Abstract
Vaccinia virus DNA polymerase catalyzes duplex-by-duplex DNA joining reactions in vitro and many features of these recombination reactions are reprised in vivo. This can explain the intimate linkage between virus replication and genetic recombination. However, it is unclear why these apparently ordinary polymerases exhibit this unusual catalytic capacity. In this study, we have used different substrates to perform a detailed investigation of the mechanism of duplex-by-duplex recombination catalyzed by vaccinia DNA polymerase. When homologous, blunt-ended linear duplex substrates are incubated with vaccinia polymerase, in the presence of Mg2+ and dNTPs, the appearance of joint molecules is preceded by the exposure of complementary single-stranded sequences by the proofreading exonuclease. These intermediates anneal to form a population of joint molecules containing hybrid regions flanked by nicks, 1–5 nt gaps, and/or short overhangs. The products are relatively resistant to exonuclease (and polymerase) activity and thus accumulate in joining reactions. Surface plasmon resonance (SPR) measurements showed the enzyme has a relative binding affinity favoring blunt-ended duplexes over molecules bearing 3′-recessed gaps. Recombinant duplexes are the least favored ligands. These data suggest that a particular combination of otherwise ordinary enzymatic and DNA-binding properties, enable poxvirus DNA polymerases to promote duplex joining reactions.
INTRODUCTION
Poxviruses are large double-stranded DNA viruses that, similar to many other virus and bacteriophage, are subjected to very high frequencies of genetic recombination during viral replication (1). Although our understanding of the recombination systems used by most phage and virus is still incomplete, many of these infectious agents encode exonucleases and the activity of these enzymes can play a key role in production of virus recombinants. For example bacteriophage lambda encodes the Red system (comprising a DNA binding protein and 5′–3′ exonuclease encoded by bet and exo genes, respectively), and it is the Red enzymes that catalyze most of the phage general homologous recombination detected in replication-permitted crosses (2). HSV-1 encodes an alkaline exonuclease that seems to serve a similar role in herpes virus recombination (3) and the reverse transcriptase ribonuclease H activity (although not strictly an exonuclease) plays a key role in catalyzing retroviral reverse transcription and recombination (4). Exonucleases are critically important enzymes in most recombination pathways, but where duplicated sequences exist they play a central role in catalyzing what are broadly called ‘single-strand annealing’ (SSA) reactions. This is a non-conservative and widely employed method for producing recombinants (5). In the simplest form of an SSA reaction, an exonuclease first attacks the exposed ends of broken double-stranded DNAs, exposing complementary regions on substrate molecules that can then anneal through Watson–Crick base pairing. This produces heteroduplex spanning the joint and the recombination of sequences flanking the site of hybrid formation. The simplicity of such systems stands in stark contrast to the more complex and often redundant recombination pathways employed by cellular organisms. However, these systems serve other functions in conservative processes such as meiotic recombination (6) and DNA repair (7) that may not be quite so critical for virus biology.
Poxvirus recombination reactions show all the molecular genetic hallmarks of simple SSA reactions including the transient production of abundant hybrid DNA and evidence of processing of joint regions by a 3′–5′ exonuclease (8,9). Poxviruses replicate in the cytoplasm of infected cells and are generally viewed as encoding most or all of the machinery necessary for virus DNA replication. Thus it is notable that the only exonuclease encoded by Chordopoxvirus genomes is that which comprises the 3′–5′ proofreading activity of poxvirus DNA polymerases. Nevertheless this seems to suffice to meet the needs of virus biology, and highly purified vaccinia virus DNA polymerase will catalyze single-strand-by-duplex and duplex-by-duplex joining reactions in vitro (10,11). This enzyme can also repair small gaps and excise 3′ ended single-stranded tails from model joint molecules in vitro (12). These observations show that poxvirus DNA polymerases are endowed with the enzymatic capacity needed to form and (where necessary) repair joint molecules. These non-covalently linked reaction products can then by converted into fully duplex recombinants by the subsequent activity of enzymes like poxvirus or cellular DNA ligases.
What is still unclear is what are the special biochemical properties of poxvirus DNA polymerases that create the capacity to catalyze duplex-by-duplex strand joining. These are seemingly typical B-family DNA polymerases, endowed with 3′–5′ proofreading and 5′–3′ DNA synthetic activities. To our knowledge these are the only members of the B-family of DNA polymerases that can catalyze recombination-like reactions. In this study we have examined this question using, as enzymatic substrates and binding partners, small oligonucleotide duplexes. This permits a fine scale analysis of the kinetics of excision and repair as it relates to the concurrent formation to joint molecules. We have also used surface plasmon resonance methods (SPR) to investigate the binding affinities exhibited by the enzyme towards these substrates and putative reaction products. Our results suggest that the 3′–5′ proofreading exonuclease has a reduced enzymatic preference or binding affinity for the products of these reactions (joint molecules bearing simple nicks or 1–2 nt gaps) relative to the blunt-ended duplexes that are used as substrates. These particular enzymatic properties can do much to explain how poxvirus DNA polymerases could serve a dual role as viral ‘recombinases’.
MATERIALS AND METHODS
Recombinant protein
Vaccinia virus DNA polymerase (>95% purity) was prepared using the procedure of McDonald and Traktman (11,13,14). The method removes the A20R processivity factor and the protein is thus expected to interact with DNA in a distributive manner (15,16).
Radioactive labeling of oligonucleotides
Oligonucleotides were purchased from Sigma-Genosys (Mississauga, Canada) or the DNA core facility, University of Alberta. These DNAs were 5′ end-labeled and purified as described previously (12). Figure 2 illustrates the sequences and structures of the DNAs used in these experiments. The 25mer and 30mer size markers have the same DNA sequence as the predicted products of a 3′-exonuclease attack on labeled 45 and 50mer strands, thus ensuring better accuracy in assigning the sizes of reaction products.
Preparation of double-stranded DNA joint substrates
Duplex substrates were prepared by mixing 0.3 nmol of labeled oligonucleotide with 0.4 nmol of complementary unlabeled oligonucleotide in MMLV reverse transcriptase buffer [50 mM Tris–HCl (pH 8.3), 75 mM KCl and 3 mM MgCl2] in a total volume of 40 μl. The mixture was boiled for 1 min and then cooled slowly to 35°C.
Only one of the two 3′ ends on each of these duplex substrates encodes the homology required by a SSA reaction. To suppress exonucleolytic attack on the other 3′ ended strands, the substrates permit incorporation of a cidofovir residue into these other 3′ ends (17). The cidofovir addition reaction contained the DNA duplexes described above, 20 U of MMLV reverse transcriptase (Invitrogen), 0.1 M dithiothreitol, 100 μM cidofovir diphosphate (synthesized by Trilink Technologies, San Diego), and 50 μM dATP or dTTP (Invitrogen). The reaction was incubated at 37°C overnight, heated to 70°C for 10 min, and the DNA was purified by passage through a Sephadex-25 spin column. In control experiments we noted that this treatment did not completely block exonuclease attack, but served the necessary purpose if reactions were limited to times <30 min (data not shown). Biotinylated oligonucleotides were prepared in the same way except we substituted 40 μM N4-modified biotin-14-dCTP (Invitrogen) for cidofovir diphosphate.
Strand joining reactions
Each 0.13 ml reaction normally contained 0.1 nmol of each duplex substrate, 230 ng vaccinia virus DNA polymerase (∼2 pmol), 15 μM dATP, 15 μM dCTP, 5 μM dGTP, 10 μM TTP, 30 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 70 mM NaCl, 1.8 mM DTT and 80 μg/ml acetylated BSA (10). These starting concentrations of dNTPs were chosen because they are thought to reflect the in vivo concentrations in vaccinia-infected cells (18). The reaction was incubated at 37°C with periodic sampling. Each aliquot was mixed with twice-concentrated gel loading buffer [40% sucrose, 0.1 μg/ml bromophenol blue, 0.1 μg/ml xylene cyanol, 0.2 ug/ml Orange G and 20 mM EDTA (pH 8.0)] to stop the reaction. The reaction products were separated by non-denaturing electrophoresis in 10% polyacrylamide gels, the gels were dried, and the distribution of the label determined using a Storm phosphoimager and ImageQuant software (Amersham).
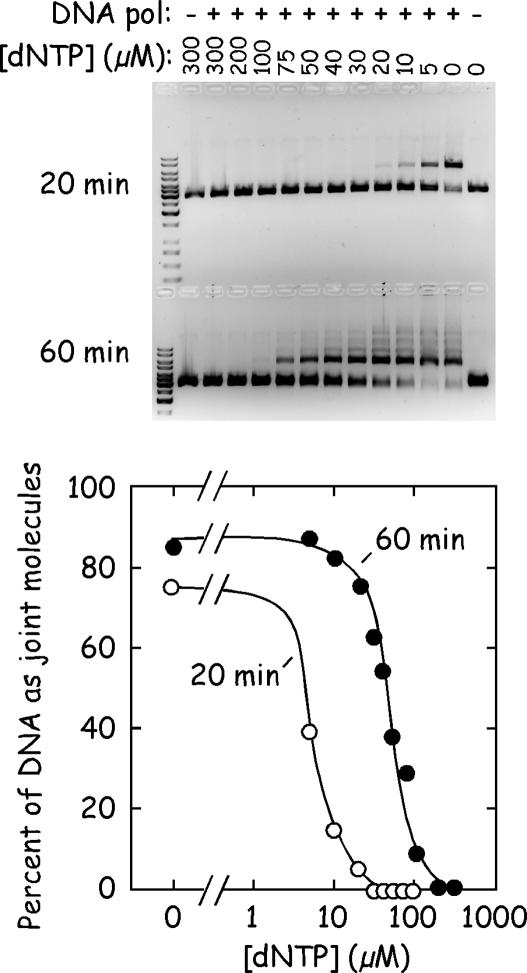
To test what effect the dNTP concentration has on the efficiency of strand joining we used our original agarose-based gel assay (10). Plasmid pBluescript II KS(+) DNAs were cut with XhoI and HindIII (which creates molecules sharing 21 bp of overlapping homology), and equimolar quantities of DNA incubated with the polymerase and single-strand binding proteins in the presence of different concentrations of dNTPs. Substrates and products were separated by agarose gel electrophoresis. These controls showed that higher concentrations of dNTPs inhibit strand joining, but this effect can be suppressed using longer incubation times (Figure 1). Under the simplified conditions used in this study [oligonucleotide substrates (Figure 2), 45 μM dNTP, no single-strand binding protein we noted that the maximum yield of joint molecules is seen after ∼20 min of incubation (Figure 3).]
Figure 1.
Effects of dNTP concentration and reaction time on DNA joining reactions. Standard reactions were prepared as described previously (10) containing an equimolar mixture of XhoI- and HindIII-cut pBluescript II DNAs, vaccinia single-strand binding protein, and vaccinia DNA polymerase. The reactions were supplemented with the indicated amounts of dNTPs. The reactions were incubated for 20 or 60 min and the products separated by agarose gel electrophoresis (upper panel) and quantified using ethidium fluorescence (lower panel). The reaction midpoints are seen at 5 and 50 μM dNTP for 20 and 60 min reactions, respectively.
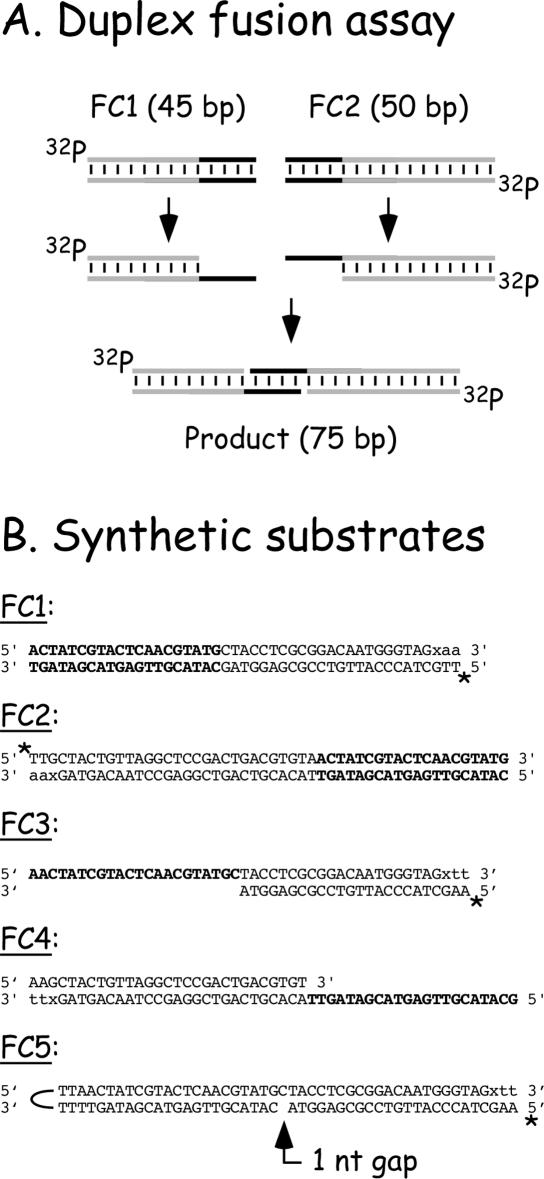
Figure 2.
Experimental scheme and oligonucleotide sequences. (A) Principle behind the joining reaction. By exposing complementary sequences, vaccinia DNA polymerase promotes duplex joint formation. (B) Sequences of the oligonucleotide substrates used in this paper. Homologous sequences are shown in boldface. The nucleotides shown in lower case are incorporated post-synthetically using reverse transcriptase, cidofovir diphosphate or N4-modified biotin-14-dCTP (‘x’), and dATP or dTTP. Note that the primer–template design, in FC3, FC4 and FC5, prevents extension from the other 3′ end by this fill-in reaction. The 32P label is indicated with an asterisk (‘*’).
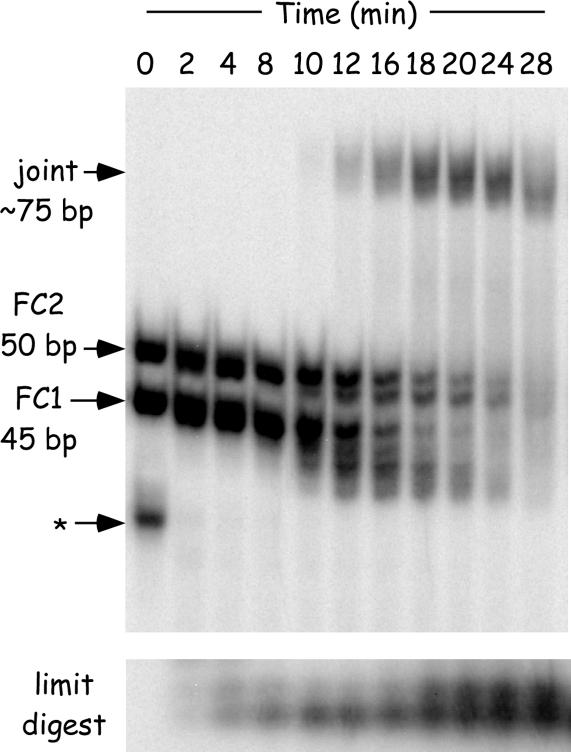
Figure 3.
Native gel analysis of the joining reaction. A joining reaction was prepared containing 32P-labeled substrates FC1 and FC2 and Mg2+ and four dNTPs, incubated at 37°C, and aliquots removed and mixed with EDTA at the indicated times to stop the reaction. Loading buffer was then added and the samples subjected to electrophoresis using a non-denaturing 10% polyacrylamide gel. After drying the gel, autoradiography was used to locate the reaction products. A trace of unannealed strand can be seen at time zero (‘*’). Vaccinia DNA polymerase degrades such single-stranded DNAs to nucleotides within seconds under these reaction conditions (12).
Second dimension analysis
Gels containing the separated products of the joining reaction were stained with methylene blue to visualize the DNA. The lane corresponding to each time point was cut from the gel, sliced into sections, and the DNA extracted by crushing each slice in 200 μl of 0.3 M sodium acetate in 10 mM Tris–HCl, 1 mM EDTA and shaking overnight. The acrylamide was removed by centrifugation through aerosol-resistant pipet tips (Axygen) and the DNA precipitated using glycogen and three volumes of 95% ethanol. The precipitated DNA was washed once with 70% ethanol, dried and resuspended in 10 μl TE. Each sample was mixed with 10 μl of loading buffer [80% deionized formamide, 0.1 μg/ml bromophenol blue, 0.1 μg/ml xylene cyanol and 20 mM EDTA (pH 8.0)]. The samples were then denatured by heating to 56°C and fractionated through a 10% polyacrylamide gel containing 8 M urea and a 19:1 ratio of acrylamide to bis-acrylamide. The gel was fixed with 10% (v/v) acetic acid and 20% (v/v) methanol in water, dried on Whatman paper, and subjected to phosphorimager analysis.
Surface plasmon resonance analysis
Measurements were taken on a Biacore 2000 SPR instrument using control software version 3.1. The sensor surface consisted of a CM5 sensor chip to which a low level [∼200 response units (RU)] of streptavidin had been amine coupled. Biotinylated oligo complexes (FC1, FC3 and FC5) were bound to flow cells 2, 3 and 4 of the sensor chip, respectively, at a density of 50–100 RU. All remaining streptavidin sites were blocked by injection of 5 μM d-biotin, including flow cell 1 (the reference flow cell). Samples were diluted from an enzyme stock into polymerase buffer containing 80 μg/ml bovine serum albumin and stored in a chilled (4°C) sample block prior to injection. Concentrations of 100, 75, 50, 40, 30, 20, 10, 5, 1 and 0.1 nM were injected in duplicate in random order for 2.5 min at a flow rate of 20 μl/min. Regeneration was with 1 M NaCl between cycles. Responses from a control flow cell containing biotin-blocked streptavidin, and a blank injection with polymerase buffer, were subtracted from each dataset to control for bulk refractive index changes. Datasets were aligned, the reference subtracted, and fit to an equilibrium-binding model using Scrubber version 1.1g, BIAevaluation 4.1, or Prism software. The data report standard errors.
RESULTS
Figure 2A shows the assay used to monitor duplex strand joining (‘fusion’) reactions catalyzed by vaccinia virus DNA polymerase. These assays originally used linearized plasmid DNAs and/or PCR amplicons [Figure 1 and Ref. (10)], but in this study we have substituted synthetic oligonucleotide duplexes because using smaller DNAs facilitates high-resolution analysis of the fusion process. We prepared two double-stranded substrates [FC1 (45 bp) and FC2 (50 bp)], which share 20 bp of sequence homology (Figure 2B, boldface text), and should be processed into a 75 bp recombinant product under strand joining conditions. In order to limit the degradation of the duplex substrates to just the ends encoding homologous sequences, the 3′ ends located distal to the regions of homology were protected from exonuclease attack by filling in those ends with molecules of cidofovir (see Materials and Methods). We have previously shown that the polymerase and exonuclease activities of vaccinia DNA polymerase are greatly inhibited by cidofovir located at the penultimate 3′ end of the primer strand (17).
Figure 3 shows the kinetics of conversion of these substrates into recombinant products. Equal amounts of 32P-labeled FC1 and FC2 were incubated with vaccinia DNA polymerase in the presence of physiological concentrations of dNTPs in vaccinia virus infected cells (15 μM dATP, 15 μM dCTP, 5 μM dGTP and 10 μM TTP) (18). The initial stages of the reaction are slow, with little visible change in the migration of the two substrates. A trace quantity of labeled residual single-stranded DNA was, however, degraded immediately. Continued processing of FC1 and FC2 was indicated by an increase in the mobility of the two substrates, followed by the label being chased into larger recombinant products. Between 16 and 24 min, the total amount of DNA migrating as a product was approximately constant, while the amount present as precursors declined by 50% as judged by phosphorimager analysis. During this time, analysis of the phosphorimager signal also showed that ∼20% of the input label was maximally converted into recombinant products under these particular conditions. The gradual blurring and eventual disappearance of these molecules seemed to correlate with the timing of a slow exonuclease attack on the unlabelled, cidofovir-containing, strand. During the later stages of the reaction, all of the DNAs were converted into what appeared to be a limit digest composed of a mixture of di- and tri-nucleotides, which migrated together near the electrophoresis front.
In order to determine the composition of the molecules that were being formed during these reactions, DNA was extracted from regions of the native gel comprising processed substrates and annealed recombinants, and subjected to further analysis by denaturing gel electrophoresis. This involved excising each lane from the native gel, cutting each lane into sections spanning the size ranges from ∼18 to 55 nt, extracting the DNA from each section, and subjecting the recovered DNA to denaturing gel electrophoresis (Figure 4A). In these experiments we also labeled either FC1 or FC2, but not both (as in Figure 3), to simplify the interpretation of the resulting autoradiographs. Figure 4B and C shows the results of this experiment. Overall, both of the labeled strands in FC1 and FC2 were processed with similar kinetics and into similar exonuclease products. There was a slow degradation of the labeled strand at early time points, followed by a more rapid degradation at later time points. A few intermediary digestion products appeared in the precursor lanes; many map near sites where pyrimidine residues are located and which may be natural pause sites for the proofreading exonuclease (12).
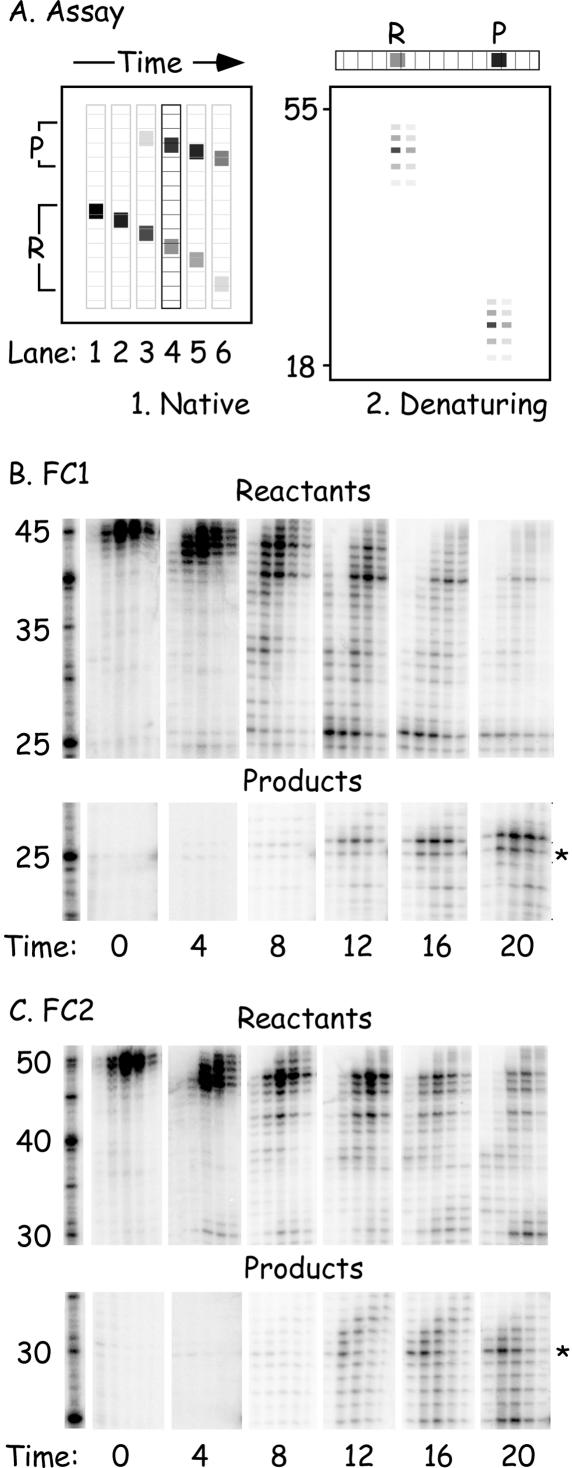
Figure 4.
Two-dimensional electrophoretic analysis of the joining reaction. (A) Shows the principles of the assay. A non-denaturing polyacrylamide gel was used to separate reactants (‘R’) and products (‘P’) and then each lane sliced into sections, the DNA recovered, and separately subjected to denaturing gel electrophoresis. (B and C) Shows portions of the resulting autoradiographs encompassing the majority of the radiolabeled material. Substrate FC1 was labeled in (B), FC2 was labeled in (C). An asterisk (‘*’) marks the location of strands, derived from a recombinant product, where the extent of excision would have permitted formation of a nicked duplex. Larger and smaller excision products would create branched and gapped duplexes, respectively.
The appearance of substantial amounts of recombinant product did not occur until there were processed precursors, which had been degraded to the extent that the homologous regions on both duplexes were nearly fully exposed. At 8 min, we began to see the presence of precursor (‘reactant’) strands that showed evidence of the excision of 19–20 nt. Product molecules started to appear shortly thereafter, about 8–12 min into the reaction. The kinds of excision products that had been incorporated into these recombinant products varied in size. We saw some molecules (marked with a ‘*’ in Figure 4B and C) that would permit the formation of duplexes bearing a simple nick at the junction point. However, other types of 3′-excision products were also present. The largest proportion (∼45%) of the FC1-derived strands were incorporated into the recombinants as 26mer (Figure 4B), which would create a 1 bp overhang when paired with the 5′ end of the FC2-derived strand. Molecules of shorter lengths were also observed, which would create recombinants bearing small (1–5 bp) gaps. The processing of the FC2-derived strand generated a similar variety of excision products, although in this case the predominant species (∼25%) were strands processed to a size that could form nicked recombinants (Figure 4C).
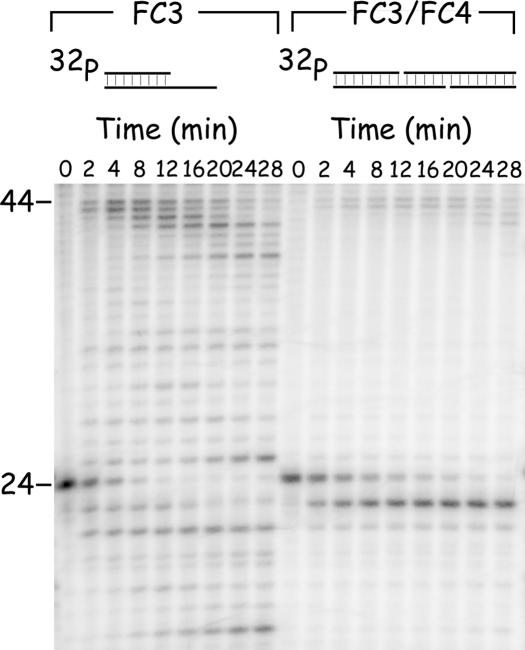
These observations suggested that vaccinia DNA polymerase may have a substrate preference which favors the enzymatic modification of molecules not incorporated into recombinant structures. To test this possibility directly, we constructed two precursor analogs, FC3 and FC4 (Figure 2B), which bear gaps similar in size to those produced during processing of FC1 and FC2. We then compared the rate of exonucleolytic attack upon one of these recessed strands, depending upon whether or not this strand was incorporated into a nicked recombinant duplex. The results are shown in Figure 5. Equimolar amounts of FC3 alone, or FC3 annealed to FC4, were incubated with vaccinia DNA polymerase and the reaction sampled at different time points. The labeled strand being tracked on the gel was the same in both substrates. We observed that the 32P-labeled strand in FC3 was rapidly processed in the absence of FC4, being either extended through DNA synthesis or degraded by the exonuclease. At later stages in the reaction, the extended products were also subjected to limited exonucleolytic attack. In contrast, the target strand in the recombinant structure comprising FC3+FC4 was far less susceptible to enzymatic modification, with most molecules losing only a single nucleotide over the length of the experiment. Similar results were obtained when FC4 was labeled (data not shown).
Figure 5.
Nicked joint products are metastable. A hypothetical gapped recombinant intermediate (FC3) and a reconstructed recombinant product (FC3 annealed to FC4) were treated with vaccinia DNA polymerase in the presence of Mg2+ and four dNTPs. Aliquots were withdrawn and mixed with loading buffer to stop the reaction. The DNA was then subjected to denaturing PAGE and the label imaged by autoradiography.
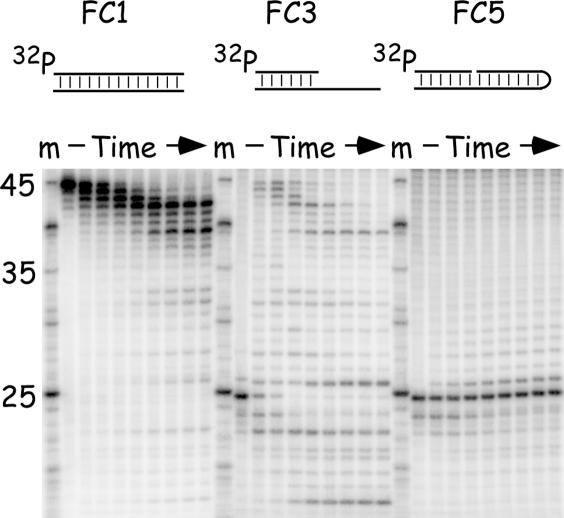
What is the fate of these gapped molecules, regardless of whether they are produced through strand annealing (Figures 3 and 4) or by subsequent exonuclease attack on nicked reaction products (Figure 5)? Liked nicked substrates, molecules bearing small gaps are also relatively stable in the presence of vaccinia DNA polymerase and dNTPs. Figure 6 shows a side-by-side comparison of the substrate properties of FC1, FC3 and FC5 where FC5 comprises a DNA duplex containing a small one-nucleotide gap (Figure 2B, the purpose of the hairpin terminus is explained below). As previously noted, the unprotected 3′ ended strand in FC1 is attacked slowly in the presence of dNTPs (Figure 3) and this 3′ end is an excellent substrate for both the 3′–5′ exonuclease and the DNA polymerase in FC3 (Figure 5). However, the one-nucleotide gap in FC5 is subjected to only limited further modification by vaccinia DNA polymerase, insofar as we detected the excision or addition of only 1–2 nt over the course of these reactions. The majority of molecules suffered no further net modification at all over the 28 min incubation period (Figure 6).
Figure 6.
Comparative substrate properties of molecules used for surface plasmon resonance (SPR) analysis. The three 32P-labeled (and cidofovir-modified) substrates were prepared as described in Materials and Methods and incubated with vaccinia DNA polymerase in the presence of Mg2+ and four dNTPs. Samples were removed at regular intervals (0, 2, 4, 8, 12, 16, 20, 24 and 28 min) and analyzed by denaturing gel electrophoresis followed by autoradiography.
The differences we saw in the experiments detailed above, suggested that vaccinia DNA polymerase has a differential affinity for substrates (linear duplex ends) versus reaction products and that this difference in affinity would effectively favor the formation and stabilization of recombinant products. To test whether this reflected absolute differences in DNA binding properties, we used SPR to directly assess the binding of vaccinia DNA polymerase to three different kinds of molecules. These molecules were FC1, FC3 and FC5. FC1 comprises one of the two reaction substrates and FC3 and FC5 model two examples of reaction products. In order to perform this analysis we had to add a hairpin to the terminus of FC5 (Figure 2B) so as to eliminate a 3′ end that would complicate the analysis of enzyme binding to a second internal 3′ end, located next to the one-nucleotide gap. In order to perform an SPR experiment a method is also required to attach the DNA to the CM5 sensor surface. To do this, the annealed substrates were labeled with biotin (substituting biotinylated dCTP for cidofovir diphosphate) by filling in one end of the duplex using N4 biotin-14-dCTP, dATP (or dTTP), and reverse transcriptase. The sensor surface was coupled with streptavidin, and the biotinylated FC1, FC3 and FC5 molecules attached to three of the four chip surfaces at a low surface density (50–100 RU). The last (fourth) section of the sensor contained no DNA and served as a negative control for non-specific binding to biotin-blocked streptavidin.
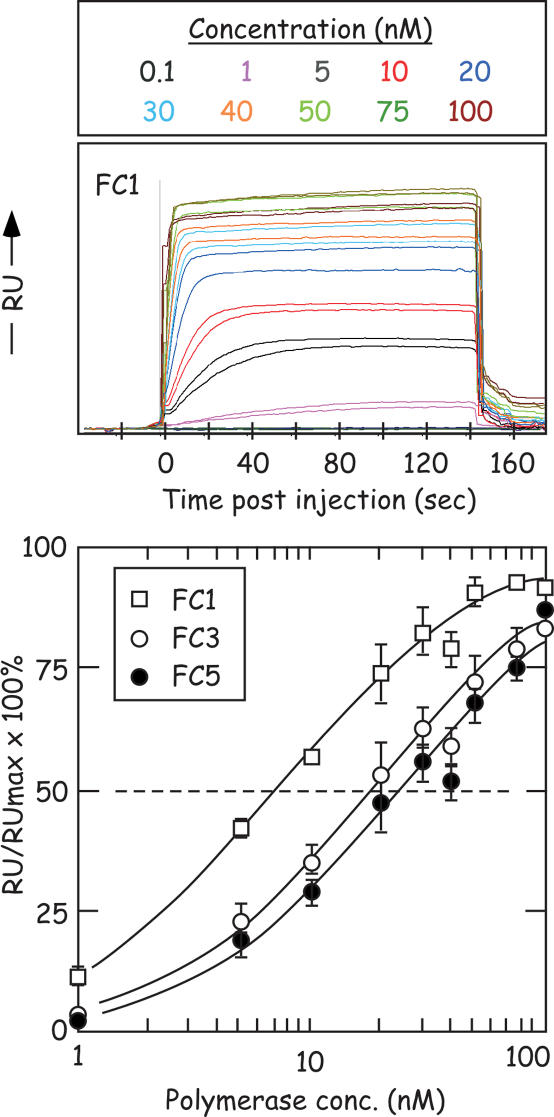
To perform the binding analysis the sensor surface was washed with Mg2+-free polymerase buffer, and then varying concentrations of vaccinia DNA polymerase were passed over the surface in the same buffer for 2.5 min to monitor an association rate. The concentrations of polymerase used ranged from 0.1 to 100 nM (∼0.01–10 μg/ml). Washing with buffer permitted monitoring the dissociation rate. Figure 7 shows the resulting duplicate binding curves, for FC1, after subtracting the reference cell and blank injection signals from the experimental data. By fitting these data to a simple 1:1 equilibrium-binding model we established apparent equilibrium-dissociation constants (KD) for FC1, FC3 and FC5 of 7.0 ± 0.7, 18 ± 2 and 25 ± 4 nM, respectively, using Prism software and BIAeval data points (Figure 7, lower panel). Similar numerical results, and the same trend, were obtained using BIAevaluation and Scrubber software. The three methods of analysis suggest that vaccinia virus DNA polymerase has a relative binding preference for FC1 > FC3 ≥ FC5.
Figure 7.
Vaccinia DNA polymerase binding to FC1, FC3 and FC5. The three different oligonucleotide substrates were fixed to three chambers of a streptavidin-coated chip using a 3′-biotin label as described in Materials and Methods. The upper panel shows the observed changes in the SPR signal [in arbitrary response units (RU)] when buffers containing different concentrations of polymerase were flowed over a chip surface coated with FC1 at 37°C. Each binding curve was measured in duplicate followed by washing with polymerase-free buffer. Magnesium and dNTPs were omitted from the reaction buffer to prevent enzymatic modification of the DNA substrates. The on–off rates are too fast to be measured with satisfactory accuracy by this method, so the KD values were calculated from a curve fit (R2 = 0.98) to the effects of enzyme concentration upon the equilibrium-binding signal (lower panel).
DISCUSSION
Vaccinia DNA polymerase can promote SSA reactions, which lead to the fusion of homologous duplex DNAs in vitro and can be used as a tool for assembling recombinant molecules. Many of the features of the in vitro reaction are also observed when linear DNAs are transfected into poxvirus-infected cells, including the fact that using cidofovir to block exonuclease activity (17) inhibits recombination in vitro and in vivo and this block can be overcome by virus encoding cidofovir-resistant (19) DNA polymerases (D. Gammon and D. Evans, unpublished data). This strongly suggests that the proofreading exonuclease activity of these polymerases is also used by poxviruses to promote double-strand break repair and recombination in vivo. This is an unusual situation, because in all of the other exonuclease-dependent recombination and repair systems characterized to date, a specialized enzyme typically provides the nuclease activity (e.g. RecBCD, RecJ, lambda Red exonuclease, HSV-1 alkaline exonuclease, WRN exonuclease, Mre11, etc.). Poxvirus genomes do not encode such specialized exonucleases and have, instead, seemingly adapted the DNA polymerase to serve this function. However, this raises questions regarding the nature of these enzymatic adaptions. In this study we have examined the behavior of the enzyme when presented with DNAs resembling the substrates, putative intermediates, and representative products of these strand joining reactions. Our observations suggest a straightforward explanation for how vaccinia DNA polymerase might catalyze this unusual reaction.
The DNA joining reactions are readily followed using gel electrophoresis and 45 and 50 bp substrates that are rapidly converted into 75 bp joint molecules (Figure 3). Using high-resolution denaturing gel electrophoresis it was observed that the 3′–5′ exonuclease activity processed the reacting strands (FC1 and FC2) into a variety of gapped molecules, and that a subset of these gapped species were then selectively incorporated into metastable reaction products (Figures 3 and 4). Close inspection of the reaction kinetics shows that exposure of most of the homology precedes the appearance of joint molecules, spontaneous annealing probably then creates joint molecules. Only a small fraction of the reaction products likely bear simple nicks on both sides of the 20 nt region of homology, due to some variability in the extent of strand excision (Figure 4B and C). However the mixture of nicks, short-unpaired extensions, and small gaps do not destabilize the joint molecules enough to inhibit joint molecule formation. Such nicks gaps and extensions would also be readily repaired by bacterial enzymes following the transfection of in vitro reaction products into E.coli, which would explain how the method can be used to produce recombinant clones in bacteria (10).
The transient stability of the duplex joints seen in Figure 3 is probably an important feature of this reaction. The effect is much better illustrated in Figure 5 where we directly compare how vaccinia DNA polymerase processes a nick-containing recombinant duplex (FC3 annealed to FC4) with the effects of the enzyme upon a gapped precursor of this molecule (FC3 alone). The joint molecule is very stable, suffering only limited attack by the exonuclease, whereas FC3 is subjected to a mixture of primer extension and modification by the exonuclease. We have previously noted that duplex molecules bearing longer unpaired 3′ extensions are also rapidly processed by vaccinia DNA polymerase into metastable structures bearing small nicks and gaps (12). These are all reaction features that greatly favor the formation, processing, and transient stabilization of recombinant products.
These observations suggest that vaccinia polymerase might exhibit binding properties that favors binding 3′ ends located within either duplex substrates (FC1) or embedded within a classic primer-template structure (FC3), and disfavors binding to nicks or small gaps. To test this hypothesis we used SPR methods and another substrate that utilizes a hairpin structure to eliminate the 3′-terminus of the duplex end (FC5). FC5 can thus provide a test of the enzyme's affinity for recombinant reaction products, if one makes the not unreasonable assumption that the hairpin structure is an unlikely alternate binding site. Like the nicks in FC3/FC4, the one base gap in FC5 is subjected to only limited enzymatic processing by the polymerase, although in this substrate the one-nucleotide gap in the primer strand mostly suffers no further net modification (Figure 6) rather than suffer a loss of 1 or 2 nt (Figure 5). FC1, FC3 and FC5 were then coupled to the surface of Biacore chip, using a biotin-streptavidin attachment method, and enzyme binding monitored using SPR. We estimate that the 14 atom-linked-biotin tether, plus the length of the attached oligonucleotide (≥22 bp), would place the 3′ end bound by the enzyme at least ∼75 Å from the streptavidin surface. Assuming vaccinia polymerase exhibits dimensions similar to a protein like RB69 DNA polymerase [where the polymerase active site is located ∼30 Å from the protein's edge, measured along the DNA (20)], this distance should well exceed that needed to avoid any steric interference between enzyme and chip surface.
The SPR method clearly showed that vaccinia DNA polymerase has a binding preference most favoring duplex ends and least favoring molecules resembling the typical products of strand joining reactions (i.e. FC1 > FC3 ≥ FC5). Where others have used similar substrates (e.g. a blunt-ended duplex molecule like FC1), but different enzymes, there is still surprisingly good agreement between measured dissociation constants. We measured a KD for vaccinia polymerase of 7.0 ± 0.7 nM using FC1 (95% confidence interval = 5.6–8.4 nM), versus 13 nM reported for human DNA polymerase β using SPR methods and a similar DNA (21). Using stopped-flow methods Otto et al. (22) measured a KD for T4 DNA polymerase of 21 nM. One notable difference between these studies is that stopped-flow methods detect on-off rates for T4 DNA polymerase that are so fast (koff = 9.3 s−1) as to suggest that DNA polymerase binding and dissociation rates should not be readily measurable using SPR methods. This fact is consistent with our observations (Figure 7), but is not consistent with what has been reported for earlier SPR measurements utilizing DNA polymerase β. These authors reported a koff= 0.0099 s−1 for duplex DNA substrates. The origin of this discrepancy in reaction rates is difficult to identify, given the many differences in experimental protocols and the different enzymes used. In particular there are significant differences in buffer composition and reaction temperatures. However, we note that in this earlier SPR study the authors also coupled up to 50-fold more DNA to the chip surface than we did [50–100 RU versus 2500 RU (21)], and this high substrate density might have had a negative effect on reaction rates while not ultimately affecting KD values. Regardless of the cause of this discrepancy, the fact that these KD measurements all cluster between 7 and 21 nM provides some confidence that these methods all measure values close to true KDs. This is actually quite remarkable given the necessity of making several methodological assumptions (including the assumption that all the enzyme is active in these preparations) and the different techniques and enzymes used to calculate dissociation constants.
When the relative binding preferences we have measured are considered within the context of how the enzyme processes homologous DNAs (Figures 3 and 4), it becomes apparent how this enzyme can promote duplex joining reactions in vitro. Vaccinia DNA polymerase has a clear preference for binding duplex ends, which are then processed by 3′–5′ exonuclease into a pool of 3′-gapped molecules. This reaction proceeds even in the presence of dNTPs and may reflect some fundamental difference in the way DNA partitions between exonuclease and polymerase active sites in poxvirus enzymes. If these gaps span complementary sequences, and are long enough to create stable heteroduplex molecules, then these DNAs can anneal spontaneously to form joint molecules. The 3′ ends that are subsumed within the resulting typical hybrid structures are ∼4-fold less-favored enzyme binding sites, compared with blunt ends, and thus this combination of reaction and binding properties will favor the production of metastable joint molecules. In contrast, the proofreading exonucleases of other DNA polymerases have only a limited appetite for properly duplex (as distinct from mismatched) DNA ends in the presence of dNTPs, and thus under our reaction conditions most other polymerases would not generate the gapped molecules from which recombinant duplexes are formed.
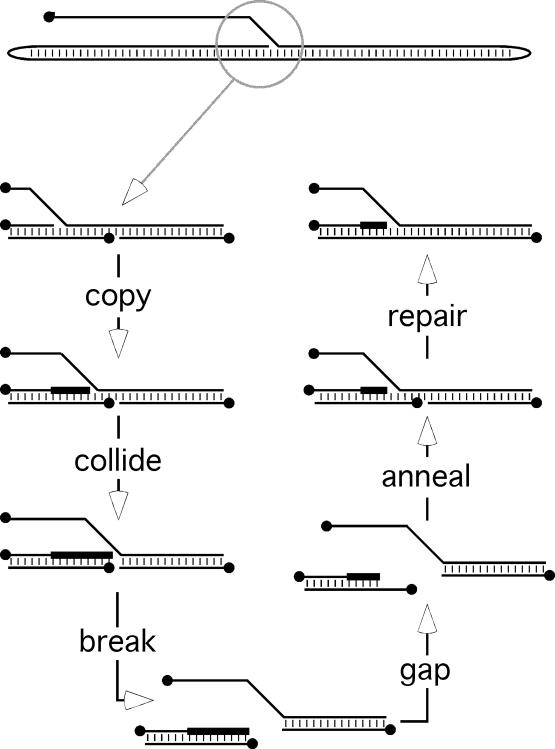
These same reaction features are also likely operative in vivo, including a relative preference for binding and exonuclease processing of duplex ends by the polymerase. However, these reactions would be affected by the activities of other viral replication factors, single-strand binding proteins, and changing dNTP pools. These effects would be difficult to predict a priori. It is clear that the exonuclease (12) and strand joining (Figure 1) activities are both modulated by dNTP concentration in vitro and so this could be a key factor regulating recombination in vivo. Unfortunately we do not understand how dNTP concentrations change temporally and spatially during infection and thus cannot predict exactly what effect these will have on virus recombination in vivo. It is also unclear what proportion of the E9 protein is incorporated into the processive form (16,23) of enzyme complex, by complexing with the A20 and D4 proteins, nor is it certain if this fraction remains stable over time. Nevertheless we would note that even when incorporated into a replication complex, there is no reason to believe the exonuclease would be rendered inactive by A20 or D4. The activity is needed for proofreading and it could also serve an essential purpose in facilitating the repair of broken replication forks (24). One of the difficulties posed by the Moyer–Graves rolling hairpin model for poxvirus replication (25) is that collision of a replication complex with a nick in the template strand would create double-strand breaks in the virus DNA (Figure 8). We have shown that molecules bearing 5′-overhangs next to the region of homology [albeit only small ones have been tested (10)] are still substrates for duplex-by-duplex joining reactions. Consequently, these breaks could be repaired by a stalled replication complex using a SSA reaction coupled with nick ligation. We are currently devising efficient ways of isolating poxvirus replication complexes to test this proposal.
Figure 8.
Production and repair of double-strand breaks formed during poxvirus DNA replication. The Moyer–Graves model proposes that replication starts near a hairpin telomere and the polymerase then copies the genome in the manner shown at the top of the figure. If this replicating complex were to collide with a nick in the template strand, in a manner best understood in the context of bacterial systems (24), it would break the DNA. Such breaks could be repaired using the method shown. Black dots indicate 5′ ended strands and thicker lines are newly synthesized DNA.
Acknowledgments
We thank J. Booth and H. Jenkins for excellent technical assistance and Dr Karl Hostetler for the gift of cidofovir diphosphate. The research was supported by funding from the AHFMR, NSERC and CIHR (to D.E.) and by NIH award N01-AI-15436 (to R.M.B.). A.N. is a recipient of an American Heart Association pre-doctoral fellowship and D.G. is an NSERC Centennial Scholar. Funding to pay the Open Access publication charges for this article were provided by CIHR (MOP-10923).
Conflict of interest statement. None declared.
REFERENCES
- 1.Evans D.H., Stuart D., McFadden G. High levels of genetic recombination among cotransfected plasmid DNAs in poxvirus-infected mammalian cells. J. Virol. 1988;62:367–375. doi: 10.1128/jvi.62.2.367-375.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karakousis G., Ye N., Li Z., Chiu S.K., Reddy G., Radding C.M. The beta protein of phage lambda binds preferentially to an intermediate in DNA renaturation. J. Mol. Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 3.Reuven N.B., Willcox S., Griffith J.D., Weller S.K. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 2004;342:57–71. doi: 10.1016/j.jmb.2004.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peliska J.A., Benkovic S.J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 5.Pastink A., Eeken J.C., Lohman P.H. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 6.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreuzer K.N. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 2005;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- 8.Fisher C., Parks R.J., Lauzon M.L., Evans D.H. Heteroduplex DNA formation is associated with replication and recombination in poxvirus-infected cells. Genetics. 1991;129:7–18. doi: 10.1093/genetics/129.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao X.D., Evans D.H. Characterization of the recombinant joints formed by single-strand annealing reactions in vaccinia virus-infected cells. Virology. 2003;308:147–156. doi: 10.1016/s0042-6822(02)00089-2. [DOI] [PubMed] [Google Scholar]
- 10.Willer D.O., Yao X.D., Mann M.J., Evans D.H. In vitro concatemer formation catalyzed by vaccinia virus DNA polymerase. Virology. 2000;278:562–569. doi: 10.1006/viro.2000.0686. [DOI] [PubMed] [Google Scholar]
- 11.Willer D.O., Mann M.J., Zhang W., Evans D.H. Vaccinia virus DNA polymerase promotes DNA pairing and strand-transfer reactions. Virology. 1999;257:511–523. doi: 10.1006/viro.1999.9705. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton M.D., Evans D.H. Enzymatic processing of replication and recombination intermediates by the vaccinia virus DNA polymerase. Nucleic Acids Res. 2005;33:2259–2268. doi: 10.1093/nar/gki525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald W.F., Traktman P. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr. Purif. 1994;5:409–421. doi: 10.1006/prep.1994.1059. [DOI] [PubMed] [Google Scholar]
- 14.Tseng M., Palaniyar N., Zhang W., Evans D.H. DNA binding, aggregation, and annealing properties of the vaccinia virus I3L gene product. J. Biol. Chem. 1999;274:21637–21644. doi: 10.1074/jbc.274.31.21637. [DOI] [PubMed] [Google Scholar]
- 15.McDonald W.F., Traktman P. Vaccinia virus DNA polymerase. In. vitro analysis of parameters affecting processivity. J. Biol. Chem. 1994;269:31190–31197. [PubMed] [Google Scholar]
- 16.Klemperer N., McDonald W., Boyle K., Unger B., Traktman P. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J. Virol. 2001;75:12298–12307. doi: 10.1128/JVI.75.24.12298-12307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magee W.C., Hostetler K.Y., Evans D.H. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 2005;49:3153–3162. doi: 10.1128/AAC.49.8.3153-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendricks S.P., Mathews C.K. Allosteric regulation of vaccinia virus ribonucleotide reductase, analyzed by simultaneous monitoring of its four activities. J. Biol. Chem. 1998;273:29512–29518. doi: 10.1074/jbc.273.45.29512. [DOI] [PubMed] [Google Scholar]
- 19.Andrei G., Gammon D.B., Fiten P., De Clercq E., Opdenakker G., Snoeck R., Evans D.H. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 2006;80:9391–9401. doi: 10.1128/JVI.00605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 21.Tsoi P.Y., Yang M. Kinetic study of various binding modes between human DNA polymerase beta and different DNA substrates by surface-plasmon-resonance biosensor. Biochem. J. 2002;361:317–325. doi: 10.1042/0264-6021:3610317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto M.R., Bloom L.B., Goodman M.F., Beechem J.M. Stopped-flow fluorescence study of precatalytic primer strand base-unstacking transitions in the exonuclease cleft of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10156–10163. doi: 10.1021/bi9800754. [DOI] [PubMed] [Google Scholar]
- 23.Stanitsa E.S., Arps L., Traktman P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- 24.Cox M.M. Historical overview: searching for replication help in all of the rec places. Proc. Natl Acad. Sci. USA. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer R.W., Graves R.L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]