Abstract
The major immunological barrier that prevents the use of wild-type pig xenografts as an alternative source of organs for human xenotransplantation is antibody-mediated rejection. In this study, we identify the immunoglobulin variable region heavy (IgVH) chain genes encoding xenoantibodies to porcine heart and fetal porcine islet xenografts in non-immunosuppressed rhesus monkeys. We sought to compare the IgVH genes encoding xenoantibodies to porcine islets and solid organ xenografts. The immunoglobulin M (IgM) and IgG xenoantibody response was analysed by enzyme-linked immunosorbent assay and cDNA libraries from peripheral blood lymphocytes were prepared and sequenced. The relative frequency of IgVH gene usage was established by colony filter hybridization. Induced xenoantibodies were encoded by the IGHV3-11 germline progenitor, the same germline gene that encodes xenoantibodies in humans mounting active xenoantibody responses. The immune response to pig xenografts presented as solid organs or isolated cells is mediated by identical IgVH genes in rhesus monkeys. These animals represent a clinically relevant model to identify the immunological basis of pig-to-human xenograft rejection.
Keywords: alpha gal carbohydrate, IgVH gene, xenoantibody response
Introduction
The use of organs from non-human species (xenografts) such as pigs represents a solution for the chronic shortage of available organs for human transplantation.1,2 Pigs are physiologically similar to humans, easily bred and are available in unlimited numbers.3,4 The carbohydrate galactose α (1,3) galactose (gal α1,3 gal epitope) expressed on pig cells initiates the humoral response and contributes to cell-mediated rejection of porcine xenografts.5–7 Humans and Old World primates do not express a functional α1,3-galactosyltransferase gene and develop high levels of pre-existing anti-gal xenoantibodies as a result of antigenic stimulation by gastrointestinal bacteria expressing the α-gal epitope.8,9 A small number of germline progenitors encode anti-gal xenoantibodies in humans and galactosyltransferase knockout mice.10–13 IgVH genes encoding the antibody response have an evolutionarily conserved structural configuration.14,15 Their possible counterpart in non-human primates which represent an important preclinical model for human xenotransplantation, has not been characterized. Since the human and rhesus variable region heavy chain family 3 (VH3) homologues defined to date show similarity, the humoral immune response in primates may be a closely related, appropriate model for studying the human immune response to porcine xenografts.16 We report the identification of the IgVH genes encoding xenoantibody responses induced following transplantation of rhesus monkeys (Macaca mulatta) with isolated fetal porcine islet cells and neonatal porcine hearts in the absence of immunosuppressive treatments. Fetal pig islet cells express high levels of the gal carbohydrate epitope, whereas adult cells do not.17 The xenoantibody responses to both hearts and fetal islet cells are encoded by a rhesus IgVH gene most like Homo sapiens IGHV3-11. The kinetics of the anti-α-gal xenoantibody responses in four monkeys exposed to porcine heart or fetal pig islet cell xenografts is similar, and is encoded by a restricted group of genes.
Materials and methods
Animals
Four colony-bred rhesus macaques (Macaca mulatta) from the California National Primate Research Center, University of California at Davis, CA were housed in accordance with The American Association for Accreditation of Laboratory Animal Care standards. All animal tests satisfied the requirements of the Animal Welfare Act, and protocols were approved by the Institutional Animal Care and Use Committee at the University of California, Davis. Two 4-year-old male rhesus monkeys (Macaca mulatta) were selected for porcine islet cell immunization and two 3-year-old monkeys were selected for porcine heart transplantation. To maximize the length of organ survival in non-immunosuppressed non-human primates, all rhesus monkeys were chosen based on low xenoantibody levels specific for the gal α1,3 gal epitope (Galα1-3Galβ1-4GlcNAc-R) by enzyme-linked immunosorbent assay (ELISA). Porcine hearts selected for transplantation were derived from neonatal porcine donors (5–9 days old). Porcine islet-like cell clusters (ICCs) were isolated from pancreata derived from fetal pigs at 66 days of gestation.
Heart transplantation
Porcine hearts were transplanted into juvenile rhesus monkeys with low pre-existing antibodies directed at the gal α1,3 gal epitope as identified by ELISA. Hyperacute rejection occurred within 2 hr. The porcine heart was removed, leaving a short segment of graft vessels (aorta and pulmonary artery) remaining in situ to provide an ongoing stimulus for xenoantibody production after heart removal. Blood samples were taken prior to transplantation (d0), 4 hr, 8 hr, 24 hr, 11 days, 21 days and monthly post-transplant. Serum samples were used to characterize the xenoantibody response by ELISA. Peripheral blood lymphocytes were isolated to produce cDNA libraries from pre- and post-transplantation samples.
Preparation and immunostaining of porcine islet-like cell clusters
Freshly isolated fetal ICCs were grown on poly-lysine-coated coverslips for immunostaining. Cells were washed with phosphate-buffered saline (PBS) containing 2 mm MgCl2, then blocked for 1 hr with 1% bovine serum albumin (BSA) in PBS. The first antibody [guinea-pig anti-human insulin, immunoglobulin G (IgG) fragment] (Linco Research Inc., St Charles, MO) was diluted 1 : 100 in 1% BSA and was added overnight at 4°. The cells were washed with PBS/0·02% Triton X-100. Texas-Red-conjugated donkey anti-guinea-pig IgG (Jackson Immuno Research, West Grove, PA) was added to detect the bound anti-insulin antibody and the BSA-Bandeiraea simplicifolia isolectin B4 conjugated to fluorescein isothiocyanate (BS-IB4 lectin-FITC) (1 mg/ml) (Sigma Aldrich, St Louis, MO), both antibodies at a dilution of 1 : 50, was added for gal carbohydrate detection.18 The cell nucleus was stained with 0·2 μl/ml 4′,6-diamidino-2-phenylindol (DAPI). Anti-fading solution (V-Laboratories Inc., Covington, LA) was used for mounting. The pictures were taken at the Image Core facility at the Children's Hospital of Los Angeles, CA.
Islet cell immunization
Porcine fetal ICCs (15 × 106 cells) were prepared by culturing collagenase-digested pancreata from fetuses at 66 days of gestation. Cells were cultured for 1–3 weeks19 and injected via an intraperitoneal route into two monkeys. ICCs were stained with BS-IB4 lectin-FITC and an anti-insulin antibody conjugated to Texas Red20 to determine whether insulin-secreting cells expressed the gal carbohydrate after culture. Blood samples were taken prior to transplantation on day 0, then at day 8, day 12, day 20 and day 39 post-transplantation. The two monkeys were re-immunized on day 45 with 15 × 106 newly prepared porcine ICCs. Samples were taken prior to the second injection, on day 45, and at days 75 and 90. These samples were used to characterize the xenoantibody response (IgM, IgG) by ELISA. Peripheral blood lymphocytes were isolated to establish IgM cDNA libraries from days 0, 8 and 12. IgG cDNA libraries were prepared from peripheral blood samples obtained at days 20 and 21. Time-points were selected based on the xenoantibody level measured in the ELISA.
Xenoantibody binding to the α-gal carbohydrate epitope identified by ELISA
Microtitre plates (Nunc, Rochester, NY) were coated with Galα1-3Galα1-4Glc-BSA (250 ng/50 μl) (V-Laboratories, Inc., Covington, LA) and blocked with 1% BSA in 0·5% Tween-20/PBS. Monkey serum (dilution: 1 : 40) was added for 1 hr followed by washing and detection with peroxidase-conjugated goat anti-human IgG (γ-chain specific) F(ab′)2 (1 : 400) (Sigma Aldrich) and peroxidase-conjugated goat anti-human IgM F(ab′)2 at a dilution of 1 : 7000 (Jackson Immuno Research) for 1 hr. SureBlue TMB Microwell peroxidase substrate (KPL, Gaithersburg, MD) was added and the reaction was stopped with sulphuric acid. Absorbance was measured at 450 nm (HTS 7000 plus) (Perkin Elmer, Wellesley, MA). The concentration (in ng/ml) of whole serum immunoglobulin IgM and IgG in the monkeys was quantified by ELISA (Quantitation Kit Bethyl Laboratories Inc., Montgomery, TX) based upon a standard curve.
Preparation of cDNA libraries from peripheral blood lymphocytes
Blood samples from the monkeys mounting active xenoantibody responses detected by ELISA were used to produce IgVH cDNA libraries. RNA from peripheral blood lymphocytes was extracted with TRIZOL (Invitrogen, Carlsbad, CA), precipitated with isopropanol, and reverse transcribed into cDNA using a cDNA synthesis kit according to the manufacturer's instructions (Roche, Indianapolis, IN). The IgVH family 3 immunoglobulin genes encoding IgM antibodies were cloned using a nested PCR as reported previously from our laboratory.10 Briefly, the first round of PCR was performed using an anchor primer 1 5′-ATTACGGCGGCCGCGGATCC-3′ and Cμ primer 3 5′-GGGAAAAGGGTTGGGGCGGATGCA-3′ in PCR-buffer containing 15 mm MgCl2, 25 mm dNTPs and 2·5 U Taq DNA polymerase. The conditions for the first PCR were 94° for 3 min, 25 cycles at 94° for 30 seconds, 48° for 30 seconds, and 72° for 1 min, followed by a 7-min extension at 72°. A second nested PCR was carried out for 25 cycles at an annealing temperature of 55° using 10 μl of the first PCR product. The primers for the nested PCR were Cμ primer 3 and VH3 FR1 5′-GAGGTGCAGCTGGTGGAGTCTGG-3′. The third PCR was performed using Cμ primer 3 and VH3 primer 2 5′-TCTGGGGGAGGCTTGGTC-3′, at an annealing temperature of 55°. Genes encoding IgG xenoantibodies from the IgVH family 3 were cloned using the primers Monk CGI 5′-GGGTTGTAGTCCTTGACCAGGCAG-3′ and primer Monk CGII 5′-GACCGATGGGCCCTTGGTGGAGGC-3′, both specific for the constant region of IgG. The annealing temperature used to amplify IgG genes was 52°. The last PCR reaction was visualized on a 2% ethidium bromide agarose gel. The band closest to the predicted size of 425 base pairs (bp) was cut, eluted by electrolysis with 0·5× TBE-buffer and cloned (Original TA Cloning ® Kit, Invitrogen, Carlsbad, CA). The reaction was transformed into INVαF′ One Shot ® competent cells (Invitrogen, Carlsbad, CA).
Colony filter hybridization to identify the relative frequency of IgVH gene usage
Colony filter hybridization was performed as previously described.10 One hundred colonies, positive for the 425-bp insert, were selected and transferred to a nylon filter. After denaturing and neutralization of the filters, the DNA was cross-linked with UV light (Stratalinker, Stratagene, La Jolla, CA), treated with 2 mg/ml Proteinase K for 1 hr at 37°, and incubated with digoxigenin-dideoxy-UTP-labelled oligonucleotides (100 pmol/μl) in a solution of 5× transferase buffer, 25 mm cobalt dichloride solution, and 25 units/μl terminal transferase (Roche, Indianapolis, IN) for 30 min at 37°. The oligonucleotides RVH11: 5′-TCACTTTCAGTGACTACATGAGCTGGA-3′ (specific for the IGHV3-11 family) and HV3FR1: 5′-GAGGTGCAGCTGGTGGAGTCTGG-3′ (specific for amplifying genes in the VH3 family) were digoxigenin-dideoxy-UTP-labelled. Filters were prehybridized with DIG Easy hybridization buffer for 1 hr at room temperature, and digoxigenin-dideoxy-UTP-labelled oligonucleotides were added to the filters at a concentration of 10 pmol/μl overnight at room temperature. The filters were washed with 2× saline sodium citrate, 0·1% sodium dodecyl sulphate at room temperature and with 0·5× saline sodium citrate, 0·1% sodium dodecyl sulphate at 37°. The chemiluminescence detection was performed according to the manufacturer's instructions (Roche, Indianapolis, IN). Briefly, filters were blocked for 1 hr, 30 μl of anti-digoxigenin-AP Fab-fragments (150 U/200 μl) (Roche, Indianapolis, IN) were added to the blocking solution, and filters were gently rotated for 30 min at room temperature. The filters were equilibrated with detection buffer and CSPD chemiluminescence substrate drops were added. Filters were exposed to Lumi-Film (chemiluminescence detection film) and positive colonies were counted to calculate the relative frequency of IgVH gene usage. The HV3FR1 probe detected the number of human VH3 family genes in the library.
Immunoglobulin gene sequencing
One hundred randomly picked cDNA clones identified in the IgM VH3 family libraries and 80 cDNA clones from IgG VH3 family libraries from monkeys exposed to porcine islet cells or heart xenografts were sequenced using the ALFexpress™ automated DNA sequencer (Amersham, Piscataway, NJ) The clones were sequenced using the M13 Universal primer 5′-(cy5) GTAAAACGACGGCCAGT-3′ and the M13 reverse primer 5′-(cy5) AACAGCTATGACCATG-3′. blast (http://www.ncbi.nlm.nih.gov/Blast/) was used to identify the closest germline gene. Results were confirmed using the software pc-gene.
Results
Insulin-secreting cells in fetal porcine islet-like cell clusters express the gal carbohydrate
Porcine fetal ICCs were isolated from pancreata obtained from piglets at 66 days of gestation. Since these cells were cultured prior to intraperitoneal injection, gal carbohydrate expression was confirmed by immunofluorescence staining. Islet cells grown on coverslips stained positive for the α-gal epitope using the BS-IB4-lectin labelled with FITC. The endocrine cells in the islet-like cell clusters stained positive for insulin secretion (Texas red staining) and expressed the gal epitope (FITC-staining). The staining demonstrated that the gal carbohydrate was present on fetal porcine islet cells following culture under the conditions described. On the basis of these results, the xenoantibody response in monkeys immunized with fetal porcine islet cells includes antibodies directed against the gal carbohydrate.
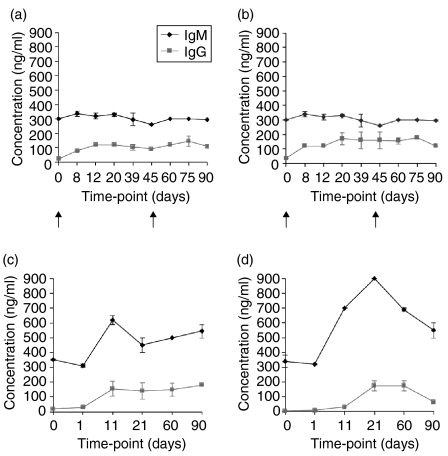
IgM and IgG anti-gal xenoantibodies are induced following transplantation with porcine hearts and fetal porcine islet cells
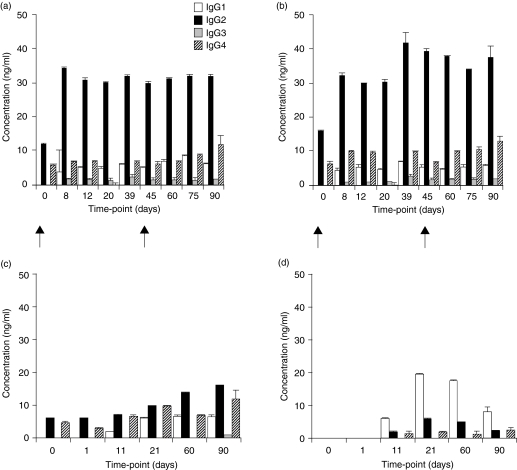
Serum samples taken from four monkeys at multiple time-points following exposure to porcine cells or solid organ xenografts were used to characterize the xenoantibody response by ELISA. IgM and IgG xenoantibodies directed at the gal carbohydrate were induced following immunization with fetal porcine islets and reached plateau levels by day 8 post-injection (Fig. 1a,b). IgM and IgG anti-gal xenoantibody levels remained high for the duration of the study (90 days). The IgG xenoantibodies induced were predominantly of the IgG2 subtype (Fig. 2a,b). We then compared these results with the kinetics, level and specificity of xenoantibodies induced following transplantation with a solid organ xenograft. Serum samples were taken prior to heart transplantation (day 0), and at 4 hr, 8 hr, 24 hr, 11 days, 21 days and monthly post-transplant. Elevated levels of anti-gal IgM and IgG xenoantibodies were demonstrated at day 11 post-transplantation in both monkeys, and reached a maximum level by day 21 (Figs 1c,d). Anti-gal xenoantibodies remained high in the first monkey (Fig. 1c), but declined in the second monkey by day 90 (Fig. 1d). Relative levels of IgG subtypes were analysed by ELISA at sequential time-points post-transplantation. High levels of IgG2 were induced following immunization with fetal ICCs, and an elevation in both IgG2 (Fig. 2c) and IgG1 (Fig. 2d) was demonstrated following porcine heart transplantation. These results indicate that the presence of porcine vessels expressing gal+ endothelium, when left in situ, provides a long-term stimulus for xenoantibody production. Fetal porcine islet cells induce much higher levels of IgG2 xenoantibodies when compared with the response noted following porcine heart transplantation.
Figure 1.
ELISA demonstrating anti-gal xenoantibodies produced following islet cell xenotransplantation (a, b) and/or solid organ (heart) (c, d) xenotransplantation over a timeframe of 90 days. ELISA was performed for each time-point in duplicate, and repeated twice. Arrows indicate the time of islet cell injection. The immune response was examined in four monkeys.
Figure 2.
Analysis of xenoantibody IgG subtypes by ELISA. The subtypes IgG1-IgG4 were analysed in the serum of four rhesus monkeys at different time-points after injection of islet cells (a, b) or after heart transplantation (c, d). An increase in IgG2 was demonstrated after islet cell injection. Arrows indicate the time of intraperitoneal injection. IgG1 levels were induced after heart xenotransplantation (c, d). Serum samples were run in duplicate, and the assay was repeated twice.
IgVH gene usage following porcine heart and fetal islet cell xenograft transplantation
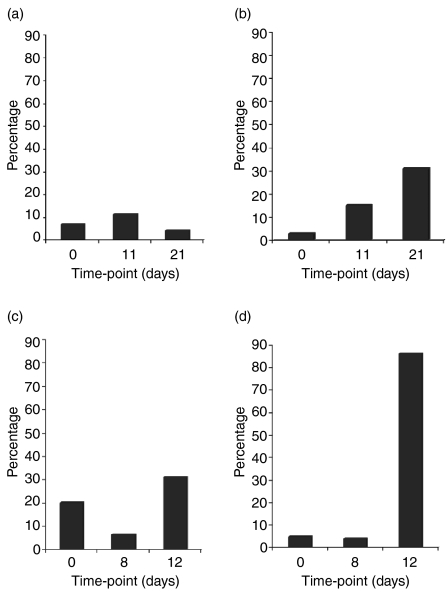
A total of 180 cDNA clones was selected randomly for sequencing to identify the IgVH genes expressed in the IgM and IgG cDNA libraries. The sequences were analysed using NCBI-Ig Blast (http://www.ncbi.nlm.nih.gov/Blast/), and the VBASE2 database (http://www.vbase2.org) to identify the closest human or Macaca mulatta germline gene. The sequence of IgM cDNA clones was obtained from days 0 and 12 following fetal islet cell injection and days 0 and 11 following porcine heart xenograft transplantation. The sequence of cDNA clones identified in IgG libraries was examined at day 20 following islet cell immunization and at day 21 after heart transplantation. The days were selected based on the increased IgM and IgG serum levels identified by ELISA (Fig. 1). By sequencing 180 clones from the cDNA libraries, we identified the IgVH genes encoding xenoantibodies in rhesus monkeys that were most closely related to the Homo sapiens IgVH gene IGHV3-11. The data were confirmed using the colony filter hybridization assay to identify the relative frequency of expression of IGHV3-11 in libraries that were each represented by a minimum of 100 colonies. The RVH11 oligonucleotide probe, specific for all functional alleles of the IGHV3-11 germline gene progenitor, was used to screen these colonies.
At day 11 following exposure to porcine heart xenografts, 11·2% of the IgVH genes were encoded by IGHV3-11, which was elevated to 31% by day 21 (Fig. 3a,b). The day 21 time-point in this animal corresponded with the time at which the highest serum level of IgM was identified by ELISA (Fig. 1d). We similarly analysed the IgM VH3 gene usage following fetal ICC injection. The relative frequency of the IgVH gene expression was evaluated by random sequencing and by colony lift hybridization assay. At day 12 after the islet cell injection, both monkeys utilized the VH gene most closely related to the IGHV3-11 gene. The frequency of usage of this IgVH gene in each experimental animal was 31% (Fig. 3c) and 86% (Fig. 3d), respectively. The data showed that the IgM heavy chain genes expressed in primates mounting active xenoantibody responses are encoded by a gene most like Homo sapiens germline gene IGHV3-11.
Figure 3.
The VH3-11 gene is expressed at increased levels in all monkeys following transplantation with pig hearts (a, b) or fetal porcine islets (c, d), as identified by colony filter hybridization.
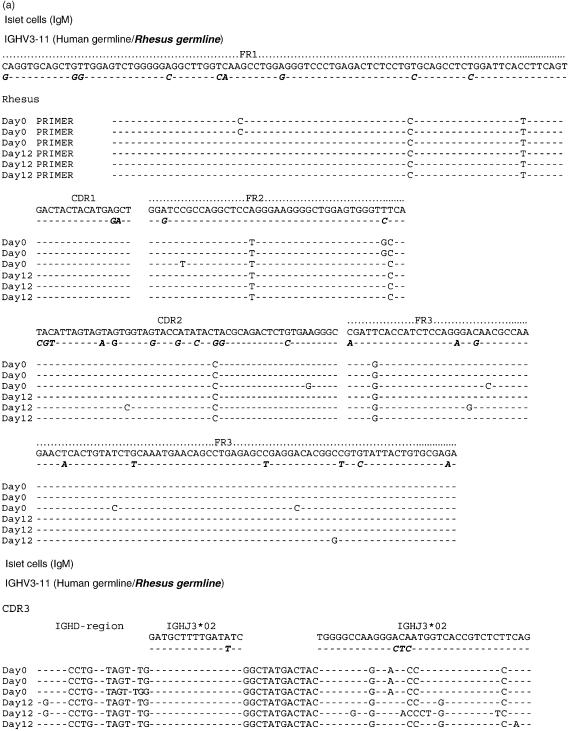
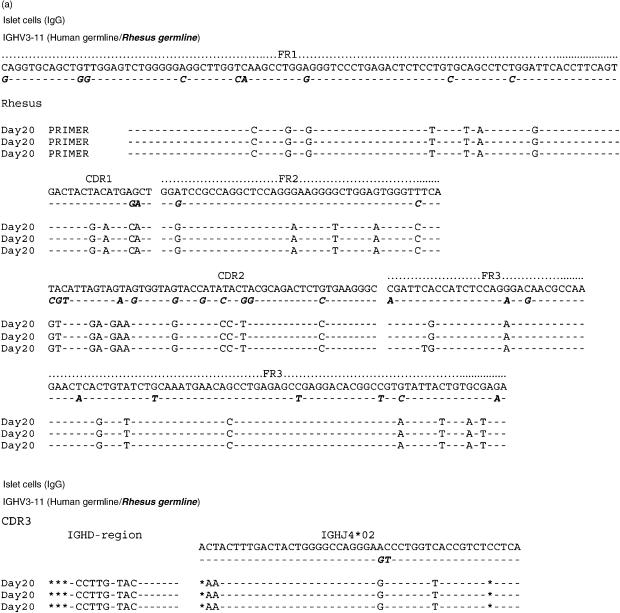
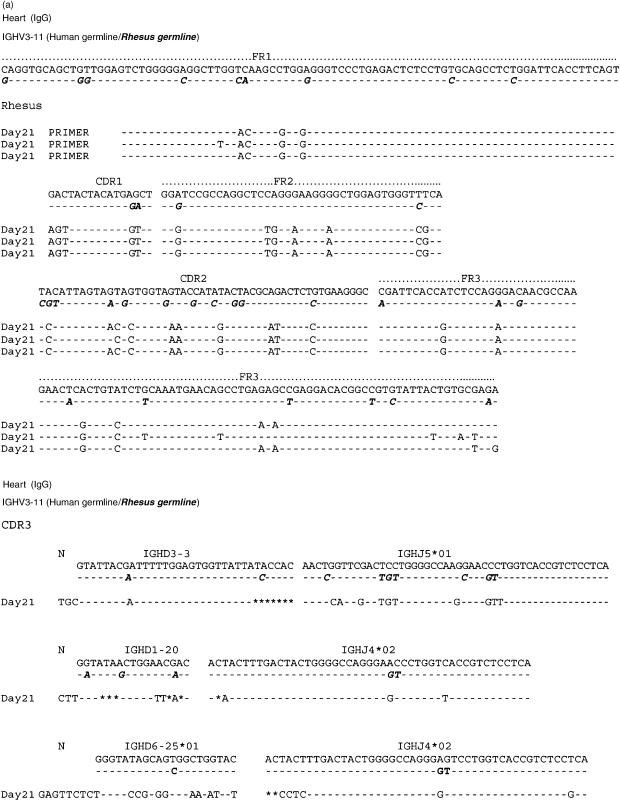
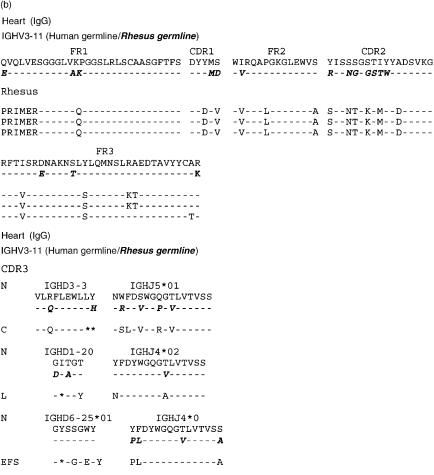
The nucleic acid sequences of six individual clones obtained from IgM cDNA libraries produced prior to and following porcine islet cell and heart xenotransplantation are shown in Figs 4(a) and 5(a). IgG clones obtained from cDNA libraries at day 20 after islet cell immunization and at day 21 after porcine heart transplantation were similarly aligned to the nucleic acid sequence of the human and rhesus germline gene IGHV3-11 (Figs 6a and 7a). Their amino acid sequence is shown in Figs 6(b) and 7(b). The results show that the IgG xenoantibodies induced following both islet cell immunization and heart transplantation demonstrate a number of mutations, and utilized different CDR3 regions (Figs 6a,b and 7a,b). The IgVH genes utilized to encode IgM xenoantibodies directed at isolated islet cells or solid organs expressed identical complementarity-determining regions (CDRs) and framework regions (FRs) following translation into amino acid sequences (Figs 4b and 5b). The immunoglobulin heavy chain diversity region (IGHD) showed little homology to be assigned. The corresponding J region was identified as IGHJ3*02 (accession number X86355). IgG cDNA clones were more diverse in D and J gene usage (Figs 6a,b and 7a,b). The D regions included IGHD5*01 (X13972), IGHD3-3*01 (accession number X13972/M37277), IGHD1-20 (X97051) and IGHD6-25*01 (X97051) (Figs 7a.b). The IGHJ regions encoding IgG xenoantibodies were identified as IGHJ4*02 (accession number X86356) and IGHJ5*01 (Figs 7a.b). The corresponding human IGHD and IGHJ regions were identified using the IMGT Database http://www.imgt.cines.fr:8104 and VBASE2.21–23
Figure 4.
Alignment of nucleic acid sequences of VH genes encoding IgM xenoantibodies in rhesus monkeys. The sequences of genes encoding antibodies expressed following exposure to islet cells are shown aligned with the closest human germline gene progenitor (IGHV3-11) and the rhesus germline IGHV3-11 (bold, italic). Each time-point represents the sequence identified in three individual representative IgM cDNA clones. Nomenclature of the germline genes is based on the list of the human IGV germline genes listed at http://www.mrc-cpe.cam.ac.uk. Amino acid translations as well as the complementarity-determining region 3 (CDR3) of IgM xenoantibodies expressed in islet cell immunized monkeys are shown compared with the human and rhesus germline gene IGHV3-11 in(b)The amino acid sequence of the constant region specific for Macaca mulatta is compared to that of Homo sapiens in(c)(bold, italic). Data from Homo sapiens and Macaca mulatta IgM heavy chain are listed with the NCBI GenBank database under accession numbers AY671780 and AF046784. The sequences identified for immunoglobulin genes encoding xenoantibodies induced following immunization with islet cells are similar to sequences of rhesus origin.
Figure 5.
(a) Alignment of the nucleic acid sequence of IgM xenoantibodies from monkeys exposed to solid organ hearts. Three individual representative cDNA clones from each time-point were aligned with the closest human germline gene progenitor (IGHV3-11) and the rhesus germline IGHV3-11 (bold, italic). Amino acid translations of the sequences are shown compared with the sequence of the human IGHV3-11 germline progenitor encoding human xenoantibodies (b). The amino acid sequence of the IgM constant region of Macaca mulatta and Homo sapiens is compared to the sequence we obtained in monkeys exposed to porcine heart xenografts (c) (bold, italic). The sequence from the constant region of Homo sapiens and Macaca mulatta IgM heavy chain is listed in the NCBI GenBank database under accession numbers AY671780 and AF046784.
Figure 6.
Nucleic acid sequence alignment of three representative cDNA clones encoding IgG xenoantibodies in rhesus monkeys at day 20 following exposure to islet cells is shown aligned with the human germline gene IGHV3-11 and the rhesus germline gene clone 18 closest to human IGHV3-1116 in (a). Nomenclature of the germline genes is based on the list of the human IGV germline genes listed at http://www.mrc-cpe.cam.ac.uk. Stars represent missing nucleotides. (b) shows the amino acid sequence of these genes.
Figure 7.
The nucleotide sequence and the amino acid translation of three individual IgG cDNA clones encoding xenoantibodies at day 21 post heart transplant were aligned with the human germline IGHV3-11 gene and the rhesus germline gene clone 18 (a, b).16 This rhesus germline gene is most similar to human IGHV3-11. Stars represent missing nucleotides. Nomenclature of the germline genes is based on the list of the human IGV germline genes listed at http://www.mrc-cpe.cam.ac.uk.
Alignment of the constant region sequence of cDNAs isolated from humans and monkeys indicates that monkey-specific regions of the DNA could be identified in several regions, including the constant region of the immunoglobulin genes (Figs 4c and 5c). This excludes the possibility that the use of identical CDR3 regions in monkeys and humans mounting active immune responses to porcine xenografts is the result of contamination of rhesus samples with human material. The IgM and IgG genes encoding xenoantibodies identified by both random sequencing and colony filter hybridization demonstrated an increased usage of the IGHV3-11 gene, irrespective of the type of xenograft (Figs 3a, 4a, 5a, 6a and 7a).
Discussion
Preformed and induced antibodies are the major obstacles in xenotransplantation. In humans, xenografts induce a vigorous humoral rejection, mediated by antibodies expressed in germline configuration.10,24 This immune response to xenoantigens is encoded by a small number of IgVH genes with a conserved structural configuration.13,15,25 Their possible counterparts in non-human primates, which represent an important preclinical model for human xenotransplantation, have not been characterized. In this study, we were interested in the characterization of xenoantibodies directed at the carbohydrate epitope that initiates rejection of vascularized organs (hearts) from porcine donors and fetal porcine islet cells in rhesus monkeys (Macaca mulatta) in the absence of immunosuppression, and/or genetic manipulation of the xenograft donor.
Porcine islet cell transplantation represents a potential treatment for diabetes, especially type 1 diabetes mellitus. Porcine islets regulate glucose levels in a physiological range similar to that of humans, and pig insulin adequately substitutes for human insulin.26,27 Porcine islet cell clusters at gestational age 66–81 days have been used in clinical trials for patients with type 1 diabetes mellitus and end-stage diabetic nephropathy.28 The transplanted fetal porcine pancreatic tissue remained viable for several months. All patients developed xenoantibody responses against porcine antigens.28,29 The ability to prevent the immune response to porcine islet xenografts could prolong the survival and function of porcine islets in diabetic patients.
In our study, we used fetal porcine islet-like clusters isolated from pancreata obtained from piglets at 66 days of gestation, similar to those used in human diabetic patients treated with porcine islet cells.28 We selected fetal porcine islet cells since they express high levels of the xenoantibody-inducing gal epitope, which adult islet cells do not.16,30,31 The rhesus monkeys produced high levels of IgM and IgG2 subclass antibodies after islet cell injection over a timeframe of 75–90 days. Patients with type 1 diabetes similarly produce high levels of IgM and IgG2 xenospecific antibodies against gal α1,3 gal following exposure to fetal porcine islet cells for extended periods of time (6–8 years) following xenotransplantation.32
We then compared the antibody response following porcine islet cell and heart xenotransplantation. Although transplanted porcine hearts are rejected within 2 hr, removal of the heart leaving a small segment of aorta and pulmonary artery in situ provides a stimulus sufficient to induce a substantial immune response. Elevated serum levels of IgM and IgG, as well as increasing IgG1 xenoantibody levels, were identified at day 11 and remained high for 90 days. The antibody response in our study was found to be comparable to studies performed by other laboratories demonstrating elevated xenoantibody levels induced by pig tissue placed in humans and baboons.33–36
Our investigation of the IgVH genes encoding xenoantibodies in non-immunosuppressed rhesus monkeys demonstrated that identical IgVH genes encode the xenoantibody response to porcine hearts and islet cells. The genes are most closely related to the IGHV3-11 germline progenitor that encodes xenoantibodies in humans. A similar restriction in IgVH gene usage was identified in gal knockout mice.11 Interestingly, the CDR3 region of the immunoglobulin heavy chain that forms the centre of the antigen binding site was also restricted in IgM xenoantibodies induced following exposure to pig cells. IgG xenoantibodies, induced later, were more diverse. Somatic mutations were identified in several sites throughout the genes encoding IgG xenoantibodies, and the CDR3 regions showed no evidence of selective expansion of specific genes.10 Similar results have been reported in humans mounting active xenoantibody responses.37 Since variation in sequence and size of the CDR3 plays a role in antibody diversity38–42 we hypothesize that the unique structure of this region contributes to optimal xenoantibody/gal carbohydrate binding in the early phase of the xenograft rejection. We believe that it is now feasible to apply structure-based drug design to identify reagents that will prevent xenoantibody/carbohydrate-binding based on the structural similarity we have noted in genes encoding xenograft responses. Since identical genes encode xenoantibodies directed at solid organs and isolated pig cells, the application of this methodology functionally to inhibit the binding of this small group of antibodies will be beneficial for preventing the induced humoral immune response directed at different types of xenografts.
Acknowledgments
This work was suppoted by National Institutes of Health grants RO1AI52079 (to M.K.J.) and R21AI49922 (to M.K.J.).
References
- 1.Cooper DK, Gollackner B, Sachs DH. Will the pig solve the transplantation backlog? Annu Rev Med. 2002;53:133–47. doi: 10.1146/annurev.med.53.082901.103900. [DOI] [PubMed] [Google Scholar]
- 2.Platt JL. Xenotransplantation: recent progress and current perspectives. Curr Opin Immunol. 1996;8:721–8. doi: 10.1016/s0952-7915(96)80091-4. [DOI] [PubMed] [Google Scholar]
- 3.Sachs DH. The pig as a potential xenograft donor. Vet Immunol Immunopathol. 1994;43:185–91. doi: 10.1016/0165-2427(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 4.Platt JL. Xenotransplantation: a potential solution to the shortage of donor organs. Transplant Proc. 1997;29:3324–6. doi: 10.1016/s0041-1345(97)00931-7. [DOI] [PubMed] [Google Scholar]
- 5.Galili U. Interaction of the natural anti-Gal antibody with alpha-galactosyl epitopes: a major obstacle for xenotransplantation in humans. Immunol Today. 1993;14:480–2. doi: 10.1016/0167-5699(93)90261-i. [DOI] [PubMed] [Google Scholar]
- 6.Sandrin MS, Vaughan HA, Dabkowski PL, McKenzie IF. Anti-pig IgM antibodies in human serum react predominantly with Gal (alpha1–3) Gal epitopes. Proc Natl Acad Sci USA. 1993;90:11391–5. doi: 10.1073/pnas.90.23.11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanemura M, Yin D, Chong A, Galili U. Differential immune response to carbohydrate epitopes on allo- and xenografts: implications for accommodation. Transplant Proc. 2000;32:991–3. doi: 10.1016/s0041-1345(00)01080-0. [DOI] [PubMed] [Google Scholar]
- 8.Galili U, Swanson K. Gene sequences suggest inactivation of alpha-1, 3-galactosyl-transferase in catarrhines after the divergence of apes from monkeys. Proc Natl Acad Sci USA. 1991;88:7401–4. doi: 10.1073/pnas.88.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galili U, Mandrell RE, Hamadeh RM, Shohet SB, Griffiss JM. Interaction between human natural anti-alpha-galactosyl immunoglobulin G and bacteria of the human flora. Infect Immun. 1988;56:1730–7. doi: 10.1128/iai.56.7.1730-1737.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearns-Jonker M, Swensson J, Ghiuzeli C, Chu W, Osame Y, Starnes V, Cramer DV. The human antibody response to porcine xenoantigens is encoded by IGHV3-11 and IGHV3-74 IgVH germline progenitors. J Immunol. 1999;163:4399–12. [PubMed] [Google Scholar]
- 11.Nozawa S, Xing PX, Wu GD, et al. Characteristics of immunoglobulin gene usage of the xenoantibody binding Gal-alpha(1,3)gal target antigens in the gal knockout mouse. Transplantation. 2001;72:147–55. doi: 10.1097/00007890-200107150-00028. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Sharma A, Chen L, et al. The structure of anti-Gal immunoglobulin genes in naive and stimulated Gal knockout mice. Transplantation. 2001;72:1817–25. doi: 10.1097/00007890-200112150-00018. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Radic MZ, Galili U. Human anti-Gal heavy chain genes. Preferential use of VH3 and the presence of somatic mutations. J Immunol. 1995;155:1276–85. [PubMed] [Google Scholar]
- 14.Cramer DV, Wu GD, Kearns-Jonker M. Synthesis of xenoantibodies at the gene and molecular level. Curr Opin Organ Transpl. 2001;6:42–6. [Google Scholar]
- 15.Cramer DV, Wu GD, Kearns-Jonker M, Gochi E, Wakiyama S, Shirwan H, Borie D. The humoral response to xenografts is controlled by restricted repertoire of immunoglobulin VH genes. Transplantation. 1998;66:1375–83. doi: 10.1097/00007890-199811270-00019. [DOI] [PubMed] [Google Scholar]
- 16.Helmuth EF, Letvin NL, Margolin DH. Germline repertoire of the immunoglobulin VH3 family in rhesus monkeys. Immunogenetics. 2000;51:519–27. doi: 10.1007/s002510000170. [DOI] [PubMed] [Google Scholar]
- 17.Bennet W, Bjorkland A, Sundberg B, Davies H, Liu J, Holgersson J, Korsgren O. A comparison of fetal and adult porcine islets with regard to. Gal Alpha(1,3)Gal expression and the role of human immunoglobulins and complement in islet cell cytotoxicity. Transplantation. 2000;69:1711–17. doi: 10.1097/00007890-200004270-00030. [DOI] [PubMed] [Google Scholar]
- 18.McKenzie IF, Xing PX, Vaughan HA, Prenzoska J, Dabkowski PL, Sandrin MS. Distribution of the major xenoantigen (gal (alpha 1–3) gal) for pig to human xenografts. Transpl Immunol. 1994 2000;2:81–6. doi: 10.1016/0966-3274(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsunoda T, Furui H, Klandorf H, Matsuo S, Soon-Shiong P, Mullen Y. Functional maturation of porcine fetal pancreatic explants. Transplant Proc. 1989;21:2667–8. [PubMed] [Google Scholar]
- 20.Todorov IT, Attaran A, Kearsey S. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–45. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz M, Pallares N, Contet V, Barbi V, Lefranc MP. The human immunoglobulin heavy diversity (IGHD) and joining (IGHJ) segments. Exp Clin Immunogenet. 1999;16:173–84. doi: 10.1159/000019109. [DOI] [PubMed] [Google Scholar]
- 22.Link JM, Hellinger MA, Schroeder HW., Jr The Rhesus monkey immunoglobulin IGHD and IGHJ germline repertoire. Immunogenetics. 2002;54:240–50. doi: 10.1007/s00251-002-0468-2. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz M, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucl Acids Res. 2000;28:219–21. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cramer DV. Natural antibodies and the host immune responses to xenografts. Xenotransplantation. 2000;7:83–92. doi: 10.1034/j.1399-3089.2000.00061.x. [DOI] [PubMed] [Google Scholar]
- 25.Kearns-Jonker M, Fraiman M, Chu W, Gochi E, Michel J, Wu GD, Cramer DV. Xenoantibodies to pig endothelium are expressed in germline configuration and share a conserved immunoglobulin VH gene structure with antibodies to common infectious agents. Transplantation. 1998;65:1515–19. doi: 10.1097/00007890-199806150-00023. [DOI] [PubMed] [Google Scholar]
- 26.Groth CG, Korsgren O, Wennberg L, Song Z, Wu G, Reinholt F, Tibell A. Pig-to-human islet transplantation. Transplant Proc. 1998;30:3809–10. doi: 10.1016/s0041-1345(98)01246-9. [DOI] [PubMed] [Google Scholar]
- 27.MacKenzie DA, Hullett DA, Sollinger HW. Xenogeneic transplantation of porcine islets: an overview. Transplantation. 2003;76:887–91. doi: 10.1097/01.TP.0000087114.18315.17. [DOI] [PubMed] [Google Scholar]
- 28.Groth CG, Korsgren O, Tibell A, et al. Transplantation of porcine fetal pancreas to diabetic patients. Lancet. 1994;344:1402–4. doi: 10.1016/s0140-6736(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 29.Groth CG, Korsgren O, Andersson A, et al. Evidence of xenograft function in a diabetic patient grafted with porcine fetal pancreas. Transplant Proc. 1992;24:972–3. [PubMed] [Google Scholar]
- 30.Rayat GR, Rajotte RV, Hering BJ, Binette TM, Korbutt GS. In vitro and in vivo expression of Galα-(1,3)Gal on porcine islet cells is age dependent. J Endocrinol. 2003;177:127–35. doi: 10.1677/joe.0.1770127. [DOI] [PubMed] [Google Scholar]
- 31.McKenzie IF, Koulmanda M, Mandel TE, Sandrin MS. Expression of Galα(1–3)Gal by porcine islet cells and its relevance to xenotransplantation. Xenotransplantation. 1995;2:139–42. [Google Scholar]
- 32.Lindeborg E, Kumagai-Braesch M, Tibell A, Moller E. Continued production of xenoimmune antibodies 6–8 years after clinical transplantation of fetal pig islet-like cell-clusters. Xenotransplantation. 2001;8:273–83. doi: 10.1034/j.1399-3089.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- 33.Baquerizo A, Mhoyan A, Kearns-Jonker M, Arnaout WS, Shackleton C, Busuttil RW, Demetriou AA, Cramer DV. Characterization of human xenoreactive antibodies in liver failure patients exposed to pig hepatocytes after bioartificial liver treatment: An ex vivo model of pig to human xenotransplantation. Transplantation. 1999;67:5–18. doi: 10.1097/00007890-199901150-00003. [DOI] [PubMed] [Google Scholar]
- 34.Baquerizo A, Mhoyan A, Shirwan H, Swensson J, Busuttil RW, Demetriou AA, Cramer DV. Xenoantibody response of patients with severe acute liver failure exposed to porcine antigens following treatment with a bioartificial liver. Transplant Proc. 1997;29:964–5. doi: 10.1016/s0041-1345(96)00330-2. [DOI] [PubMed] [Google Scholar]
- 35.Dehoux JP, de la Parra B, Latinne D, Bazin H, Gianello P. Characterization of baboon anti-porcine IgG antibodies during acute vascular rejection of porcine kidney xenograft. Xenotransplantation. 2002;9:338–49. doi: 10.1034/j.1399-3089.2002.01090.x. [DOI] [PubMed] [Google Scholar]
- 36.McCurry KR, Parker W, Cotterell AH, et al. Humoral responses to pig-to-baboon cardiac transplantation: Implications for the pathogenesis and treatment of acute vascular rejection and for accommodation. Hum Immunol. 1997;58:91–105. doi: 10.1016/s0198-8859(97)00229-2. [DOI] [PubMed] [Google Scholar]
- 37.Williams GT, Jolly CJ, Kohler J, Neuberger MS. The contribution of somatic hypermutation to the diversity of serum immunoglobulin: dramatic increase with age. Immunity. 2000;13:409–17. doi: 10.1016/s1074-7613(00)00040-6. [DOI] [PubMed] [Google Scholar]
- 38.Abergel C, Tipper JP, Padlan EA. Structural significance of sequence variability in antibody complementarity-determining regions. Res Immunol. 1994;145:49–53. doi: 10.1016/s0923-2494(94)80043-x. [DOI] [PubMed] [Google Scholar]
- 39.Sing GK, Li D, Chen X, Macnaughton T, Lichanska AM, Butterworth L, Ladhams A, Cooksley G. A molecular comparison of T lymphocyte populations infiltrating the liver and circulating in the blood of patients with chronic hepatitis B. Evidence for antigen-driven selection of a public complementarity-determining region 3 (CDR3) motif. Hepatology. 2001;33:1288–98. doi: 10.1053/jhep.2001.24026. [DOI] [PubMed] [Google Scholar]
- 40.Lohr HF, Pingel S, Weyer S, Fritz T, Galle PR. Individual and common antigen-recognition sites of liver-derived T cells in patients with autoimmune hepatitis. Scand J Immunol. 2003;57:384–90. doi: 10.1046/j.1365-3083.2003.01236.x. [DOI] [PubMed] [Google Scholar]
- 41.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 42.Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720–9. [PubMed] [Google Scholar]