Abstract
Cytokines are the most important inducers of T helper (Th) cell differentiation. Interleukin-12 (IL-12) and interferon-α (IFN-α) are responsible for human Th1-cell differentiation, while IL-4 is the critical cytokine promoting Th2-cell development. These two subsets of cells co-ordinate immunological responses to pathogens as well as autoimmune or allergic reactions. The pim family of proto-oncogenes encodes serine/threonine-specific kinases involved in cytokine-mediated signalling pathways in haematopoietic cells. Here we demonstrate that expression of pim-1 and pim-2 mRNAs is selectively up- or down-regulated in human cord-blood-derived CD4+ cells freshly induced to polarize towards Th1 or Th2 cells, respectively, whereas their expression is inhibited in both cell types by the immunosuppressive transforming growth factor β (TGF-β). Moreover, the Th1-specific cytokines IL-12 and IFN-α, but not the Th2-specific cytokine IL-4, transiently up-regulate pim-1 and pim-2 mRNA expression in human peripheral blood T cells and natural killer cells. In addition, the Pim-1 protein levels are strongly up-regulated by Th1-specific cytokines in all of these cell types. Taken together, our results suggest that pim genes and their protein products are involved in the early differentiation process of T helper cells.
Keywords: cytokines, natural killer cells, Pim kinases, T helper cells, transforming growth factor-β
Introduction
When undifferentiated naive CD4+ T-lymphocyte precursors encounter antigens, several signalling cascades are activated and a complex developmental programme is initiated, which results in differentiation of these cells into distinct effector T helper (Th) cell populations. The functional differences between fully differentiated Th-cell subsets are primarily explained by the cytokines they secrete (reviewed in ref. 1). Th1 cells produce interferon-γ (IFN-γ), interleukin-2 (IL-2) and tumour necrosis factor-β (TNF-β), and are responsible for cell-mediated/inflammatory immunity, whereas Th2 cells secrete IL-4, IL-5, IL-6, IL-10 and IL-13, and are responsible for humoral response. Cytokines also have important roles in regulation of the differentiation process itself. The critical Th2-inducing cytokine is IL-4, which mediates its effects through the signal transducer and activator of transcription 6 (STAT6) signalling pathway. IL-12 acting via the STAT4 signalling pathway is in turn the primary inducer of Th1 development in synergy with IFN-α.
Several transcription factors involved in regulation of Th1 or Th2 differentiation have been identified, such as T-bet for Th1 cells2 and GATA33 and c-Maf4 for Th2 cells. These factors mediate the effects of signalling pathways initiated by T-cell receptors and cytokine receptors together with other less specific transcription factors such as nuclear factor of activated T cells (NFATc) family members5 and activator protein 1 (AP-1).6 Recently, oligonucleotide array approaches have been used to identify genes that are differentially expressed in human T cells polarized towards the Th1 or Th2 direction.7,8 Hundreds of such genes have been identified, but more detailed functional analyses will be needed to obtain information about how the two different Th-cell subsets have evolved and are maintained.
Natural killer (NK) cells are an essential early component of the innate immune response and are recruited to the site of infection within minutes following pathogen invasion.9 NK cells express receptors for proinflammatory cytokines such as TNF-α, IL-12 and IL-18, the activation of which leads to production of IFN-γ. This amplifies the local inflammatory response and provides an initial source of IFN-γ for the adaptive immune response. Similar to Th1 cells, also in NK cells IL-12 and STAT4 play important roles in IFN-γ production.10,11
The pim gene family consists of three members, pim-1, pim-2 and pim-3 (reviewed in ref. 12). In haematopoietic cells, expression of pim-1 mRNA can be induced by multiple cytokines including IL-2, IL-3, IL-12, IL-15 and IFN-α13–15 and pim-2 mRNA has been suggested to be regulated in a similar fashion.16 The pim genes encode two to three alternatively translated small serine/threonine-specific kinases.17 While the precise functions of Pim kinases remain to be fully elucidated, we and others have previously shown that Pim-1 is involved in cytokine signalling via its interactions with the p100 coactivator,18 the NFATc1 transcription factor19 and the suppressor of cytokine signalling (SOCS) family proteins.20,21 Pim-1 also targets several regulators of the cell cycle and both Pim-1 and Pim-2 can protect haematopoietic cells from apoptosis.12
When overexpressed, pim genes are able to synergize with myc, or gfi-1 oncogenes to promote lymphomagenesis.12 Interestingly, studies with transgenic mice overexpressing either gfi-1 or pim-1 have revealed that these two genes act in opposite ways in β-selection, which is a critical step in thymocyte differentiation.22 In addition, gfi-1 and pim-1 are both expressed in peripheral T cells after antigenic activation.23,24 Recently Gfi-1 was shown to be induced by IL-4 and to promote Th2-cell expansion in synergy with GATA3.25 We have previously demonstrated that pim-1 expression is in turn up-regulated, when peripheral blood T cells are stimulated with Th1-specific cytokines.15 Here the mRNA and protein expression profiles of pim-1 and its close homologue pim-2 were more carefully examined in human cord blood CD4+ T cells induced to polarize towards a Th1 or Th2 direction as well as in human peripheral blood T cells and NK cells treated with Th1- or Th2-specific cytokines.
Materials and methods
Cytokines
IL-2, IL-4, IL-12, transforming growth factor-β (TGF-β) and anti-IL-12 were purchased from R & D Systems (Abingdon, UK or Minneapolis, MN) and IFN-α was obtained from the Finnish Red Cross Blood Transfusion Service (Helsinki, Finland).
Cell culture
Naive CD4+ T cells were isolated as previously described26 from human cord blood of healthy neonates, cultured in Yssel's medium (Irvine Scientific, Santa Ana, CA) supplemented with 1% AB serum (Finnish Red Cross Blood Transfusion Service) and activated either with phytohaemagglutinin (100 ng/ml; Murex Diagnostics, Chatillon, France) and irradiated B7-CD32-transfected mouse L fibroblasts or with plate-bound anti-CD3 and soluble anti-CD28 antibodies (0·5 μg/ml; Immunotech, Marseille, France) under Th-cell polarization conditions: Th1 medium contained 2·5 ng/ml IL-12 and Th2 medium contained 10 μg/ml anti-IL-12 and 10 ng/ml IL-4 (all from R & D systems). After 2 days the cultures were supplemented with 40 U/ml IL-2 (R & D Systems). The polarized phenotypes were controlled at day 7, when cells were stimulated for 16–20 hr with 1 ng/ml phorbol 12-myristate 13-acetate (Sigma, Steinheim, Germany), after which secretion of IFN-γ or IL-4 into the supernatant was measured by enzyme-linked immunosorbent assay (Genzyme, Cambridge, MA). The expression patterns for known differentially expressed marker genes such as SLAM, IFN-γRβ and IL-12Rβ26,27 were also determined to further confirm the phenotypes.
Human peripheral blood T cells comprising both CD8+ (60%) and CD4+ (40%) were isolated as previously described15 from leucocyte-rich buffy coats obtained from healthy blood donors (Finnish Red Cross Blood Transfusion Service). Purified T cells were activated with anti-CD3 and anti-CD28 monoclonal antibodies (0·5 μg/ml; R & D Systems) and cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS) and 100 IU/ml human recombinant IL-2 for 5–6 days, after which they were further propagated for 5–6 days without FCS. In each experiment, T cells from two to four donors were used. The human NK-92 natural killer cells were maintained in continuous culture as previously described.28 Activated human peripheral blood T cells or NK-92 cells were stimulated with IL-12 (5 ng/ml), IFN-α (100 IU/ml) and/or IL-4 (10 ng/ml) for the time-points indicated.
Real-time quantitative polymerase chain reaction
Total cellular RNA from cord blood-derived CD4+ T cells was isolated using an RNeasy minikit (Qiagen, Valencia, CA). Real-time quantitative polymerase chain reaction (PCR) was performed to measure gene expression levels using Taqman ABI Prism 7700 (Applied Biosystems, Foster City, CA) as described before.27 Elongation factor 1α (EF1α) was used as a reference because expression of this housekeeping gene remains stable during Th1 and Th2 differentiation.29primer express software (Applied Biosystems) was used to design primers and probes (Medprobe, Oslo, Norway). EF1α:29, pim-1: probe 5′-6(FAM)-CATCCGCGTCTCCGACAACTTGC-(TAMRA)-3′, forward primer 5′-GCTTCGGCTGGTCTACTCA-3′, reverse primer 5′-CCACCGGTAGTTTGTGCAC-3′. The fold differences in expression were calculated as previously described.27
Oligonucleotide array studies
Total cellular RNA from cord blood-derived CD4+ T cells pooled from up to six donors was isolated using the TRIzol method (Invitrogen, Carlsbad, CA) and RNeasy minikit (Qiagen), and oligonucleotide array studies were performed as previously described.8 Briefly, 5-μg aliquots of total RNA were hybridized on Affymetrix Human Genome U95Av2 arrays (Affymetrix, Santa Clara, CA) containing probes for ∼9300 genes. Two biological repeats for each microarray experiment were performed with two distinct sets of probes for each gene. The gene transcript levels were determined from data images with genechip microarray suite software (Affymetrix MAS5).
Northern blotting
Total cellular RNA from peripheral blood T cells and NK-92 cells was isolated by guanidium isothiocyanate lysis followed by centrifugation through CsCl cushion as previously described.30 Equal amounts of RNA were size-fractionated on formaldehyde/agarose gel, transferred to nylon membranes (Hybond, Amersham Biosciences, Buckinghamshire, UK) and hybridized with cDNA probes for human pim-115 or pim-2 genes. The probe for pim-2 was cloned by reverse transcription-PCR with 5′-CAGAGTGGATCCCTCGACACCAGTACC-3′ sense and 5′-GTCCATGGATCCCTGTGACATGGCCAT-3′ antisense primers from total RNA obtained from IL-15-induced NK-92 cells. The PCR product was subcloned into a pGEM3-Zf(+) vector (Promega, Madison, WI) and sequenced. Ethidium bromide staining of rRNA bands was used to ensure equal RNA loading. The probes were labelled with [α-32P]-dCTP and the membranes were hybridized as previously described.15
Western blotting
Cells were harvested and lysed by three freeze–thaw cycles in 0·1 m KPO4 buffer, pH 7·8 supplemented with protease inhibitor mix (Sigma-Aldrich, St Louis, MO) and 1 mm dithiothreitol. One-hundred-microgram aliquots of protein lysates were mixed with equal volumes of Laemmli sample buffer, boiled, separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred onto Immobilon-P (Millipore, Bedford, MA) or Hybond-P (Amersham Biosciences) membranes. Membranes were stained with anti-Pim-1 (19F7) or anti-Pim-2 (C-20) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) followed by horseradish peroxidase-linked secondary antibody (Zymed, San Francisco, CA) and enhanced chemiluminescence (ECL) + Plus reagents (Amersham Biosciences). Equal protein loading was verified by blotting stripped membranes with anti-β-actin antibody (Sigma-Aldrich).
Results
pim-1 and pim-2 mRNAs are selectively up-regulated in human cord-blood-derived CD4+ cells induced to polarize towards Th1 cells
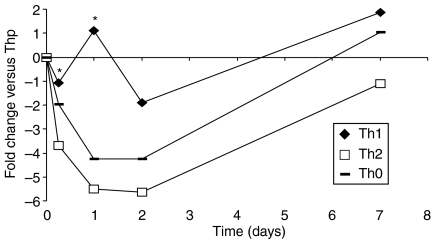
We had previously shown that pim-1 expression can be up-regulated by the Th1-specific cytokines IL-12 and IFN-α in human peripheral blood T cells.15 To determine differentiation-associated pim-1 mRNA levels from naive Th cells that had not previously encountered any antigen, we isolated CD4+ T cells from human cord blood. These cells were either activated (Th0) or simultaneously polarized with either IL-12 or IL-4 for Th1 or Th2 cell differentiation, respectively, and real-time PCR was used to measure pim-1 expression. When the expression values were compared with those obtained at the 0 hr time-point from naïve CD4+ Thp (T helper precursor) cells, it became evident that pim-1 is expressed more efficiently in cord-blood-derived T cells polarized by IL-12 towards the Th1 direction than in cells that were treated with either IL-4 or activated without polarization (Fig. 1). Even though there was some variation between samples from the four analysed individuals, pim-1 expression was always higher in Th1-polarized cells at every time-point measured, with statistically significant differences at the earliest 6- and 24-hr time-points (P ≤ 0·05). Under Th2-polarizing conditions, pim-1 was strongly down-regulated at the 24- or 48-hr time-points, suggesting a selective role for pim-1 during the early steps of Th-cell differentiation. By the 7-day time-point, the differences in pim-1 mRNA levels had mostly disappeared, which could be the result of induction of pim-1 expression by IL-2 added at the 48-hr time-point to promote cell proliferation.
Figure 1.
Expression of pim-1 mRNA is differentially regulated during early steps of Th1- and Th2-cell differentiation. Human cord-blood-derived CD4+ T cells isolated from four individuals were activated with phytohaemagglutinin and irradiated B7-CD32-transfected mouse L fibroblasts (Th0) or simultaneously activated and polarized towards Th1 (2·5 ng/ml IL-12) or Th2 (10 μg/ml anti-IL-12 and 10 ng/ml IL-4) cells. After 48 hr, IL-2 (40 IU/ml) was added. Samples were collected at 0-hr, 6-hr, 24-hr, 48-hr or 7-day time-points during polarization. The average pim-1 expression levels determined by real-time quantitative PCR are shown. The fold differences were calculated by comparing the expression values to those obtained at the 0-hr time-point from Thp cells. The statistical significance of the differences in the expression levels between Th1 and Th2 conditions was determined using unpaired t-test (*P ≤ 0·05).
We also collected data from oligonucleotide array studies with the cord-blood-derived CD4+ T cells. Cells activated for 48 hr with anti-CD3 and anti-CD28 antibodies together with IL-12 (Th1) expressed slightly more pim-1 and pim-2 as compared to cells that were only activated in the absence of polarizing cytokines (Th0), whereas cells cotreated with IL-4 (Th2) showed no statistically significant difference in their expression (Table 1). By contrast, addition of the immunosuppressive agent TGF-β was able to counteract the Th1-specific up-regulation of both pim-1 and pim-2 and to down-regulate pim expression also in cells polarized towards Th2 direction for 48 hr.
Table 1.
Expression changes in pim mRNA levels in human cord-blood-derived CD4+ T cells obtained from Affymetrix hybridizations
| Th1 versus Th0 | Th2 versus Th0 | Th1 + TGF-β versus Th1 | Th2 + TGF-β versus Th2 | |
|---|---|---|---|---|
| pim-1 | 1·6 ± 0·31 | no change | −1·4 ± 0·1 | −1·4 ± 0·1 |
| pim-2 | 1·7 ± 0·6 | no change | −2·1 ± 0·5 | −1·8 ± 0·4 |
Average fold change. Data represent means and standard deviations from two biological replicates analysed with two distinct probe sets. Only statistically significant values are shown, as determined by the analysis software.
IL-12 and IFN-α, but not IL-4, up-regulate pim-1 and pim-2 mRNA expression in human peripheral blood T cells and in NK cells
Next we wanted to study in more detail whether pim-1 and also pim-2 expression is induced only by cytokines promoting Th1-cell differentiation and not by Th2-specific cytokines. Therefore, purified human peripheral blood T cells comprising both CD8+ (60%) and CD4+ (40%) cell populations were activated with anti-CD3 and anti-CD28 antibodies and were grown in the presence of IL-2 for several days to expand the population. Cells were then treated with different cytokines and collected at the indicated time-points. The Th1-specific cytokines IL-12 and IFN-α both increased pim-1 as well as pim-2 mRNA levels (Fig. 2a). However, treatment with IL-12 resulted in more persistent pim expression than treatment with IFN-α. IL-4 had no effect on expression of either pim gene, indicating that both pim-1 and pim-2 are indeed selectively induced by Th1-specific cytokines (Fig. 2a and data not shown). Very similar results to confirm this conclusion were obtained, when pim-1 and pim-2 mRNA levels were measured from cytokine-treated NK-92 cells that have been widely used as a model for activated human NK cells28 (Fig. 2b). However, it should be noted that in the NK-92 cells (Fig. 2b), the cytokine-dependent responses were more sustained than in the more heterogeneous population of peripheral blood cells (Fig. 2a). Furthermore, the basal levels of pim-2 mRNA were higher than those for pim-1, especially in NK-92 cells. When either type of cells was simultaneously treated with both Th1- and Th2-specific cytokines, it became evident that IL-4 is not only unable to support pim-1 or pim-2 expression, but also cannot antagonize their up-regulation by Th1-specific cytokines (Fig. 2a,b). By contrast, IL-4 even slightly down-regulated the high basal levels of pim-2 mRNA in NK cells (Fig. 2b). We also measured pim expression levels from preactivated peripheral blood T cells polarized for up to two weeks in the presence of Th1- or Th2-specific cytokines, and observed more prominent pim-1 and pim-2 levels in cells differentiated towards the Th1 direction than towards Th2 (data not shown).
Figure 2.
pim-1 and pim-2 mRNAs are up- or down-regulated in T and NK cells treated with Th1- or Th2-specific cytokines, respectively. Activated human peripheral blood T cells (a) or NK-92 cells (b) were stimulated with IL-12 (5 ng/ml), IFN-α (100 IU/ml) and/or IL-4 (10 ng/ml), or were left untreated for the indicated time-points. Total cellular RNA was isolated, and 10-μg aliquots were used for Northern blotting with probes specific for either pim-1 or pim-2. The representative results from cells pooled from two to four donors are shown.
pim-1 expression is also up-regulated at the protein level in cells treated with Th1-specific cytokines
When we analysed Pim-1 protein expression by Western blotting with anti-Pim-1 antibodies, we observed significant steady-state levels of Pim-1 in cord-blood-derived T cells polarized for 48 hr in a Th1 direction, but not in cells cotreated with TGF-β or differentiated towards Th2 in the absence or presence of TGF-β(Fig. 3a). More strikingly, in peripheral blood T cells and NK-92 cells, both IL-12 and IFN-α, but not IL-4, were able to increase Pim-1 protein expression (Fig. 3b,c). Indeed, the Pim-1 protein levels correlated well with the mRNA levels because their up-regulation was stronger and more sustained in cells treated with IL-12 as compared to those treated with IFN-α. Likewise, the IFN-α-induced levels of Pim-1 were maintained longer in NK cells (6 hr) than in peripheral blood T cells (3 hr).
Figure 3.
Pim-1 protein levels are elevated by Th1-specific cytokines in human T and NK cells. Human cord-blood-derived CD4+ T cells (a), peripheral blood T cells (b) or NK-92 cells (c) were activated and stimulated as for the experiments shown in Figs 1 and 2. Protein samples were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, and analysed by Western blotting with anti-Pim-1 or anti-β-actin antibodies.
Discussion
Our results indicate that the mRNA levels of pim family genes are selectively up-regulated by Th1-specific, but not Th2-specific, cytokines in cord-blood-derived CD4+ T helper cells, in peripheral blood CD8+/CD4+ T lymphocytes as well as in the NK-92 line of NK cells. These results may be explained by differential action of the cytokine-inducible STAT family members. IL-12 has been shown to stimulate binding of STAT4 to the GAS element of the pim-1 promoter, while IFN-α stimulates STAT1, STAT3 and STAT4 binding there.15 By contrast, it is unlikely that IL-4 would induce STAT6 binding to the pim-1 promoter because the spacing of the symmetrical GAA half-sites distinguish STAT6 binding sites from those for other STAT proteins.31
Sustained IL-12 signalling was recently demonstrated to be required for human Th1 development, while the effects of IFN-α are much weaker, correlating with its transient nature of action.32 Moreover, IL-2 is able to activate pim genes via STAT5,33 to potentiate sustained IL-12/Stat4 responses through up-regulation of IL-12R and to synergize with IL-12 in driving Th1-cell development. These results fit well with our data indicating that pim mRNA expression is up-regulated only transiently by IFN-α, but in a more persistent fashion by IL-12.
TGF-β has a crucial role in T-cell regulation. Besides its antiproliferative effects, which can be largely explained by its ability to inhibit IL-2 production, TGF-β is able to suppress differentiation of both Th1 and Th2 lineages (reviewed in ref. 34). While TGF-β has been shown to inhibit the expression of GATA-3 and thereby inhibit Th2 development, the mechanisms involved in inhibition of Th1 differentiation are less clear. A promising candidate for the TGF-β target in Th1 cells is T-bet, the functional Th1-cell analogue to GATA-3.4 This hypothesis is supported by observations according to which T-bet can induce and TGF-β inhibit the expression of IL-12Rβ2.35,36 According to our data, expression of both pim-1 and pim-2 genes are down-regulated by TGF-β in cells polarized to either the Th1 or Th2 direction. These results are of interest because very few genes are so far known to be regulated by TGF-β during differentiation of human cord-blood-derived T cells.8
Our Western blot analyses confirmed that the Pim-1 protein levels correlate relatively well with its mRNA levels. We would have carried out such analyses with the Pim-2 protein also, but unfortunately the Pim-2 antibodies tested were not specific enough (data not shown). However, similar results would have been expected because the pim-1 and pim-2 mRNAs were coregulated during Th-cell differentiation, and because expression of the Pim-2 protein has been shown to follow its mRNA levels in murine haematopoietic cells.37 Furthermore, recent crystallization studies have revealed that Pim kinases adopt a constitutively active conformation,38 suggesting that also their activity is mostly regulated at the level of expression.
While the functional relevance of the selective induction of pim-1 and pim-2 genes by Th1-specific cytokines remains to be elucidated, one intriguing possibility (based on the observations by us and others) is that Pim proteins regulate cytokine-dependent signalling in differentiating Th-cell subsets via their interactions with the SOCS-1 and SOCS-3 proteins.20,21 Pim-1 is able to stabilize SOCS-120 and possibly also SOCS-3. As observed in Socs-1-deficient mice,39 thymocytes of mice lacking both pim-1 and pim-2 genes show prolonged STAT6 phosphorylation upon IL-4 signalling.20 In humans, both SOCS-1 and SOCS-3 are preferentially expressed in Th2-polarized cells.8,40 Thus, the strong down-regulation of pim genes in cells induced to differentiate towards Th2 cells may prevent the stabilization of SOCS family proteins and thus also prevent the negative feedback regulation of IL-4-induced STAT6 activation. Since Th1 and Th2 cells have been reported to be differentially susceptible for apoptotic inducers,41 another interesting possibility is that pim expression is essential for the survival of Th1 cells, but not of Th2 cells. Finally, it was very recently demonstrated that T cells of mice deficient for all the three pim family genes develop normally in the thymus but do not properly respond to antigen or cytokine stimulation.42 Since Th-cell differentiation is largely dependent on such responses, our future experiments will be directed to determine whether Th1-cell versus Th2-cell polarization is impaired in such human T cells where pim gene levels are up- or down-regulated.
Acknowledgments
We thank Kaija-Liisa Laine and Outi Melin for expert technical assistance and Helena Ahlfors for help in the data analyses. This work was supported by grants from the Academy of Finland (to P.J.K., R.L. and S.M.), Turku University Hospital Fund (to R.L.) and Finnish Cultural Foundation (to T.L.T.A.).
References
- 1.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nature Rev. 2002;2:933–44. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 2.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 3.Zheng W-P, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 4.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–83. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 5.Rinćon M, Flavell RA. Transcription mediated by NFAT is highly inducible in effector CD4+ T helper 2 (Th2) cells but not in Th1 cells. Mol Cell Biol. 1997;17:1522–34. doi: 10.1128/mcb.17.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinćon M, Derijard B, Chow CW, Davis RJ, Flavell RA. Reprogramming the signalling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T-cells into effector Th1 and Th2 cells. Genes Funct. 1997;1:51–68. doi: 10.1046/j.1365-4624.1997.00007.x. [DOI] [PubMed] [Google Scholar]
- 7.Rogge L, Bianchi E, Biffi M, et al. Transcript imaging of the development of human T helper cells using oligonucleotide arrays. Nat Genet. 2000;25:96–101. doi: 10.1038/75671. [DOI] [PubMed] [Google Scholar]
- 8.Lund R, Aittokallio T, Nevalainen O, Lahesmaa R. Identification of novel genes regulated by IL-12, IL-4, or TGF-beta during the early polarization of CD4+ lymphocytes. J Immunol. 2003;171:5328–36. doi: 10.4049/jimmunol.171.10.5328. [DOI] [PubMed] [Google Scholar]
- 9.Glas R, Franksson L, Une C, Eloranta ML, Ohlen C, Orn A, Kärre K. Recruitment and activation of natural killer (NK) cells in vivo determined by the target cell phenotype. An adaptive component of NK cell-mediated responses. J Exp Med. 2000;191:129–38. doi: 10.1084/jem.191.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–7. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 11.Thierfelder WE, van Deursen JM, Yamamoto K, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–4. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 12.Bachmann M, Möröy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37:726–30. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Dautry F, Weil DYuJ, Dautry-Varsat A. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J Biol Chem. 1988;263:17615–20. [PubMed] [Google Scholar]
- 14.Lilly M, Le T, Holland P, Hendrickson SL. Sustained expression of the pim-1 kinase is specifically induced in myeloid cells by cytokines whose receptors are structurally related. Oncogene. 1992;7:727–32. [PubMed] [Google Scholar]
- 15.Matikainen S, Saraneva T, Rönni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-α activates multiple STAT proteins and up-regulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–91. [PubMed] [Google Scholar]
- 16.Allen JD, Verhoeven E, Domen J, van der Valk M, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15:1133–41. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- 17.Saris CJM, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–64. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leverson JD, Koskinen PJ, Orrico FC, Rainio EM, Jalkanen KJ, Dash AB, Eisenman RN, Ness SA. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell. 1998;2:417–25. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 19.Rainio EM, Sandholm J, Koskinen PJ. Cutting edge: Transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase. J Immunol. 2002;168:1524–7. doi: 10.4049/jimmunol.168.4.1524. [DOI] [PubMed] [Google Scholar]
- 20.Chen XP, Losman JA, Cowan S, et al. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc Natl Acad Sci USA. 2002;99:2175–80. doi: 10.1073/pnas.042035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peltola KJ, Paukku K, Aho TLT, Ruuska M, Silvennoinen O, Koskinen PJ. Pim-1 kinase inhibits STAT5-dependent transcription via its interactions with SOCS1 and SOCS3. Blood. 2004;103:3744–50. doi: 10.1182/blood-2003-09-3126. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt T, Karsunky H, Rödel B, Zevnik B, Elsässer HP, Möröy T. Evidence implicating Gfi-1 and Pim-1 in pre-T-cell differentiation steps associated with β-selection. EMBO J. 1998b;17:5349–59. doi: 10.1093/emboj/17.18.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2) -dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–68. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wingett D, Long A, Kelleher D, Magnuson NS. pim-1 proto-oncogene expression in anti-CD3-mediated T cell activation is associated with protein kinase C activation and is independent of Raf-1. J Immunol. 1996;156:549–57. [PubMed] [Google Scholar]
- 25.Zhu J, Guo L, Min B, Watson CJ, Hu-Li J, Young HA, Tsichlis PN, Paul WE. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16:733–44. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 26.Lund R, Ylikoski E, Aittokallio T, Nevalainen O, Lahesmaa R. Kinetics and STAT4- or STAT6-mediated regulation of genes involved in lymphocyte polarization to Th1 and Th2 cells. Eur J Immunol. 2003;33:1105–16. doi: 10.1002/eji.200323899. [DOI] [PubMed] [Google Scholar]
- 27.Hämäläinen H, Meissner S, Lahesmaa R. Signalling lymphocytic activation molecule (SLAM) is differentially expressed in human Th1 and Th2 cells. J Immunol Meth. 2000;242:9–19. doi: 10.1016/s0022-1759(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 28.Strengell M, Matikainen S, Siren J, Lehtonen A, Foster D, Julkunen I, Sareneva T. IL-21 in synergy with IL-15 or IL-18 enhances IFN-γ production in human NK and T cells. J Immunol. 2003;170:5464–9. doi: 10.4049/jimmunol.170.11.5464. [DOI] [PubMed] [Google Scholar]
- 29.Hämäläinen HK, Tubman JC, Vikman S, Kyrölä T, Ylikoski E, Warrington JA, Lahesmaa R. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem. 2001;299:63–70. doi: 10.1006/abio.2001.5369. [DOI] [PubMed] [Google Scholar]
- 30.Sareneva T, Julkunen I, Matikainen S. IFN-α and IL-12 induce IL-18 receptor gene expression in human NK and T cells. J Immunol. 2000;165:1933–8. doi: 10.4049/jimmunol.165.4.1933. [DOI] [PubMed] [Google Scholar]
- 31.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 32.Athie-Morales V, Smits HH, Cantrell DA, Hilkens CMU. Sustained IL-12 signaling is required for Th1 development. J Immunol. 2004;172:61–9. doi: 10.4049/jimmunol.172.1.61. [DOI] [PubMed] [Google Scholar]
- 33.Mui AL, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated STAT5: role of STAT5 in proliferation. EMBO J. 1996;15:2425–33. [PMC free article] [PubMed] [Google Scholar]
- 34.Gorelik L, Flavell RA. Transforming growth factor-beta in T-cell biology. Nat Rev Immunol. 2002;2:46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 35.Mullen AC, High FA, Hutchins AS, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–10. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 36.Gorham JD, Guler ML, Fenoglio D, Gubler U, Murphy KM. Low dose TGF-beta attenuates IL-12 responsiveness in murine Th cells. J Immunol. 1998;161:1664–70. [PubMed] [Google Scholar]
- 37.Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–54. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian KC, Wang L, Hickey ER, et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human pim-1 kinase. J Biol Chem. 2005;280:6130–7. doi: 10.1074/jbc.M409123200. [DOI] [PubMed] [Google Scholar]
- 39.Naka T, Matsumoto T, Narazaki M, et al. Accelerated apoptosis of lymphocytes by augmented induction of Bax in SSI-1 (STAT-induced STAT inhibitor-1) deficient mice. Proc Natl Acad Sci USA. 1998;95:15577–82. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seki Y, Inoue H, Nagata N, et al. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat Med. 2003;9:1047–54. doi: 10.1038/nm896. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XR, Zhang LY, Devadas S, Li L, Keegan AD, Shi YF. Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ. 2003;10:203–10. doi: 10.1038/sj.cdd.4401138. [DOI] [PubMed] [Google Scholar]
- 42.Mikkers H, Nawjin M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM-kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–15. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]