Abstract
Phyto-oestrogens are polyphenolic non-steroidal plant compounds with oestrogen-like biological activity. Phyto-oestrogens have many biological effects including oestrogen agonist/antagonist properties. However, the effect of phyto-oestrogens on allergic responses remains unclear. In this study we investigated whether formononetin, a phyto-oestrogen, and its metabolites, daidzein and equol, affect production of interleukin-4 (IL-4), a pro-inflammatory cytokine closely associated with allergic immune response, in primary CD4+ T cells and EL4 T lymphoma cells. Formononetin, daidzein and equol significantly enhanced IL-4 production from both CD4+ T cells and EL4 cells in a dose-dependent manner. Formononetin, daidzein and equol also enhanced IL-4 gene promoter activity in EL4 cells transiently transfected with IL-4 gene promoter constructs, but this effect was impaired in EL4 cells transfected with an IL-4 promoter construct deleted of P4 site carrying nuclear factor of activated T cells (NF-AT) and activator protein-1 (AP-1) binding sites. In addition, formononetin, daidzein and equol increased AP-1 DNA binding activities while did not affect NF-AT DNA binding activities. The enhancing effects on IL-4 production and AP-1 DNA binding activities were abrogated by specific inhibitors for phosphatidylinositol-3-kinase (PI3K), protein kinase C (PKC) and p38 mitogen-activated protein kinase (MAPK), indicating that formononetin, daidzein and equol might enhance IL-4 production by increased activation of AP-1 through the PI3-K/PKC/p38 MAPK signalling pathway. These results suggest that phyto-oestrogens and some of their metabolites may increase allergic responses via the enhancement of IL-4 production in T cells.
Keywords: phyto-oestrogen, interleukin-4, allergy, AP-1, T cell
Introduction
Phyto-oestrogens are secondary plant metabolites that can mimic the effect of the female sex hormone, oestrogen.1 Experimental findings indicate that phyto-oestrogens may play a significant inhibitory role during the initiation and promotional phases of cancer development.2–4 Soy isoflavonoids, which are phyto-oestrogens, act as antioxidants and also influence cell signalling, cell division and growth, and gene expression.5–7 In contrast, soybean products are known as common inducers of food allergy. Soy is a food that is most likely to induce food-specific immunoglobulin E (IgE) sensitization in infancy and childhood.8,9 The major soybean allergen in children with soy allergy is an IgE-binding epitope on a seed vacuole protein, P34.10,11 These reports indicate that soy proteins are immunogenic and allergenic. However, the mechanism by which soy isoflavonoids cause these allergic responses is unknown. In this study, we determined if allergic responses induced by formononetin, a soy isoflavonoid, and its metabolites, daidzein and equol, were associated with an increase in IL-4 production by T cells.
Interleukin-4 (IL-4), a pleiotropic cytokine produced by activated T cells, basophils and mast cells, regulates many cellular and humoral immune responses.12 Dysregulation of IL-4 expression results in uncontrolled allergic inflammation including asthma and aberrant immune responses to pathogens.13–15 To date, direct binding to and/or activation of the IL-4 promoter has been demonstrated for nuclear factor of activated T cells (NF-AT), c-Maf, and JunB.16–18 Both the mouse and human IL-4 promoters contain five binding elements (designated P0 to P4) for the NF-AT family of transcription factors.19,20 Both P1 and P4 are immediately flanked by sequences with affinity for activator protein-1 (AP-1) family proteins, allowing for cooperative binding with NF-AT proteins.21 Although all P elements to some extent contribute to IL-4 gene control, the major positive regulatory P elements seem to be P1 and P4.22 AP-1 consists of a dimer of Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra1, and Fra2) family members. Jun family members form homo- and heterodimers that recognize a TGAGTCA consensus DNA sequence. Fos family members, which are unable to dimerize with each other, augment transcriptional activation by association with Jun family members.23 AP-1 is inducible by a variety of cytokines and growth factors.24 AP-1 binding sites are found in the promoter region of many proinflammatory genes including T helper 2 (Th2) cytokines, adhesion molecules and cell proliferation growth factors.25,26
In this report we studied the effects of formononetin, daidzein and equol on IL-4 production in activated primary CD4+ T cells and EL4 cells. We also investigated the enhancing mechanisms of formononetin, daidzein and equol on IL-4 production in activated T cells.
Materials and methods
Materials and cell culture
Daidzein and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma (St. Louis, MO). Formononetin and equol were purchased from Fluka (Milwaukee, WI). Anti-mouse IL-4 monoclonal antibody (mAb) 11B11 and BVD6 were from M. Howard, DNAX Research Institute (Palo Alto, CA) and recombinant murine IL-4 was purchased from PharMingen (San Diego, CA). 1-(5-isoquinolines-ulfonyl)-2-methylpiperazine dihydrochloiride (H7), chelerythrine, bisindolylmaleimide (GF109203X) and 2-(2-amino-3-methoxyphenyl)-oxanaphthalen-4-one (PD98059) were purchased from Tocris Cookson Inc. (Ellisville, MO). Wortmannin, 2-[4-morpholinyl]-8-phenyl-1[4H]-benzophyran-4-one (LY294002) and other reagents were purchased from the Sigma. Cultures of EL4 thymoma cells were maintained in RPMI-1640 medium (Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY) and 1% penicillin/streptomycin at 37° in a 5% CO2 humidified air atmosphere.
Isolation of CD4+ T cells
CD4+ T cells were positively purified from lymph node cells of the immunized mice by incubating with anti-mouse CD4 (L3T4) conjugated microbeads (Miltenyi Biotech., Germany) for 20 min at 4°. After incubation, the magnetically labelled cells were applied into the magnetic-activated cell sorting (MACS) positive selection column in a VarioMACS (Miltenyi Biotech.) according to the manufacturer's protocol. After washing with a buffer (1× PBS containing 2 mm ethylenediaminetetraacetic acid (EDTA) and 0·5% bovine serum albumin), the CD4+ T cells were eluted and washed with serum-free RPMI-1640 medium, and stimulated for 4 days with keyhole limpet haemocyanin (KLH) in the absence or presence of FN, DA and EQ. The levels of IL-4 and interferon-γ (IFN-γ) were determined by a sandwich enzyme-linked immunosorbent assay (ELISA) and reverse transcription–polymerase chain reaction (RT–PCR).
Cytokine assay
The levels of IL-4 and IFN-γ in the culture supernatants were determined by a sandwich ELISA using mAbs for mouse IL-4 and IFN-γ as previously described.27 Murine recombinant IL-4 and IFN-γ (PharMingen) were used as standard for quantitation of IL-4 and IFN-γ levels in the supernatants. The mAbs for coating the plates and the biotinylated second mAbs were as follows: for IL-4, 11B11 and BVD6; for IFN-γ, HB170 and XMG1·2. The lower limits of detection were 3 pg/ml for IL-4 and 25 pg/ml for IFN-γ.
RT–PCR
Total RNA was prepared from the cells and reverse transcribed into cDNA. PCR amplification of the cDNA was preformed as previously described.28 Total cellular RNA was isolated by the single-step method using the TRIzol reagent (Sigma). The sequences of PCR primers are as follows: mouse IL-4 (sense, 5′-ATGGGTCTCAACCCCCAGCTAGT-3′; antisense, 5′-GCTCTTTACGCTTTCCAGGAAGTC-3′), mouse IFN-γ (sense, 5′-AGTTATATCTTGGCTTTTCA-3′; antisense, 5′-ACCGAATAATTAGTCAGCTT-3′) and β-actin (sense, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′; antisense, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′). The PCR reactions were run for 35 cycles for 94° (30 s), 58° (45 s), 72° (30 s) using a PCR Thermal Cycler (MJ Research, Watertown, MA). After the amplification, the RT–PCR products were separated in 1·5% (w/v) agarose gels and stained with ethidium bromide. The sizes of PCR products for IL-4 and β-actin genes were 397 bp and 349 bp, respectively.
IL-4 promoter constructs, transient transfection and AP-1 minimal promoter assay
The −741/+56 fragment of murine IL-4 promoter was generated by PCR from genomic DNA of DBA/2 mice. The PCR product was cloned into the BamHI/EcoRI sites of the pGEM-7Z and then subcloned into the SacI/XhoI sites of the pGL3-basic luciferase vector (Promega Co., Madison, WI). All the deletion mutants were generated by PCR using an upstream primer containing BamHI site. For transfections, EL4 cells were cultured in RPMI-1640 medium and transfected with indicated plasmid in the presence of Superfectam according to the manufacturer's protocol (Qiagen, Hilden, Germany). The cells were stimulated with PMA (1 ng/ml) in the absence or presence of phyto-oestrogens. The cells were harvested 24 hr later, and luciferase activity was assayed. Results are shown as relative fold induction compared to the unstimulated EL4 cells. AP-1 minimal promoter and stimulator, c-fos, were obtained from J.W. Lee (Baylor College of Medicine, TX). For transfections, HeLa cells were cultured in DMEM medium and transfected with the indicated plasmid in the presence of Superfectam according to the manufacturer's protocol (Qiagen). The cells were stimulated with PMA (1 ng/ml) in the absence or presence of phyto-oestrogens. The cells were harvested 24 hr later, and luciferase activity was assayed.
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA)
EL4 cells were stimulated in the presence of PMA (1 ng/ml), and incubated for 1 hr with varying amounts of phyto-oestrogens. Nuclear extracts were prepared from the cells as previously described,29 and aliquots were frozen at −80°. Protein concentrations in nuclear extracts were determined by BCA protein assay kit (Pierce Biotech., Rockford, IL). Synthetic oligonucleotides were annealed and end-labelled using [α-32P] dCTP (250 µCi; Amersham) and Klenow enzyme. For binding assays, 10 µg of total nuclear extract was incubated with 32P-labelled oligonucleotide in the presence of a reaction mixture containing 20 mm dithiothreitol, poly(dI-dC), 10× gel retardation assay buffer for 30 min at room temperature. For competition experiments, the extracts were preincubated with a 50-fold excess of labelled specific or non-specific probes. Protein/DNA binding complexes were separated from free probes using a 4% polyacrylamide gel in 0·5× Tris-borate-EDTA buffer at 200–250 V for 45 min. Dried gels were exposed to an X-ray film at −80°. The base sequences of oligonucleotides used in binding and competition assays were as follows: NF-AT, 5′-CGCCAAAGACGCCAAAGAGGAAAATTTGTTTCATA-3′; AP-1, 5′-GATCTGCATGAGTCAGACACACA-3′; NF-κB, 5′-CCGGTTAACAGAGGGGGCTTTCCGAG-3′; CRE, 5′-GATCCGAGCCCGTGACGTTTACACTCATTCT-3′; P4, 5′-GATCTATGGTGTAATTTCCTATGCTGAAA-3′; mut P4, 5′-GATCTATGGTGTAGTCGACTATGCTGAAA-3′.
Preparation of cell lysates and western blot analysis
The stimulated cells were washed twice with ice-cold phosphate-buffered saline solution and harvested with a plastic scraper. The cells were lysed in lysis buffer (100 mm NaCl, 50 mm Tris-Cl, 1 m EDTA, 10% glycerol, 1% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 50 µg/ml phenylmethylsulphonyl fluoride) by incubation on ice. Cell lysates were then centrifuged at 20 000 g at 4°, and the supernatants were transferred to fresh tubes and stored at −80° until required. Protein concentration of the lysates was determined using the BCA protein assay reagent (Pierce Biotec.). Proteins of 10 µg were resolved in 10% sodium dodecyl sulphate (SDS) polyacrylamide gels at 30 mA and 200–250 V for 1 hr in a Mini-Protein II gel apparatus (Bio-Rad, Richmond, CA). After electrophoresis, the proteins were blotted to the prewetting nitrocellulose membrane using a Semi-Phor (Hoefer Scientific Instrument, San Francisco, CA). The membrane was then incubated with washing buffer containing 5% non-fat milk for at least 1 hr to block non-specific protein binding. Primary mAb was diluted up to 1 : 5000 in washing buffer and applied to the membrane for 1 hr at room temperature. Following washing, the blots were incubated with the appropriate biotinylated secondary mAb for 1 hr at room temperature. Immunoreactive bands were visualized by the enhanced chemiluminescence system (Amersham, Amersham, UK).
Immunoprecipitation and PKC assay
Cell lysates were incubated with PKC antibody-coated protein A beads (Pharmacia, Piscataway, NJ) for 2 hr at 4° by gentle rocking. Immunocomplexed beads were washed twice with cell lysis buffer and twice with reaction buffer (0·5 mm ethyleneglycoltetraacetic acid, 10 mm MgCl2, 20 mm HEPES, 50 µm ATP, 2 mm dithiothreitol, 2 mm NaF, and 2 mm Na3VO4). The precipitated proteins were incubated with γ-32P-ATP in the presence of a reaction buffer and 5 µg of myelin basic protein. The precipitated proteins were incubated with γ-32P-ATP in the presence of a reaction buffer and 5 µg of myelin basic protein for 30 min at room temperature and resolved in 12% SDS polyacrylamide gels at 30 mA and 200 V for 40 min in a Mini-Protein II gel apparatus (Bio-Rad, Richmond, CA). After electrophoresis, dried gels were exposed to an X-ray film.
Statistical analyses
Student's t-test and anova were used to determine the statistical differences between the values of various experimental and control groups. P-values < 0·05 were considered significant.
Results
Enhancement of IL-4 production in activated T cells by formononetin and its metabolites
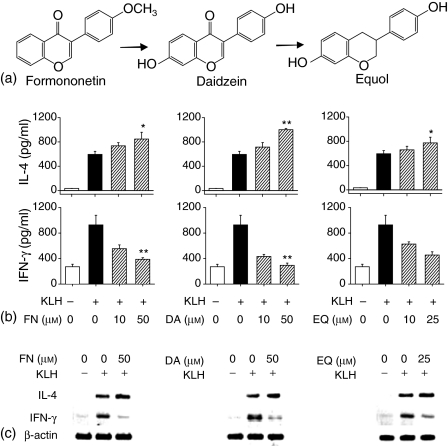
To determine whether formononetin, daidzein and equol (hereafter abbreviated as FN, DA and EQ, respectively) affected production of IL-4 by CD4+ T cells primed with KLH, BALB/c mice received first injection into the footpad with KLH (100 µg/mouse) in alum. Seven days later, lymph node cells from the immunized mice were purified and stimulated for 4 days in vitro with KLH in the absence or presence of FN, DA and EQ, and the levels of IL-4 and IFN-γ in the supernatants were determined. As shown in Fig. 1(b), FN, DA and EQ significantly increased IL-4 production in a concentration-dependent manner. In contrast, FN, DA and EQ inhibited the production of IFN-γ, a Th1 cytokine, in KLH-stimulated CD4+ T cells. The enhanced IL-4 production by FN, DA and EQ did not result from increased cell proliferation because the cell proliferation remained constant throughout the incubation period in the presence of FN, DA and EQ, as demonstrated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium (MTT) assay (data not shown). IL-4 mRNA levels were analysed in KLH-primed CD4+ T cells in the presence of FN, DA and EQ, to determine if changes in IL-4 production were accompanied by changes in the expression of IL-4 mRNA. As shown in Fig. 1(c), FN, DA and EQ significantly enhanced IL-4 mRNA levels in KLH-primed CD4+ T cells, indicating that the changes in IL-4 production were present at the transcriptional level.
Figure 1.
FN, DA and EQ enhance IL-4 production and mRNA expression in KLH-stimulated CD4+ T cells. (a) Chemical structures of formononetin and its metabolites. (b) Isolated CD4+ T cells, as described in the Materials and methods, were re-stimulated with KLH (100 µg/ml) in the presence of FN (10, 50 µm), DA (10, 50 µm) and EQ (10, 25 µm) for 4 days. The cell culture supernatants were harvested and assayed for IL-4 and IFN-γ levels by ELISA. (c) Isolated CD4+ T cells were re-stimulated with KLH (100 µg/ml) in the absence or presence of FN (50 µm), DA (50 µm) and EQ (25 µm) for 6 hr. Cellular RNA from each treatment was extracted and the mRNA expression for IL-4, IFN-γ and β-actin was analysed by RT–PCR. The values represent the means ± SE of three independent experiments. *P < 0·05 and **P < 0·005, relative to the corresponding control treated with KLH alone.
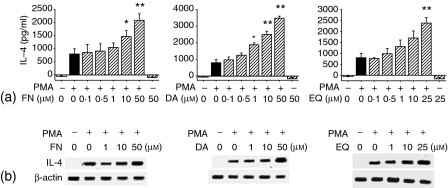
To determine whether FN and its metabolites also enhance IL-4 production in a T-cell line, EL4 T lymphoma cells were incubated with PMA in the absence or presence of FN, DA or EQ. As shown in Fig. 2, FN, DA and EQ also increased the levels of IL-4 production and IL-4 mRNA in PMA-activated EL4 cells. FN, DA or EQ itself did not induce IL-4 production by unstimulated EL4 cells. Treatment with FN, DA and EQ did not affect the expression of β-actin mRNA by both KLH-primed CD4+ T cells and PMA-activated EL4 cells, suggesting that the enhancing effect of IL-4 by FN, DA and EQ was not the result of generalized activation of these cells. In addition, the cells' viability remained constant throughout the incubation period in the presence of FN and its metabolites concentrations, as demonstrated by trypan blue exclusion test (data not shown).
Figure 2.
Enhancement of IL-4 production in activated EL4 cells by formononetin and its metabolites. (a) EL4 cells were stimulated with PMA (1 ng/ml) in the presence of varying amounts of FN, DA and EQ for 2 days. The cell culture supernatants were harvested and assayed for IL-4 levels by ELISA. (b) EL4 cells were stimulated with PMA (1 ng/ml) in the absence or presence of FN, DA and EQ for 6 hr. Cellular RNA from each treatment was extracted and the mRNA expression for IL-4 and β-actin was analysed by RT–PCR. The values represent the means ± SE of three independent experiments. *P < 0·05 and **P < 0·01, relative to the corresponding control treated with PMA alone.
Effects of FN, DA and EQ on PMA-induced IL-4 promoter activity by targeting a transcription factor AP-1
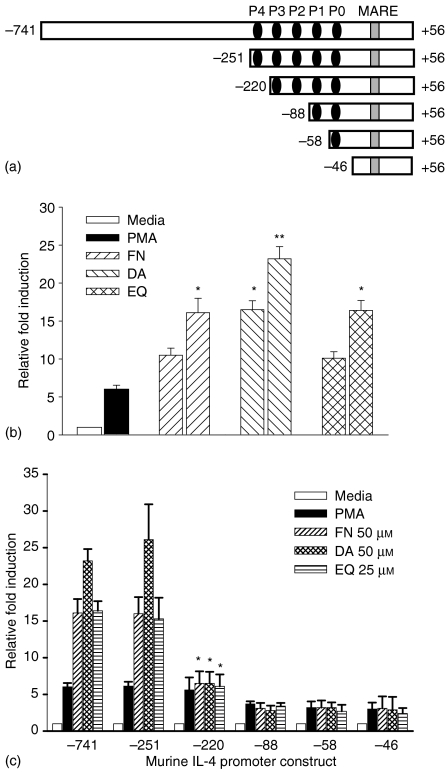
In T cells, IL-4 expression is tightly controlled at the level of transcription by multiple regulatory elements located within a proximal promoter region. The IL-4 promoter contains multiple binding sites for members of the NF-AT family of transcription factors, termed the P elements P0–P4. To identify the region involved in actions of FN, DA and EQ, a series of luciferase reporter constructs were generated containing the IL-4 promoter sequences from positions −741, −251, −220, −88, −58, and −46 to +71 (represented as −741/+71, −251/+71, −220/+71, −88/+71, −58/+71, and −46/+71) relative to the transcription initiation site (Fig. 3a). EL4 cells were transfected with each of these constructs and stimulated with PMA in the absence or presence of FN, DA and EQ, and the luciferase activity was determined. As shown in Fig. 3(b), full promoter constructs (−741/+71) showed strong stimulation with PMA in the absence of FN, DA and EQ, and enhanced stimulation in the presence of FN, DA and EQ. In particular, deleting sequences to −251 (−251/+71) did not diminish the PMA-dependent promoter activities and the enhancing effects of FN, DA and EQ was still observed. However, deleting sequence to −220 (−220/+71) eliminated the PMA-induced promoter activities enhanced by FN, DA and EQ, indicating that the target site for FN, DA and EQ resided within this region (Fig. 3c). The P4 site includes NF-AT and AP-1 binding sites.
Figure 3.
FN, DA and EQ enhance IL-4 gene promoter activity induced by PMA. (a) The schematic diagram of the proximal 750 base pairs of murine IL-4 gene promoter. The P and MARE represent NF-AT binding sites and c-Maf binding sites, respectively. P1 and P4 sites include AP-1 binding site. (b) EL4 T cells were transiently transfected with the IL-4 full promoter construct, followed by stimulation with PMA (1 ng/ml) in the absence or presence of FN (10, 50 µm), DA (10, 50 µm) and EQ (10, 25 µm). The results are represented as the induction fold over the value obtained with unstimulated EL4 cells transfected with the promoter construct with given an arbitrary value of 1. (c) EL4 cells were transiently transfected with the serially deleted IL-4 promoter constructs, followed by stimulation with PMA (1 ng/ml) in the absence or presence of FN (50 µm), DA (50 µm) and EQ (25 µm). The values represent the means ± SD of triplicate determinations. The data are representative of three independent experiments. *P < 0·01 and **P < 0·001, compared with each of IL-4 promoter construct treated with PMA alone.
These results suggest that the enhancing effect of FN, DA and EQ on IL-4 production might be mediated through NF-AT and/or AP-1 binding sites. Transcription factor NF-AT has been widely established to be the most critical transcription factor for the regulation of IL-4. Nuclear factor AP-1, allowing for co-operative binding with NF-AT proteins, is a complex transcription factor consisting of members of the c-Jun and c-Fos family of proteins.
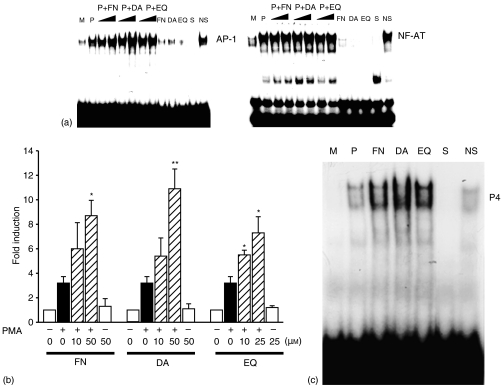
To further determine the involvement of AP-1 and/or NF-AT in IL-4 gene regulation by FN, DA and EQ, EL4 cells were stimulated with PMA in the absence or presence of FN, DA and EQ, and nuclear extracts from each treated cells were isolated and analysed in EMSA using each consensus sequence corresponding to AP-1 and NF-AT sites of P4 region. As shown in Fig. 4, unstimulated EL4 cells showed no detectable binding activity for NF-AT and AP-1. EL4 cells treated with PMA effectively induced NF-AT and AP-1 binding activity. Treatment with FN, DA and EQ enhanced the PMA-induced AP-1 DNA binding activities in EL4 cells (Fig. 4). In contrast, FN, DA and EQ did not affect NF-AT DNA binding activities. The involvement of AP-1 in the enhanced IL-4 production was further confirmed in EL4 cells transfected with AP-1 minimal promoter construct, followed by treatment with PMA in the absence or presence of FN, DA and EQ. As shown in Fig. 4(b), FN, DA and EQ enhanced the PMA-induced AP-1 minimal promoter activities. These results indicate that the enhancing effects of FN, DA and EQ on IL-4 production were mediated through the increased DNA binding activities of AP-1 transcription factor.
Figure 4.
FN, DA and EQ enhance AP-1 activation induced with PMA. (a) FN, DA and EQ enhance AP-1 DNA binding activity. Nuclear extracts from EL4 cells stimulated by PMA in the presence of FN, DA and EQ were examined for AP-1 and NF-AT DNA binding activity in the EMSA, using radiolabelled oligonucleotides containing a AP-1 site or NF-AT site, respectively. S and NS indicate the presence of an unlabelled, specific oligonucleotide (NF-AT and AP-1) and non-specific oligonucleotide (CRE or NF-κB), respectively. (b) Transfection of EL4 cells with AP-1 minimal promoter construct, followed by stimulation with PMA (1 ng/ml) in the absence or presence of FN, DA and EQ. The results are represented as the induction fold over the value obtained with unstimulated EL4 cells transfected with the promoter construct with given an arbitrary value of 1. The data are representative of three independent experiments. (c) Nuclear extracts from EL4 cells stimulated by PMA in the presence of FN (50 µm), DA (50 µm) and EQ (25 µm) were examined for P4 region DNA binding activity in the EMSA, using P4 radiolabelled oligonucleotides containing an AP-1 site. S and NS indicate the presence of an unlabelled, specific oligonucleotide (P4) and non-specific oligonucleotide (mutP4), respectively. *P < 0·005 and **P < 0·0005, compared with the EL4 cells treated with PMA alone.
To confirm these results, EL4 cells were stimulated with PMA in the absence or presence of FN, DA and EQ, and nuclear extracts from each treated cells were isolated and analysed in EMSA using a P4 region probe carrying an AP-1 site. As shown in Fig. 4(c), unstimulated EL4 cells showed no detectable binding activity for P4 region. Treatment of EL4 cells with PMA effectively induced P4 region DNA binding activity. Treatment with FN, DA and EQ enhanced the PMA-induced P4 region DNA binding activities in EL4 cells.
Involvement of PI3-K, PKC and p38 mitogen-activated protein kinase (MAPK) in IL-4 production enhanced by FN, DA and EQ in EL4 T cells
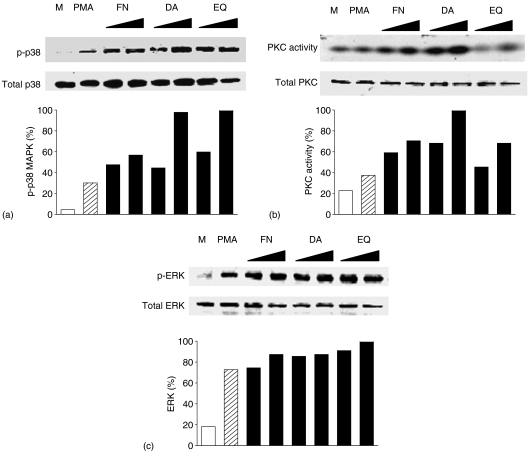
To investigate molecular requirements for the enhancement of IL-4 production by FN, DA and EQ, we performed Western blot analysis to determine protein levels of PKC, p38 MAPK and extracellular signal-related kinase (ERK). As shown in Fig. 5, FN, DA and EQ did not affect the protein levels of total PKC and total p38 MAPK in PMA-activated EL4 T cells. To assess the involvement of activated form of p38 MAPK in the enhancement of IL-4 production the lysates of each treated cells by FN, DA and EQ were immunoprecipitated using antiphospholylated tyrosine mAb and the levels of phosphorylated p38 MAPK (p-p38) were determined by anti-p38 MAPK mAb. In addition PKC activity was determined by immunoprecipitation using anti-PKC polyclonal antibody and PKC kinase assay using γ-32P-ATP. As shown in Fig. 5(a, b), PKC kinase activities and protein levels of phosphorylated form of p38 MAPK were enhanced by FN, DA and EQ. In contrast, FN, DA and EQ did not affect the ERK protein level (Fig. 5c).
Figure 5.
Enhanced p38 MAPK phosphorylation and PKC activity in PMA-stimulated EL4 T cells in the presence of FN, DA and EQ.EL4 cells were incubated with medium alone (M) or PMA in the absence or presence of FN (10, 50 µm), DA (10, 50 µm) and EQ (10, 25 µm) for 40 min. Proteins were extracted by cell lysis buffer. (a) The levels of p-p38 MAPK were assessed by immunoprecipitation using antiphosphorylated tyrosine mAb and blotted with anti-p38 MAPK mAb. (b) The protein levels of total PKC were assessed by Western blot analysis using anti-total PKC mAb. PKC activities were assessed by PKC assay using γ-32P-ATP, as described in Materials and methods. (c) The levels of ERK and p-ERK were assessed by Western blot analysis using anti-ERK mAb or anti-p-ERK mAb. Quantitative analysis of phospho-p38 MAPK expression level and PKC activity was performed by Tina 2.0 program, normalizing on the basis of total p38 MAPK level and total PKC level, respectively. The data are representative of three independent experiments.
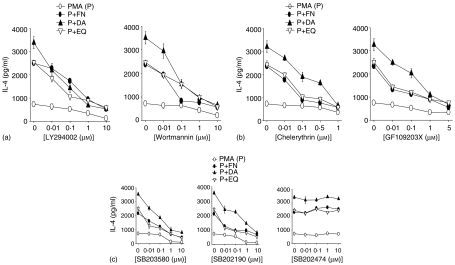
To further determine whether PKC and p38 MAPK were required for the enhanced IL-4 production, EL4 T cells were treated with specific PKC inhibitors, GF109203X or chelerythrine, or specific p38 MAPK inhibitors, SB203580 and SB202190, or SB202474, a chemical used as a negative control for p38 MAP kinase inhibition studies. SB202474 is structurally related to two well-known pyridinyl inhibitors of p38 MAPK, SB203580 and SB202190, but does not inhibit p38 MAPK.30 As shown in Fig. 6(a, c), all of PKC inhibitors and p38 MAPK inhibitors significantly suppressed IL-4 production enhanced by FN, DA and EQ.
Figure 6.
Involvement of PI3-K, PKC and p38 MAPK in the enhanced IL-4 production by FN, DA and EQ.EL4 cells were treated with varying concentrations of PKC inhibitors (a), PI3-K inhibitors (b), or p38 MAPK inhibitors (SB203580, SB202190) and a negative control of p38 MAPK inhibitors (SB202474)(c)for 40 min, followed by incubation with PMA in the presence of FN (50 µm), DA (50 µm) and EQ (25 µm). IL-4 production was assessed by ELISA. The data represent the mean ± SD of triplicate determination. The data are representative of four independent experiments.
Recent reports have indicated that PI3-K might activate PKC isoforms both in vitro and in vivo.31,32 To determine any roles of PI3-K in the PMA-induced IL-4 production enhanced by FN, DA and EQ, EL4 cells were treated with specific PI3-K inhibitors, wortmannin or LY294002, followed by stimulation with PMA in the absence or presence of FN, DA and EQ. As shown in Fig. 6(b), both PI3-K inhibitors significantly suppressed IL-4 production enhanced by FN, DA and EQ. These results indicate that the enhancing effects of FN, DA and EQ on PMA-induced IL-4 production might be mediated by PI3-K, PKC and p38 MAPK. Suppression of IL-4 production by the kinase inhibitors did not result from a general cytotoxic effect because the cells' viability in all cultures remained constant throughout the incubation period in the presence of inhibitors, as demonstrated by trypan blue exclusion test (data not shown).
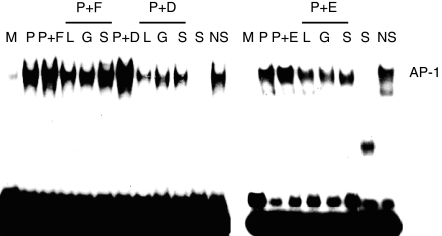
Involvement of PI3-K, PKC and p38 MAPK on AP-1 DNA binding activities enhanced by FN, DA and EQ
The activation of AP-1 requires various signals containing MAPK proteins and protein kinase C (PKC). To investigate the effects of PI3-K, PKC and p38 MAPK inhibitors on AP-1 DNA binding activity enhanced by FN, DA and EQ, EL4 T cells were treated with PI3-K inhibitor (LY294002), PKC inhibitor (GF109203X) or p38 MAPK inhibitor (SB203580), followed by stimulation with PMA in the absence or presence of FN, DA and EQ. As shown in Fig. 7, PI3-K, PKC and p38 MAPK inhibitors significantly decreased the AP-1 DNA binding activities enhanced by FN, DA and EQ. These results suggest that FN, DA and EQ might enhance IL-4 production by increased activation of AP-1 through PI3-K/PKC/p38 MAPK signalling pathway.
Figure 7.
Involvement of PI3-K, PKC and p38 MAPK in the enhanced AP-1 binding activities by FN, DA and EQ.EL4 cells were treated with 10 µm LY294002 (L), 1 µm GF109203X (G) and 10 µm SB203580 (S) for 40 min, followed by stimulation with PMA (1 ng/ml) in the absence or presence of FN (50 µm), DA (50 µm) and EQ (25 µm). DNA binding activities were assessed by EMSA. In EMSA α-32P-end labelled DNA probe that included the AP-1 binding domain of murine IL-4 promoter was incubated with nuclear extracts (10 µg/lane). Specificity of binding was determined by the use of a 50-fold excess of specific or unrelated oligonucleotide (nuclear factor-κB). The data are representative of three independent experiments.
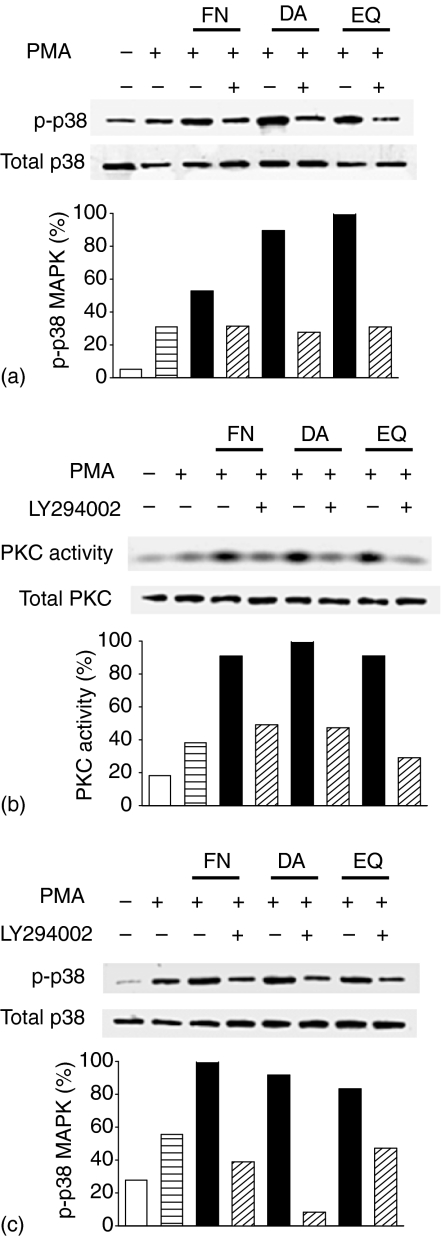
To further determine signalling pathways of IL-4 production enhanced by FN, DA and EQ, EL4 cells were treated with PI3-K and PKC inhibitors, followed by treatment with PMA in the absence or presence of FN, DA and EQ. Afterwards, p-p38 MAPK protein levels and PKC activities were analysed by Western blot or PKC kinase assay using γ-32P-ATP, respectively. As shown in Fig. 8, PKC inhibitor (GF109203X) suppressed the protein levels of p-p38 MAPK enhanced by FN, DA and EQ, suggesting the involvement of PKC as an upstream component of p38 MAPK activation by FN, DA and EQ. The enhanced PKC activities by FN, DA and EQ were inhibited by a PI3-K inhibitor (LY294002), also suggesting the involvement of PI3-K as an upstream component of PKC activation by FN, DA and EQ (Fig. 8b). PI3-K inhibitor also decreased the protein levels of phosphorylated form of p38 MAPK in PMA-activated EL4 cells (Fig. 8c). These data indicate that FN, DA and EQ enhanced AP-1 DNA binding activity through PI3-K/PKC/p38 MAPK signalling pathway.
Figure 8.
Effects of specific inhibitors for PI3-K and PKC on p38 MAPK phosphorylation and PKC activity enhanced by FN, DA and EQ. (a) Effect of PKC inhibitor on protein levels of p38 MAPK and p-p38 MAPK. EL4 T cells were treated with 1 µm GF109203X (GF) for 40 min, followed by incubation with PMA in the absence or presence of FN (50 µm), DA (50 µm) and EQ (25 µm). p-p38 MAPK were assessed by immunoprecipitation using antiphosphorylated tyrosine mAb and blotted with anti-p38 MAPK mAb. (b) Effect of PI3-K inhibitor on protein level of total PKC and PKC activity. EL4 T cells were treated with 10 µm LY294002 for 40 min, followed by incubation with PMA in the absence or presence of FN, DA and EQ. PKC activities were analysed by PKC kinase assay using γ-32P-ATP. (c) Effect of PI3-K inhibitor on protein levels of p38 MAPK and p-p38 MAPK. EL4 T cells were treated with 10 µm LY294002 for 40 min p38 MAPK were assessed by immunoprecipitation using antiphosphorylated tyrosine mAb and blotted with anti-p38 MAPK mAb. The data are representative of three independent experiments.
Discussion
In this report, we have demonstrated that FN, DA and EQ, soy phyto-oestrogens, significantly increased IL-4 production in CD4+ T cells and EL4 T cells. A series of the transient transfection assay and EMSA experiments demonstrated that FN, DA and EQ enhanced IL-4 gene expression through up-regulation of AP-1 binding activities. These results suggest that FN, DA and EQ may, at least in part, increase allergic responses via the enhancement of IL-4 production by T cells.
IL-4, mainly produced in CD4+ Th2 cells, is an important stimulus for the switching of antibody isotype to IgE in both mice and humans.33,34 Higher than normal serum IgE is often found in patients with allergic diseases, including allergic asthma. Reduction of IgE is considered as one strategy in the treatment of asthma.35 Han et al. (2002) reported that oestrogenic compounds increased the levels of immunoglobulin, such as IgM and IgE.36 In this study, the increased levels of IL-4 production in FN-, DA-, or EQ-treated EL4 cells might result in an enhancement of the IgE level in sera, leading to the enhancement of the allergic response.
The mean daily intake of soy products in Asian populations ranges between 10 and 50 g versus 1–3 g in European countries and the United States.37 Soybeans contain high concentrations of isoflavones with concentrations ranging from 0·5 to 2 mg/g.38 Industrial foods (soy milk, bean-cured, bean sprouts) contain similar form of isoflavones with soybean. Furthermore, plasma levels of phyto-oestrogens in populations of European countries and the United States are 2000 ng/ml to 8100 ng/ml, indicating that the concentrations of FN (50 µm, 13·4 ng/ml), DA (50 µm, 12·7 ng/ml) and EQ (25 µm, 6·0 ng/ml) used in the experiments are in the range of physiological doses in human. Ingested phyto-oestrogens are hydrolysed by intestinal bacteria. FN is metabolized to DA and then metabolized to EQ. Products of isoflavone metabolism may be excreted or absorbed. If absorbed, the phyto-oestrogens undergo conjugation in the liver with glucuronic acid, or to a lesser extent, sulphate, and are excreted in the urine or in the bile. Some intestinal bacteria produce glucuronidases, enzymes that can deconjugate phyto-oestrogen metabolites when they pass through the intestine, setting the stage for their recirculation through the body.39 Industrial foods and fermentation foods, especially, contain more DA than the others in high levels.
The mechanism by which FN, DA and EQ enhance IL-4 production at the molecular level is uncertain. We found that FN, DA and EQ significantly enhanced IL-4 mRNA levels in a dose-dependent manner (Fig. 2). They also enhanced the activation of the IL-4 gene promoter (Fig. 3), indicating that the enhancement of IL-4 production by FN, DA and EQ occurred at the transcriptional level. In our study, the enhancing effects of FN, DA and EQ on the PMA-activated IL-4 gene promoter disappeared in EL4 T cells transfect with a deletion construct of P4 site containing NF-AT and AP-1 binding sites (Fig. 3), suggesting that the enhancing effects of FN, DA and EQ on IL-4 production might be mediated through the increased binding activities of NF-AT or AP-1. The transcription factor NF-AT is well-known to play an essential role in the inducible transcription of IL-4 gene during T-cell activation since human and murine IL-4 gene promoters contain at least four NF-AT sites that control their induction in T cells.40,41
Interestingly, in this study FN, DA and EQ were found to significantly inhibit IFN-γ production and mRNA expression levels in CD4+ T cells (Fig. 1). IFN-γ, a cytokine preferentially secreted by activated T cells and natural killer cells, is an important regulatory molecule of the immune system.42 Recent reports have showed that IFN-γ has been implicated in the pathogenesis of several immunological diseases, especially Th1-mediated diseases, including autoimmune diseases and inflammatory responses.43 The mechanism why FN, DA and EQ differentially affected production of IL-4 and IFN-γ, respectively, is under investigation.
AP-1 is necessary for the full high-levels of IL-4 production in atopic Th2 cells and IL-4 gene promoter contain at least two AP-1 sites, P1 and P4 (Fig. 3). In our study, FN, DA and EQ enhanced AP-1 DNA binding activities but not NF-AT DNA binding activities. These data suggest that the activities of AP-1 enhanced by FN, DA and EQ co-ordinately regulate the transcription of IL-4 genes. AP-1, the proinflammatory transcriptional element, is also an important contributor to the expression of Th2 cytokines, IL-5 and IL-13 as well as IL-4. p38 MAPK has been reported to be involved in many signalling pathways of biological functions.44,45 In this study, FN, DA and EQ enhanced the protein levels of phosphorylated p38 MAPK and the PKC activity. Specific inhibitors for PI3-K, PKC and p38 MAPK significantly decreased the AP-1 DNA binding activity and IL-4 production enhanced by FN, DA and EQ, clearly indicating that they may enhance IL-4 production by the increased AP-1 DNA binding activity through PI3-K/PKC/p38 MAPK signalling pathway.
In conclusion, the present study describes that FN, DA and EQ significantly enhanced IL-4 production in activated EL4 T cells. This effect may explain the proallergic effects of FN, DA and EQ. Because the ratio of Th1 and Th2 cells is closely correlated with the outcome of many diseases,46,47 controlling exposure to soy isoflavonoids such as FN, DA and EQ may protect patients from developing diseases caused by unwanted Th2-dominated responses.
BIND identifiers
One BIND identifier (http://www.bind.ca) is associated with this manuscript: 295563.
Acknowledgments
We thank Drs M. Howard, J. W. Lee and Y. K. Choe for their generous gift of valuable reagents, and Drs E. P. Cohen and H. J. Han for helpful discussions and critical reading of the manuscript. This study was supported by Korea Research Foundation Grant (KRF-2003-005 E00012).
Abbreviations
- AP-1
activating protein-1
- DA
daidzein
- ELISA
enzyme-linked immunosorbent assay
- EMSA
electrophoretic mobility shift assay
- EQ
equol
- ERK
extracellular signal-regulated kinase
- FN
formononetin
- IL
interleukin
- MAPK
mitogen-activated protein kinase
- NF-AT
nuclear factor of activated T cell
- PI3-K
phosphoinositol-3-kinase
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
References
- 1.Sabine EK, Leane L, Manfred M. Oxidative metabolism and genotoxic potential of major isoflavone phytoestrogens. J Chromatogr B. 2002;777:211–8. doi: 10.1016/s1570-0232(02)00215-5. 10.1016/S1570-0232(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 2.Fustier P, Le Corre L, Chalabi N, Vissac-Sabatier C, Communal Y, Bignon YJ, Bernard-Gallon DJ. Resveratrol increases BRCA1 and BRCA2 mRNA expression in breast tumour cell lines. Br J Cancer. 2003;89:168–72. doi: 10.1038/sj.bjc.6600983. 10.1038/sj.bjc.6600983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H, Yun CW, Park WK, Kong JY, Kim KS, Park Y, Lee S, Kim BK. Modulation of the activity of pro-inflammatory enzymes, COX-2 and iNOS, by chrysin derivatives. Pharmacol Res. 2004;49:37–43. doi: 10.1016/s1043-6618(03)00248-2. [DOI] [PubMed] [Google Scholar]
- 4.Mishra SI, Dickerson V, Najm W. Phytoestrogens and breast cancer prevention: what is the evidence? Am J Obstet Gynecol. 2003;188:S66–70. doi: 10.1067/mob.2003.405. 10.1067/mob.2003.405. [DOI] [PubMed] [Google Scholar]
- 5.Ungar Y, Osundahunsi OF, Shimoni E. Thermal stability of genistein and daidzein and its effect on their antioxidant activity. J Agric Food Chem. 2003;51:4394–9. doi: 10.1021/jf034021z. 10.1021/jf034021z. [DOI] [PubMed] [Google Scholar]
- 6.Gong L, Li Y, Nedeljkovic-Kurepa A, Sarkar FH. Inactivation of NF-κB by genistein is mediated via Akt signaling pathway in breast cancer cells. Oncogene. 2003;22:4702–9. doi: 10.1038/sj.onc.1206583. 10.1038/sj.onc.1206583. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell JH, Duthie SJ, Collins AR. Effects of phytoestrogens on growth and DNA integrity in human prostate tumor cell lines: PC-3 and LNCaP. Nutr Cancer. 2000;38:223–8. doi: 10.1207/S15327914NC382_12. 10.1207/S15327914NC382_12. [DOI] [PubMed] [Google Scholar]
- 8.Zeiger RS. Food allergen avoidance in the prevention of food allergy in infants and children. Pediatrics. 2003;111:1662–71. [PubMed] [Google Scholar]
- 9.Lin XP, Magnusson J, Ahlstedt S, Dahlman-Hoglund A, Hanson LLA, Magnusson O, Bengtsson U, Telemo E. Local allergic reaction in food-hypersensitive adults despite a lack of systemic food-specific IgE. J Allergy Clin Immunol. 2002;109:879–87. doi: 10.1067/mai.2002.123238. 10.1067/mai.2002.123238. [DOI] [PubMed] [Google Scholar]
- 10.Helm R, Cockrell G, Herman E, Burks A, Sampson H, Bannon G. Cellular and molecular characterization of a major soybean allergen. Int Arch Allergy Immunol. 1998;117:29–37. doi: 10.1159/000023987. 10.1159/000023987. [DOI] [PubMed] [Google Scholar]
- 11.Herman EM, Helm RM, Jung R, Kinney AJ. Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 2003;132:36–43. doi: 10.1104/pp.103.021865. 10.1104/pp.103.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown MA, Hural J. Functions of IL-4 and control of its expression. Crit Rev Immunol. 1997;17:1–32. doi: 10.1615/critrevimmunol.v17.i1.10. [DOI] [PubMed] [Google Scholar]
- 13.Hahn C, Teufel M, Herz U, et al. Inhibition of the IL-4/IL-13 receptor system prevents allergic sensitization without affecting established allergy in a mouse model for allergic asthma. J Allergy Clin Immunol. 2003;111:1361–9. doi: 10.1067/mai.2003.1527. 10.1067/mai.2003.1527. [DOI] [PubMed] [Google Scholar]
- 14.Kay AB. (2003) Immunomodulation in asthma: mechanisms and possible pitfalls. Curr Opin Pharmacol. 2003;3:220–6. doi: 10.1016/s1471-4892(03)00038-9. 10.1016/S1471-4892(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 15.O'Garra A. Cytokines induce the development of functionally heterogenous T helper cell subsets. Immunity. 1998;8:275–83. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Burke T, Cumberland J, Brummet M, Beck L, Casolaro V, Georas S. Glucocorticoids inhibit calcium- and calcineurin-dependent activation of the human IL-4 promoter. J Immunol. 2000;164:825–32. doi: 10.4049/jimmunol.164.2.825. [DOI] [PubMed] [Google Scholar]
- 17.Ho IC, Hodge MR, Rooney JW, Glimsher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–83. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Tournier C, Davids RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–32. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuvpilo S, Serfling E. The inducible transcription factor interleukin-4 promoter. Immunobiology. 1995;193:268–72. doi: 10.1016/s0171-2985(11)80554-1. [DOI] [PubMed] [Google Scholar]
- 20.Li-Weber MP, Salgame C, Hu IV, Davydov O, Krammer PH. Differential interaction of nuclear factors with the PRE-1enhancer element of the human IL-4 promoter in different T cell subsets. J Immunol. 1997;158:1194–200. [PubMed] [Google Scholar]
- 21.Rooney JW, Hoey T, Glimcher LH. Coordinate and cooperative roles for NF-AT and AP-1 in the regulation of the murine IL-4 gene. Immunity. 1995;2:473–83. doi: 10.1016/1074-7613(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 22.Li-Weber MP, Salgame C, Hu IV, Davydov O, Krammer PH. Th2-specific protein/DNA interaction at the proximal nuclear factor-AT site contribute to the functional activity of the human IL-4 promoter. J Immunol. 1998;161:1380–9. [PubMed] [Google Scholar]
- 23.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–57. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 24.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interact with TPA-inducible enhancer elements. Cell. 1987;49:741–52. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 25.Dalton TP, Shertzer HG, Puga A. Regulation of gene expression by reactive oxygen. Annu Rev Pharmacol Toxicol. 1999;39:67–101. doi: 10.1146/annurev.pharmtox.39.1.67. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura H, Nakamura K, Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;135:351–69. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 27.Kim TS, Dekruyff RH, Rupper R, Maecker HT, Levy S, Umetsu DT. An ovalbumin IL-12 fusion protein is more effective than ovalbumin plus recombinant IL-12 in inducing a T helper cell type 1-dominanted immune response and inhibiting antigen-specific IgE production. J Immunol. 1997;158:4137–44. [PubMed] [Google Scholar]
- 28.Lee MH, Chung SW, Kang BY, Park J, Lee CH, Hwang SY, Kim TS. Enhanced interleukin-4 production in CD4+ T cells and elevated immunoglobulin E levels in antigen-primed mice by bisphenol A and nonylphenol, endocrine disruptors: involvement of nuclear factor-AT and Ca2+ Immunology. 2003;109:76–86. doi: 10.1046/j.1365-2567.2003.01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucl Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahti A, Kankaanranta H, Moilanen E. p38 mitogen-activated protein kinase inhibitor SB203580 has a bi-directional effect on iNOS expression and NO production. Eur J Pharmacol. 2002;454:115–23. doi: 10.1016/s0014-2999(02)02490-1. [DOI] [PubMed] [Google Scholar]
- 31.Altman A, Villalba M. Protein kinase C-theta (PKC theta). A key enzyme in T cell life and death. J Biochem. 2002;132:841–6. doi: 10.1093/oxfordjournals.jbchem.a003295. [DOI] [PubMed] [Google Scholar]
- 32.Romanelli A, Martin KA, Toker A, Blenis J. p70, S6 kinase is regulated by protein kinase Czeta and participates in a phosphoinositide 3-kinase-regulated signalling complex. Mol Cell Biol. 1999;19::2921–8. doi: 10.1128/mcb.19.4.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda A, Chandswang N, Ovary Z. The action of interleukin-4 on antigen-specific IgG1 and IgE production by interaction of in vivo primed B cells and carrier-specific cloned Th2 cells. Cell Immunol. 1990;128:31–40. doi: 10.1016/0008-8749(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 34.Shimoda K, van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 35.Hamelmann E, Rolinck-Werninghaus C, Wahn U. Is there a role for anti-IgE in combination with specific allergen immunotherapy? Curr Opin Allergy Clin Immunol. 2003;3:501–10. doi: 10.1097/00130832-200312000-00013. [DOI] [PubMed] [Google Scholar]
- 36.Han DH, Denison MS, Tachibana H, Yamada K. Effects of estrogenic compounds on immunoglobulin production by mouse splenocytes. Biol Pharm Bull. 2002;25:1263–7. doi: 10.1248/bpb.25.1263. [DOI] [PubMed] [Google Scholar]
- 37.Messina MJ, Persky V, Setchell KD. Soy intake and cancer risk; a review of the in vitro and in vivo data. Nutr Cancer. 1994;21:113–31. doi: 10.1080/01635589409514310. [DOI] [PubMed] [Google Scholar]
- 38.Barnes S. Phytoestrogens and breast cancer. Baillières Clin Endocrinol Metab. 1998;12:559–79. doi: 10.1016/s0950-351x(98)80004-9. [DOI] [PubMed] [Google Scholar]
- 39.Barrett J. Phytoestrogens. Friends or foes? Environ Health Perspect. 1996;104:478–82. doi: 10.1289/ehp.96104478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takemoto N, Koyano-Nakagawa N, Arai N, Arai K, Yokota T. Four P-like elements are required for optimal transcription of the mouse IL-4 gene: involvement of a distinct set of nuclear factor of activated T cells and activator protein-1 family proteins. Int Immunol. 1997;9:1329–38. doi: 10.1093/intimm/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 41.Szabo SJ, Gold JS, Murphy TL, Murphy KM. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells. Roles for NF-Y and NF-ATc. Mol Cell Biol. 1993;13:4793–805. doi: 10.1128/mcb.13.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 43.Balashov KE, Olek MJ, Smith DR, Khoury SJ, Weiner HL. Seasonal variation of interferon-γ production in progressive multiple sclerosis. Ann Neurol. 1998;44:824–8. doi: 10.1002/ana.410440519. [DOI] [PubMed] [Google Scholar]
- 44.Schafer PH, Wadsworth SA, Wang L, Siekierka JJ. p38 alpha mitogen-activated protein kinase is activated by CD28-mediated signaling and is required for IL-4 production by human CD4+CD45RO+ T cells and Th2 effector cells. J Immunol. 1999;162:7110–9. [PubMed] [Google Scholar]
- 45.Jun CD, Oh CD, Kwak HJ, et al. Overexpression of protein kinase C isoforms protects RAW 264.7 macrophages from nitric oxide-induced apoptosis: involvement of c-Jun N-terminal kinase/stress-activated protein kinase, p38 kinase, and CPP-32 protease pathways. J Immunol. 1999;162:3395–401. [PubMed] [Google Scholar]
- 46.Romagnani S. Lymphokine production by human T cells in disease states. Annu Rev Immunol. 1994;12:227–57. doi: 10.1146/annurev.iy.12.040194.001303. [DOI] [PubMed] [Google Scholar]
- 47.Dragana J, Zhugong L, William CG. Th1- and Th2-cell commitment during infectious disease: asymmetry in divergent pathways. Trends Immunol. 2001;22:450–7. doi: 10.1016/s1471-4906(01)01975-5. [DOI] [PubMed] [Google Scholar]