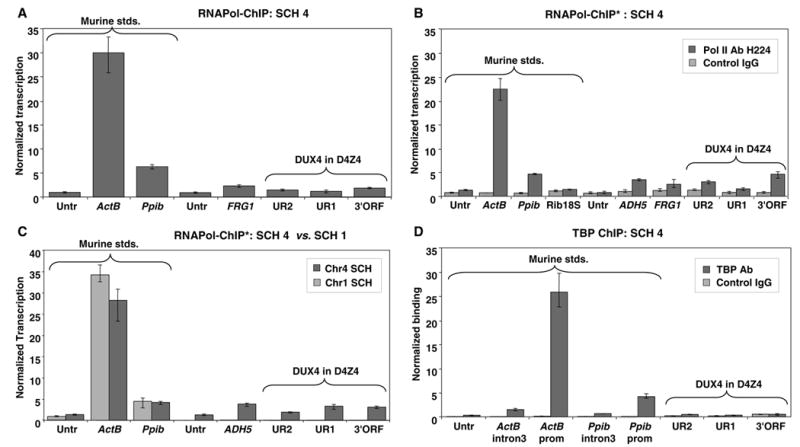
Transcriptional analysis of D4Z4 in human-rodent somatic cell hybrids. ChIP analyses of a SCH with a single human chromosome (Chr4) in a murine genetic background were done using the ab5095 RNA Pol II antibody (A), the H224 RNA Pol II antibody (B and C, indicated by an asterisk), or the TBP antibody (D). ChIP analysis in (C) included testing a SCH containing human Chr1. In (B) and (D), a rabbit control IgG reaction was run in parallel. For TBP binding, the promoter regions of the murine ActB and Ppib standard genes as well as the intron 3 regions of both genes present >2 kb downstream from the transcription start sites were analyzed. As expected with this antibody, the promoter regions gave much stronger signals. Normalized transcription values are presented as described in Fig. 2. Murine standards (Murine stds.) are indicated by a bracket; all other genomic regions tested are human sequences from Chr4. In Panel A, the normalized transcription values for the twoUntr standards, FRG1, UR2, UR1 and 3′ORF were as follows: 0.93, 2.29, 1.2, 1.47, and 1.88. VASSILI, PLEASE ADD THE MURINE UNTR AND CHECK ALL BAR HEIGHTS VS. THESE NUMBERS.
We also performed ChIP on the Chr4 SCH with an antibody against the general transcription factor TBP. We observed the expected enrichment of TBP in the promoter regions of ACTB and PPIB but the normalized binding levels for TBP minus control IgG binding for the D4Z4 sequences were no greater than for the untranscribed control sequence (Fig.5D). Therefore, TBP-binding by ChIP was not informative for detecting low-level transcription from D4Z4.
We also tested two human monochromosomal SCHs containing acrocentric human chromosomes that harbor D4Z4-like sequences amplified by Homeo and DUX4 UR1 primer-pairs upon PCR testing of monochromosomal human-rodent SCHs (data not shown). Evidence for transcription was seen for the Chr15 SCH, but not the Chr14 SCH, in RNAPol-ChIP assays with only one of the two RNA Pol II antibodies (H224). DUX4 UR1, UR2, and 3′ORF showed normalized transcription levels of 2.2, 1.2, and 1.5 times the average background level of two untranscribed control sequences, respectively (data not shown). The two negative control sequences exhibited a similar background of “normalized transcription” signals that differed from each other by only 8%.
3.5. Testing for transcription of D4Z4 arrays and satellite DNA in human cancers
Because there are major perturbations in the epigenetics of human cancers that can negatively or positively affect transcription, we tested two diverse types of cancers, ovarian epithelial carcinomas and Wilms tumors, for transcription of satellite DNA and D4Z4 by RNAPol-ChIP. Several of these cancers exhibited hypomethylation of D4Z4 relative to various control somatic tissues (data not shown). Nonetheless, no consistent evidence for transcription of D4Z4 or related sequences was observed in five ovarian carcinomas, eight Wilms tumors, and a normal spleen sample (Fig. 6 and Table 2). However, RNAPol-ChIP did reveal low-level transcription of Sat2 in many of the cancers (e.g., Fig. 6B–F). Eleven of the thirteen samples gave a normalized transcription ratio for Sat2 vs. the untranscribed control sequence of 1.4–2.0 (Table 2). Four of the cancers also gave similar ratios for Satα vs. the untranscribed control sequence.
Fig. 6.
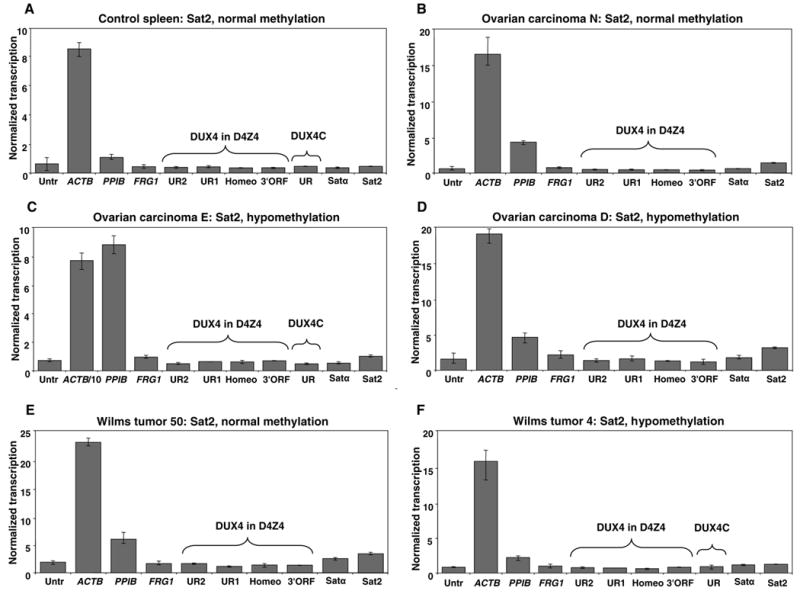
Transcriptional analysis of D4Z4 and satellite sequences in control spleen, ovarian carcinoma, and Wilms tumor samples. ChIP analyses were performed using the ab5095 antibody for RNA Pol II. Normalized transcription is shown as described in Fig. 2. The methylation status of Sat2 DNA in the samples, which was previously determined by Southern blot analysis with a CpG methylation-sensitive restriction endonuclease (Table 2), is indicated on the top of each graph. The normalized transcription levels for Untr, Satα, and Sat2 were as follows (with the panel letter in the parenthesis): (A), 0.58, 0.34, 0.42; (B), 0.66, 0.69,1.56; (C), 0.58, 0.43,0.81; (D), 1.64,1.85, 3.3; (E), 2.07, 2.81, 3.75; (F), 0.83, 1.13,1.23.
We used sqRT-PCR, with oligo(dT) priming, to look for Sat2 transcripts (Table 2). None of six control tissues was positive for Sat2 RNA. RT-dependent Sat2 transcripts were seen for four of the seven tested cancers with ChIP-determined ratios ≥1.4 for Sat2 normalized transcription divided by the background for the negative control (Fig. 4 and Table 2). Similar results were obtained by sqRT-PCR with oligo(dT) priming and by semi-quantitative or real-time RT-PCR with random priming. We had previously determined that most, but not all, of these cancer samples were hypomethylated in Sat2 and Satα sequences relative to a variety of postnatal somatic controls [29, 30]. Little correlation between this type of DNA hypomethylation and transcription of these sequences was observed (Table 2).
4. Discussion
It was hypothesized that the DUX4 ORF in abnormally short D4Z4 arrays in FSHD muscle cells is upregulated to give an unstable transcript encoding a toxic, homeodomain-containing protein, thereby resulting in FSHD [19, 45]. To measure the levels of transcription from DUX4 and DUX4-like sequences, and not only transcripts, we used a newly developed RNAPol-ChIP assay [10, 12] that measures the association of RNA Pol II with genomic sequences, preferably avoiding amplicons in or near the promoter or 5′ region of the gene. By qPCR analysis, a genomic sequence of interest can be quantified in the ChIP DNA relative to the input chromatin, as a measure of its level of transcription..A negative control of an untranscribed gene region establishes the background level. A constitutively expressed gene is included to check the quality of the immunoprecipitate but is not necessary for determining the normalized transcription level of the test genes. Among hundreds of genes assayed by us (Genpathway) commercially since 2004, those found to be transcribed (or differentially transcribed) by the RNAPol-ChIP generally displayed similar qualitative results from RNA measurement by qRT-PCR. However, transcription rates may be high while accumulated levels of RNA are low, and vice versa. Because the RNAPol-ChIP assay measures the transcriptome at the DNA level, the results are highly reproducible and not subject to the variability typically observed with RNA. In one validation assay, levels of transcription induction determined from normalized transcription values for six genes in six replicate immunoprecipitations of the same chromatin from treated vs. untreated cells differed by an average of only 21% (unpub. data). Importantly for the present study, RNAPol-ChIP assay of transcription circumvents possible problems due to highly unstable transcripts.
By RNAPol-ChIP with primer-pairs within or upstream of DUX4, we saw no evidence of D4Z4 transcription in FSHD or control myoblasts; in LCLs, including one from an ICF patient with strong hypomethylation of D4Z4; and in two types of cancers, several of which were also strongly hypomethylated in D4Z4 (unpub. data). Similarly, qRT-PCR involving random-primed cDNA synthesis did not reveal transcription of D4Z4 in these cells, with the exception of a consistent, low RT-PCR signal above background from the promoter-like region [19] upstream of DUX4 in the LCLs. Several cDNAs from an unclassified 6-week fetus contained this particular sequence and part of the DUX4 ORF [18]. However, these cDNAs, like all other reported D4Z4-related cDNAs, exhibited many mismatches to the very highly conserved D4Z4 sequence and appear to be derived from cross-hybridizing sequences in chromosomes other than 4 and 10 present as scattered repeats partially homologous to the entire 3.3-kb unit or as subrepeats [18].
It may be difficult to detect transcripts from DUX4 and related sequences because of inherent instability of the RNA due to lack of introns and polyadenylation [19]. However, RNAPol-ChIP is not subject to this limitation. Our finding of no consistent positive signal for D4Z4 in human cells by RNAPol-ChIP indicates that the level of transcription from these sequences is, at best, very low in the tested cell populations, although we did find evidence for transcription of D4Z4 sequences in three RNAPol-ChIP experiments on a human-mouse SCH containing Chr4 as the only human chromosome. Our results do not support the hypothesis of increased D4Z4 transcription in FSHD cells. We also measured transcription of two longer tandem arrays of DNA repeats, namely, Sat2 and Satα, present in pericentromeric or centromeric, which are mostly [44] constitutive heterochromatin. . In fission yeast, small non-coding RNAs originating from larger, short-lived centromeric transcripts are critical for centromere function and silencing inserted genes [2]. However, RNAi is not necessary for all heterochromatin formation involving tandem repeats, e.g., transgene arrays in rodent cells [46]. Murine satellite DNA-derived transcripts were detected in certain, but not all, cell populations [4, 47]. Centromeric and pericentromeric RNA transcripts were observed by RT-PCR from randomly or and oligo(dT) primed cDNA derived from murine embryonic stem cells [5, 6]. Satellite III transcription appears to be more easily triggered than Satα or Sat2 transcription [8, -48].
Half of the eight ovarian carcinomas or Wilms tumors that we tested by RNAPol-ChIP and sqRT-PCR displayed evidence for Sat2 transcription by both assays. There was not a strong correlation between methylation of the satellite DNA and its transcription. Similarly, no differences were seen in levels of satellite transcripts in DNA methyltransferase 1 or 3a/3b knockout embryonal stem cells and wild-type cells [6]. We also observed a low level of Sat2 transcription in LCLs by RNAPol-ChIP, and in some of the LCLs by sqRT-PCR. In addition, Satα showed evidence of transcription in a few of the cancers and in LCLs. In samples exhibiting satellite DNA transcription, we do not know where transcription initiates. It is possible that it begins in nearby gene-like sequences that contain within them short regions of satellite DNA [49]. However, dispersed copies of Sat2 and Satα sequences are likely to be a very small proportion of the total and below the limit of detection of our RNAPol-ChIP assays, which compare the level of immunoprecipitated Sat2 or Satα DNA to the total Sat2 or Satα DNA. Moreover, in other studies, Northern blotting and RNA FISH revealed long satellite III transcripts derived from the pericentromeric heterochromatin of Chr9 in HeLa cells, although only in response to heat shock [7, 8].
In an RNAi pathway-proficient SCH containing human chromosome 21, transcription of centromeric human satellite DNA was detectedb by Northern blot analysis of the small RNA fraction (20-to-30 nt) but not by RNase protection assays [3]. When the RNAi-associated Dicer nuclease was knocked down, large and small transcripts from pericentromeric and centromeric satellite DNA and chromosomal abnormalities accumulated. In the present study, from the normal tissues and myoblast cell strains tested, only one myoblast cell strain displayed evidence of transcripts or transcription from satellite DNA (Sat2). In certain cancer and LCL samples, we confirmed that Sat2 is transcribed but at a much lower level than the ACTB and PPIB standards. If transcription of long arrays of tandem DNA repeats is necessary to maintain the heterochromatic structure of centromeric, pericentromeric, and subtelomeric regions in human cells, only a very small percentage of the corresponding satellite DNA sequence might need to be transcribed at any given time, even in rapidly dividing cells. Alternatively, a small number of RNA Pol II molecules might produce very long transcripts that are processed to give many siRNA molecules. The RNAi pathway is implicated in heterochromatinization by the recruitment of H3-K9 and probably H3-K20 [6] trimethylating enzymes indirectly by siRNAs [50]. Maintenance of the condensed state of constitutive heterochromatin with only limited transcription of satellite DNA might be facilitated by the long life of the heterochromatin-associated histone modification marks of H3-K9 and H3-K20 trimethylation [6]. In addition, even in cycling cells, there may be some conservation of previously established heterochromatic histone modification patterns, even in the absence of the corresponding siRNA.
Acknowledgments
We thank Dr. Takeo Kubota for sharing two ICF LCLs with us and Renae Romero for preparation of the figures. Supported in part by NIH grant R01 NS048859 (to M.E.) and Muscular Dystrophy Association grant MDA3709 (to M.W.), and Genpathway.
References
- 1.Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 3.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki M, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 4.Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- 5.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 6.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandoval J, Rodriguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, Lopez-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, Scherer SW, Nakabayashi K, Marsden PA. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279:37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 12.Labhart P, Karmakar S, Salicru EM, Egan BS, Alexiadis V, O’Malley BW, Smith CL. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci U S A. 2005;102:1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iype T, Francis J, Garmey JC, Schisler JC, Nesher R, Weir GC, Becker TC, Newgard CB, Griffen SC, Mirmira RG. Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J Biol Chem. 2005;280:16798–16807. doi: 10.1074/jbc.M414381200. [DOI] [PubMed] [Google Scholar]
- 14.Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 17.Upadhyaya M, Cooper DN. Molecular diagnosis of facioscapulohumeral muscular dystrophy. Expert Rev Mol Diagn. 2002;2:160–171. doi: 10.1586/14737159.2.2.160. [DOI] [PubMed] [Google Scholar]
- 18.Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 19.Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 20.Ehrlich M. In: FSHD Facioscapulohumeral Muscular Dystrophy: Molecular Cell Biology &Clinical Medicine. Cooper DN, Upadhyaya M, editors. BIOS Scientific Pub; New York, NY: 2004. pp. 253–276. [Google Scholar]
- 21.Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, Frants RF, Williamson R. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 22.Wijmenga C, van Deutekom JC, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. Pulsed-field gel electrophoresis of the D4F104S1 locus reveals the size and the parental origin of the facioscapulohumeral muscular dystrophy (FSHD)-associated deletions. Genomics. 1994;19:21–26. doi: 10.1006/geno.1994.1006. [DOI] [PubMed] [Google Scholar]
- 23.van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding H, Beckers MC, Plaisance S, Marynen P, Collen D, Belayew A. Characterization of a double homeodomain protein (DUX1) encoded by a cDNA homologous to 3.3 kb dispersed repeated elements. Hum Mol Genet. 1998;7:1681–1694. doi: 10.1093/hmg/7.11.1681. [DOI] [PubMed] [Google Scholar]
- 25.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc’his D, Viegas-Pequignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- 27.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 28.van Overveld PG, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, Winokur ST, Frants RR, Padberg GW, van der Maarel SM. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol. 2005;58:569–576. doi: 10.1002/ana.20625. [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich M, Hopkins N, Jiang G, Dome JS, Yu MS, Woods CB, Tomlinson GE, Chintagumpala M, Champagne M, Diller L, Parham DM, Sawyer J. Satellite hypomethylation in karyotyped Wilms tumors. Cancer Genet Cytogenet. 2002;141:97–105. doi: 10.1016/s0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- 30.Ehrlich M, Dubeau L, Woods C, Fiala E, Youn B, Long TI, Laird P. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlich M, Buchanan K, Tsien F, Jiang G, Sun B, Uicker W, Weemaes C, Smeets D, Sperling K, Belohradsky B, Tommerup N, Misek D, Rouillard JM, Kuick R, Hanash S. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphocyte migration, activation, and survival genes. Hum Mol Genet. 2001;10:2917–2931. doi: 10.1093/hmg/10.25.2917. [DOI] [PubMed] [Google Scholar]
- 32.Shirohzu H, Kubota T, Kumazawa A, Sado T, Chijiwa T, Inagaki K, Suetake I, Tajima S, Wakui K, Miki Y, Hayashi M, Fukushima Y, Sasaki H. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. Am J Med Genet. 2002;112:31–37. doi: 10.1002/ajmg.10658. [DOI] [PubMed] [Google Scholar]
- 33.Jiang G, Yang F, Van Overveld PG, Vedanarayanan V, Van Der Maarel S, Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 34.Jeanpierre M. Human satellites 2 and 3. Ann Genet. 1994;37:163–171. [PubMed] [Google Scholar]
- 35.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 36.Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung SA, Masny PS, Figlewicz DA. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord. 2003;13:322–333. doi: 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 37.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 38.Gabellini D, D’Antona G, Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino MA, Bottinelli R, Green MR, Tupler R. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 39.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 40.van Deutekom JC, Lemmers RJ, Grewal PK, van Geel M, Romberg S, Dauwerse HG, Wright TJ, Padberg GW, Hofker MH, Hewitt JE, Frants RR. Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum Mol Genet. 1996;5:581–590. doi: 10.1093/hmg/5.5.581. [DOI] [PubMed] [Google Scholar]
- 41.Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas-Pequignot E. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 42.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, Schutte BC. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 43.Lee JH, Goto K, Matsuda C, Arahata K. Characterization of a tandemly repeated 3.3-kb KpnI unit in the facioscapulohumeral muscular dystrophy (FSHD) gene region on chromosome 4q35. Muscle & Nerve. 1995;2:S6–S13. [PubMed] [Google Scholar]
- 44.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostlund C, Garcia-Carrasquillo RM, Belayew A, Worman HJ. Intracellular trafficking and dynamics of double homeodomain proteins. Biochemistry. 2005;44:2378–2384. doi: 10.1021/bi047992w. [DOI] [PubMed] [Google Scholar]
- 46.Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol Cell Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mudge JM, Jackson MS. Evolutionary implications of pericentromeric gene expression in humans. Cytogenet Genome Res. 2005;108:47–57. doi: 10.1159/000080801. [DOI] [PubMed] [Google Scholar]
- 50.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]


