Abstract
Fragile X syndrome, the most frequent form of inherited mental retardation, is due to the absence of expression of the Fragile X Mental Retardation Protein (FMRP), an RNA binding protein with high specificity for G-quartet RNA structure. FMRP is involved in several steps of mRNA metabolism: nucleocytoplasmic trafficking, translational control and transport along dendrites in neurons. Fragile X Related Protein 1 (FXR1P), a homologue and interactor of FMRP, has been postulated to have a function similar to FMRP, leading to the hypothesis that it can compensate for the absence of FMRP in Fragile X patients. Here we analyze the ability of three isoforms of FXR1P, expressed in different tissues, to bind G-quartet RNA structure specifically. Only the longest FXR1P isoform was found to be able to bind specifically the G-quartet RNA, albeit with a lower affinity as compared to FMRP, whereas the other two isoforms negatively regulate the affinity of FMRP for G-quartet RNA. This result is important to decipher the molecular basis of fragile X syndrome, through the understanding of FMRP action in the context of its multimolecular complex in different tissues. In addition, we show that the action of FXR1P is synergistic rather than compensatory for FMRP function.
INTRODUCTION
Fragile X related genes are members of a small gene family whose founding member is the Fragile X Mental Retardation 1 gene (FMR1). Inactivation of FMR1 causes Fragile X syndrome, the most common cause of inherited mental retardation (1,2). The other members of this family, FXR1 and FXR2, are autosomal and have not been associated so far with any human disease (2–4). Animal models have been generated for Fmr1 deficiency, recapitulating the phenotype of Fragile X syndrome (5,6). Fxr2 null mice are viable and show some behavioral phenotypes, such as hyperactivity, similar to those observed in Fmr1 knockout mice (7). Fxr1 null mice die shortly after birth most likely because of heart and/or respiratory failure due to alterations in muscle development (8). In Xenopus, complete or partial inactivation of xFxr1 expression has dramatic muscle-specific effects (9). In vertebrates, members of the FXR protein family are structurally very similar and share a high degree of sequence homology in clustered regions corresponding to functional domains (2–4). Like FMRP, FXR1P contains several RNA binding domains: two KH domains and one RGG box. It also contains a nuclear localization signal (NLS), a nuclear export signal (NES) and a protein–protein interaction domain (2,10). They also share the same gene structure, derived from their common ancestor in Drosophila melanogaster (11). FXR proteins are able to bind RNA (3,4), but binding specificity has been studied in detail only for FMRP. Indeed, even if a few hundreds of different RNAs have been proposed to be putative targets of FMRP, only two structures are specifically bound by this protein, the G-quartet and the kissing complex (12–14) and one sequence, a poly(U) stretch (15). FXR1P has been reported to bind AU rich element (ARE) and, through the interaction with this element, to regulate the expression of the proinflammatory cytokine tumor necrosis factor (TNFα) in macrophages (16). In the cytoplasm the three FXR proteins are associated with polyribosomes (17), while they share only two interacting proteins, CYFIP2 and MSP58, with FMRP (2,18,19). The FXR1 primary transcript is alternatively spliced, with the possibility to generate upto 15 isoforms (20), see also www.ncbi.nlm.nih.gov/IEB/Research/Acembly/. Of notice, some of these isoforms are differentially expressed in various tissues (21). Up to date, the ability of full-length FXR1P and FXR2P to bind a G-quartet RNA structure in a specific manner has not been reported. We analyzed here the RNA binding properties of the three most abundantly expressed FXR1P isoforms and show that they have different affinities for the G-quartet RNA structure. Since all protein members of the FXR family are able to heterodimerize with FMRP, they are believed to act together (4). In the present study we determined that, when complexed to FMRP, FXR1P isoforms can modulate its affinity for G-quartet RNA and also the dynamics of this complex. Our data demonstrate that FXR1P has a synergistic molecular function with FMRP rather than a redundant role.
MATERIALS AND METHODS
Purification of recombinant proteins
Glutathione S-transferase (GST)-FMRP produced in the baculovirus system was purified as described previously (22). pET21a/FMRP (ISO1) vector was described previously (23). To construct pGex-4T-1/FMRP, ISO1 cDNA was excised from pTL1/FMRP ISO1 and subcloned into the EcoRI/NotI sites of pGex-4T-1 (Amersham). To construct pET21a/FXR1P, Isoe, Isod and Isoa isoforms were amplified by PCR using the primers (Eurogentec): EcoRI forward-5′-GGCGAATTCATGGCGGACGTGACGGTG-3′; XhoI reverse-5′-GCCCTCGAGTTATGAAACACCATTCAGGAC -3′, the PCR consisted of 1 cycle at 94°C for 4 min, 30 cycles of three steps each, 94°C for 30 s followed by 60°C for 30 s and 68°C for 2 min using the Pfx polymerase (Invitrogen). PCR fragments were purified, digested and cloned into the EcoRI/XhoI sites of pET21a (Novagen). The sequences of the cDNAs corresponding to the different FXR1P isoforms were verified by sequencing. The proteins were produced in bacteria and purified following the manufacturer's protocol. GST-MSP58 was produced and purified as described previously (19).
GST-pull down
GST-pull down assays were performed as described previously (22). Briefly, an increasing amount of recombinant His-FXR1P (1, 2 or 4 μM) was mixed with 4 μM of GST-FMRP. Pull down assays were carried out in the following buffer: [50 mM Tris–HCl (pH 7.4) at 4°C, 1 mM MgCl2, 1 mM EDTA, 150 mM KCl, 1 mM DTT], as described (22). After washing with the same buffer, the proteins bound to the beads and their interactors were eluted using 30 mM glutathione and separated by electrophoresis on 8% SDS–polyacrylamide gels. FMRP was visualized by immunoblot using the 1C3 monoclonal antibody (24), FXR1P was revealed by the 3FX monoclonal antibody (21). The proteins were also visualized on gel by Coomassie staining.
RNA binding assays
The different RNA fragments used in this study, N19 [RNA sequence derived from FMR1 cDNA and containing a G-quartet forming structure (13)] and N8 [RNA sequence not containing G-quartet structures and corresponding to the 3′-untranslated region (3′-UTR) of PP2Ac (25)], were cloned in pTL1 plasmid. For filter binding assay, pTL1 plasmids linearized with PstI were in vitro transcripbed with T7 RNA polymerase (Promega) (13). The RNAs were purified using the NucAway Spin columns (Ambion). RNAs were then ethanol precipitated and resuspended in a appropriate buffer. For binding experiments, N19 was labeled co-transcriptionally by incorporation of [α-32P]ATP. Labeled RNAs were purified on a 1% low-melting agarose gel (Ambion). Labeled RNAs (80 000 c.p.m., 5 fmol) were renatured for 10 min at 40°C in 4 μl of binding buffer [50 mM Tris–HCl (pH 7.4) at 4°C, 1 mM MgCl2, 1 mM EDTA, 150 mM KCl, 1 mM DTT] in the presence of 8 U RNasin (Invitrogen), 0, 1 μg of Escherichia coli total tRNA and 0.01% BSA. An increasing amount of protein was then added to the RNA. RNA–protein complexes were formed for 10 min on ice. After incubation, binding solutions were passed through MF-membrane filters (0.45 HA, Millipore) and washed with 2 ml binding buffer. Filters were air dried and the amount of radioactivity was measured by Cerenkov counting. Data were plotted as percentage of total RNA bound versus the protein concentration. Competition experiments to determine the relative binding strength of the different proteins to G-quartet RNA were carried out using labeled N19 RNA incubated with 1 pmol of protein in the presence of increasing concentrations of unlabeled competitors. FMRP was used as an internal positive control. For association rate determination, 5 fmol of labeled N19 were incubated with 1 pmol of the appropriate protein in the binding buffer between 10 and 300 min on ice. For dissociation rate determination, 5 fmol of labeled N19 were incubated with 1 pmol of the appropriate protein in the binding buffer for 10 min on ice, 10−6 M of competitor RNA (N19 or N8) were then added to the mixture and incubated between 10 and 300 min. Each binding curve is the result of at least three independent experiments performed with three replicates for each binding point. All data obtained for the different experiments of RNA binding, calculating the standard deviation for each binding point, are shown in Supplementary Data. All the values and curves were analyzed using the PRISM Graphpad version 4 Software.
RESULTS
Our first aim was to assess whether FXR1P is able to bind G-quartet RNA structure, which is considered to be a frequent structure recognized by FMRP and present in many of its mRNA targets (12,13,26). Due to extensive alternative splicing of FXR1 mRNA, at least seven isoforms of FXR1P are differentially expressed in various tissues (20). We decided to study the RNA binding properties of three FXR1P isoforms: Isod and Isoa (Figure 1), the two isoforms most highly expressed in brain (3), and Isoe (Figure 1) that is a FXR1P isoform highly expressed during myogenesis and in adult cardiac and skeletal muscle (21). The FXR1P-Isod and Isoa isoforms both lack exon 12 and 15 and only differ in their C-terminus due to the choice of a different splicing acceptor site in the mRNA of the FXR1P-Isoa isoform, resulting in a frameshift that induces an early stop codon (Figure 1A). On the other side, it is interesting to underline that the only differences between FXR1P-Isod and Isoe isoforms are the insertion of 28 amino acid encoded by exon 12 and the presence of 27 amino acid encoded by exon 15 (20,21) (Figure 1A). This 27 amino acid stretch is strongly recognized as a putative RNA binding motif by two different predictive programs available online [http://bindr.gdcb.iastate.edu/RNABindR/main.aspx (27) and http://129.130.115.77/cgi-bin/bindn.pl (28)], whereas the sequence of exon 12 does not apparently display such properties. The presence or absence of exon 15 raises then the possibility that the 3 isoforms share different RNA binding abilities.
Figure 1.
The FXR1P isoforms. (A) Schematic representation of the C-terminal region of the three FXR1P isoforms analyzed: Isoe 84 kDa, Isod 78 kDa, Isoa 70 kDa. In the upper part of the figure the alternatively spliced sequences are indicated. A (+) under each amino acid indicates the predicted ability of the sequence to bind RNA accordingly to the algorithm described by Terribilini and coworkers (28). (B) Production of recombinant proteins. Equal amounts of Hist-FMRP, His-FXR1P-Isoe, Isod, Isoa and GST-MSP58 were loaded on a 10% SDS–PAGE gel and revealed by Coomassie blue staining.
As the tissue distribution of FXR1P isoforms had not been investigated completely, we performed RT–PCR on various RNA samples extracted from cell lines and tissues. FXR1P containing exon 15 RNA was detected at very low level in brain, and in particular in the cerebellum, cortex and hippocampus, as well as in the neuroblastoma cell line NG108, together with FXR1P isoforms lacking exon 15 (data not shown).
To investigate the G-quartet binding properties of the three FXR1P isoforms, we generated in a bacterial system recombinant FXR1P isoforms: Isoe, Isod and Isoa (Figure 1A) (3,21), FMRP ISO1 (29) and as a control MSP58, a recently described G-quartet binding protein (19), tagged with His or GST (Figure 1B).
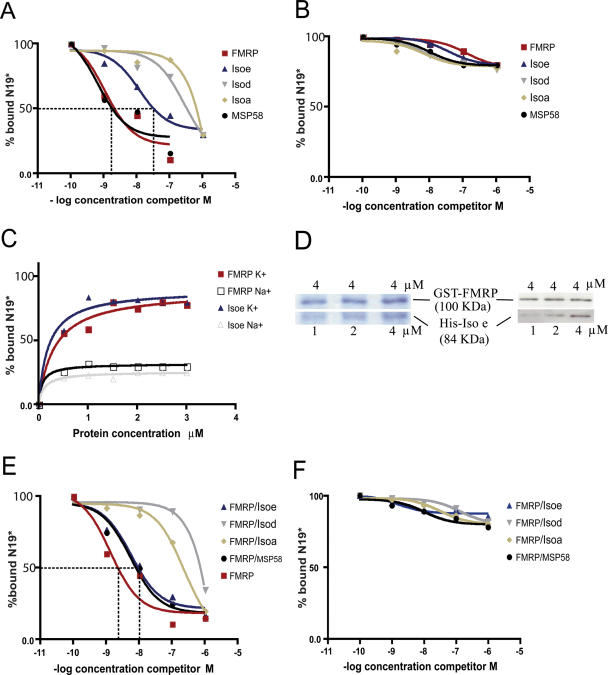
In a filter binding assay, recombinant FMRP protein produced in bacteria displays the same affinity for RNA containing a G-quartet structure as recombinant FMRP produced in an insect cell system (data not shown), confirming that the system of production does not change FMRP affinity for G-quartet RNA, in agreement with studies by Darnell and colleagues (30). Also it has been shown that FMRP acts as a nucleic acids chaperone in low-salt binding conditions (31) and is also able to bind RNA non-specifically, raising the possibility of introduction of a bias in the assesment of its binding affinities, as already suggested (13,32). Considering the high level of homology that exists between the FXR proteins (4), we reasoned that FXR1P could also display the same properties of aspecific binding to RNA. As a result, to assess the RNA binding properties of FXR1P isoforms, we used the rigorous and sensitive RNA competition assays, which alleviate the contribution of aspecific binding (13,25). Using the previously described filter binding assay (30), we observed that FXR1P-Isod and Isoa isoforms do not bind specifically G-quartet RNA structure since the amount of bound G-quartet radiolabeled probe is not competed by either the unlabeled G-quartet RNA [N19, corresponding to the portion of the FMR1 transcript containing the G-quartet structure (13)] or another RNA not containing G-quartet structures and not binding FMRP [N8, corresponding to the 3′-UTR of the PP2Ac transcript (25)] (Figure 2B). Indeed, at the equilibrium state, the dissociation constant (Kd) is around 5 μM for FXR1P-Isod isoform and 0.8 μM for FXR1P-Isoa. Conversely, FXR1P-Isoe binds G-quartet RNA but with a lower affinity compared to FMRP or MSP58 (Figure 2A). As little as 1 nM of competitor RNA is able to displace 50% of FMRP from G-quartet labeled probe, whereas ∼10 nM are necessary for FXR1P-Isoe (Figure 2A). When we used as competitor the N8 probe, that does not bind FMRP (25), no binding was observed for all proteins analyzed here (Figure 2B). To confirm that FXR1P-Isoe interaction with G-quartet RNA is specific for the structure, we performed the binding assay either in the presence of K+ or in the presence of Na+. Indeed, FXR1P-Isoe, like FMRP and MSP58 (13,19) is unable to bind G-quartet containing FMR1 RNA in the presence of Na+, a cation destabilizing the G-quartet structure (Figure 2C). This finding suggests that the effect observed is not due to the recognition of a specific RNA sequence, but to the G-quartet structure localized in the assayed RNAs (13). In addition, we repeated the same analysis by competing the binding of 32P-labeled N19 probe with the G-quartet forming RNA structures obtained from the 5′-UTR of PP2Ac (that contains four G-quartet forming structures) (25) and obtained the same results (data not shown) that are described in Figure 2A using N19 RNA competition.
Figure 2.
RNA binding properties of FMRP and FXR1P isoforms. (A) Filter binding assay using FMRP, FXR1P-Isoe, Isod, Isoa and MSP58. The RNA probe used is 32P-labeled N19 RNA, and competition was performed using the same unlabeled RNA. (B) The same experiment was repeated using as competitor the N8 RNA sequence, not containaing any G-quartet forming structure. (C) Filter binding assay was repeated with an increasing amount of FMRP and Isoe in the presence of Na+ or K+. (D) GST-pull down was performed as described in Materials and Methods. On the right part of (D), proteins used in GST-pull down assay were revealed by immunoblot. FMRP was detected by monoclonal 1C3 antibody, FXR1P by the monoclonal 3FX antibody. Lane 1: 4 μM GST-FMRP complexed with 1 μM His-FXR1P, Lane 2: 4 μM GST-FMRP complexed with 2 μM His-FXR1P, Lane 3: 4 μM GST-FMRP complexed with 4 μM His-FXR1P. On the left part of (D), proteins used in GST-pull down experiment were revealed by Coomassie stained gel. (E) Competition assay to determine the Kd at the equilibrium state binding FMRP, the heterodimers FMRP/Isoe, FMRP/Isod, FMRP/Isoa and the complex FMRP/MSP58, with the 32P-labeled N19 probe and competed with unlabeled N19. (F) The same experiment described in (E) was repeated using the N8 RNA as unlabeled competitor.
Since FXR1P isoforms display different G-quartet binding specificity, we asked whether they would affect differently FMRP binding affinity to G-quartet. First, we verified the amounts of FXR1P and FMRP that integrate the heterodimer complex. To this purpose, we performed GST-pull down experiments by mixing 4 μM of GST-FMRP with increasing amounts (1, 2, 4 μM ) of His-FXR1P-Isoe, Isod or Isoa (In Figure 2D, the results are shown only for interaction between FMRP and FXR1P-Isoe). The beads were then treated with glutathione and the eluted proteins were revealed by immunoblot using the two monoclonal antibodies 1C3 (24) and 3FX (21) recognizing FMRP and FXR1P, respectively. As shown in Figure 2D, the ratio of released FXR1P and FMRP is around 1:1 when mixed in stochiometric amounts. This result shows that when the two proteins are mixed in vitro their association is dose-dependent and, on the other side, also shows that our results are not due to an unbalanced ratio of the two interacting proteins in the FXR heterodimers.
Subsequently, we tested the ability of the FXR1P-Isoe/FMRP heterodimer to bind G-quartet RNA. Indeed, the FXR1P-Isoe/FMRP complex binds G-quartet RNA with a comparable affinity as the FMRP homodimer at different concentrations of competitor RNA (Figure 2E). Surprisingly, FXR1P-Isod and Isoa inhibit FMRP binding to G-quartet RNA when these form a heterodimer with the latter protein (Figure 2E). As a control, MSP58 protein, that binds G-quartet RNA in a specific manner (19), was used (Figure 2E), and its binding to FMRP leads to the same results as when FMRP is complexed to FXR1P-Isoe. When we used the N8 probe as a negative control, no displacement of the equilibrium was observed, as shown in Figure 2F.
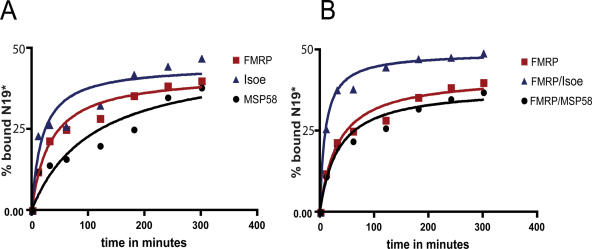
In view of these results, we decided to better dissect the dynamics of FXR1P/G-quartet RNA and FXR1P/FMRP/G-quartet RNA interactions. We evaluated the velocity of interaction of the two FXR proteins with G-quartet RNA. For this purpose 1 pmol of each protein was mixed with 5 fmol of labeled N19 RNA. At different time points (10, 30, 60, 120, 240 or 300 min), the assay was stopped and the amount of radioactivity bound by the proteins evaluated by the filter binding assay. The time necessary for FMRP/FXR1P-Isoe heterodimer to bind the half amount of total bound RNA ligand was estimated to be 9.93 min, which is lower than the time employed by FMRP or FXR1P homodimers (33.65 and 19.04 min, respectively) to bind the same amount of RNA probe (Figure 3A and B ). This result indicates that the Kon for the heterodimer is higher than the Kon for both homodimers. Conversely, the presence of MSP58 complexed with FMRP did not influence its binding to G-quartet RNA.
Figure 3.
Association rate of FMRP, Isoe and MSP58. (A) Each protein was mixed with 32P-labeled N19 RNA probe for a time lapse of 10, 30, 60, 120, 180, 240 and 300 min and then each reaction was filtered and the amount of retained radioactivity evaluated. (B) The same experiment described in (A) was repeated with the complex FMRP/MSP58 and FMRP/Isoe as indicated in the figure.
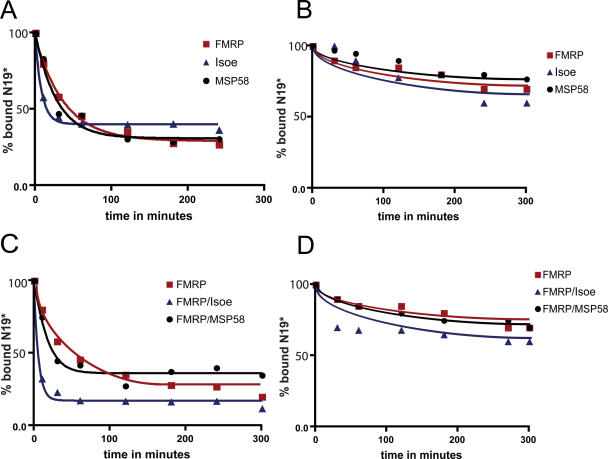
We then investigated the kinetics of FXR1P-Isoe and FMRP dissociation from the G-quartet RNA structure, when both bind to it as homodimers or as heterodimers. FMRP was mixed with 32P-labeled N19 RNA and the complex was allowed to form for 10 min on ice. Then 1 μM of cold RNA [N19 or N8, as a negative control (25)] was added, the reaction was stopped after 10, 30, 60, 120, 240 or 300 min and the amounts of retained radioactivity evaluated by filter binding assay. In this experiment, it is interesting to observe (Figure 4A) that the FXR1P-Isoe homodimer releases 50% of total bound RNA after only 15 min. As expected from its higher affinity, the FMRP homodimer releases the same amount of bound RNA after a longer lapse of time, around 45 min. The negative controls are shown in Figure 4B. In this case the binding of recombinant proteins to G-quartet containing RNA was competed using the N8 probe. Indeed, only 20–25% of the binding is competed after more than 4 h of incubation.
Figure 4.
Dissociation rate of FMRP, Isoe, MSP58 and the two complexes FMRP/Isoe and FMRP/MSP58. (A) Each protein was mixed with 32P-labeled N19 RNA probe for 10 min on ice and then an equal amount of unlabeled N19 RNA was added as competitor to each reaction, which was then filtered after a precise time lapse of 10, 30, 60, 120, 180, 240 and 300 min. The radioactivity retained on the filter was evaluated. (B) The same experiment described in (A) was repeated using unlabeled N8 RNA as competitor. (C) The same experiment described in (A) was performed using the protein complexes FMRP/Isoe and FMRP/MSP58. (D) The same experiment described in (C) was performed using the cold N8 RNA as competitor.
Finally, the heterodimer FXR1P-Isoe/FMRP and the complex MSP58/FMRP were analyzed. The effect of the heterodimer was dramatic: 50% of the labeled RNA was released after 5 min and 65% of labeled RNA was released after 10 min (Figure 4C), while the interaction with MSP58 does not change significantly the dynamics of the interaction between the G-quartet RNA and FMRP. In Figure 4D the negative controls are shown, confirming the specificity of the action described in Figure 4C. This data shows that formation of the FXR1P-Isoe/FMRP heterodimer increases the dynamics of protein–RNA interaction, favoring the release of bound mRNA.
DISCUSSION
FMRP is a component of multimolecular complexes involved in different steps of mRNA metabolism (2,12). A growing list of proteins interacting with FMRP has been described, most of them being RNA binding proteins (19,33). In addition, several hundreds of mRNAs have been described as putative targets of FMRP (12,26), however the functional significance of most of the multiple interactions established by FMRP is still elusive (12,34). A widely accepted hypothesis proposes that FMRP can transport mRNA in mRNPs shuttling between structures where RNA translation is repressed and polyribosomes (2). In the absence of FMRP, the equilibrium in mRNP normally containing FMRP is perturbed, resulting in the deregulation of the expression and localization of a subset of its target mRNAs (12,26). Based on these considerations, we reasoned that RNA binding proteins belonging to the same mRNP complex, FMRP and its interacting proteins may enter in contact with the same mRNAs and decided to test the ability of FXR1P to bind the same mRNA targets and to influence its affinity for them. Up to date, the functions of FXR1P (or FXR2P) were inferred to be, by homology and analogy, similar to that of FMRP (1). Here we propose that it is not the case, at least for FXR1P.
First, we tested FXR1P affinity for G-quartet forming RNA structures. It is surprising that among the three isoforms analyzed only one, the Isoe is able to bind a G-quartet RNA forming structure, present in a large amount of putative target RNAs of FMRP (26). The three FXR1P isoforms share the same RGG box domain. However, it has been reported that, even though a peptide corresponding to the RGG box of FMRP binds specifically G-quartet forming RNA (30), the corresponding peptide of the RGG box of FXR1P does not (35), strongly suggesting that the RGG box of FXR1P is not sufficient per se to bind the G-quartet structure. The only difference between the two isoforms Isoe and Isod are two short sequences of 28 (exon 12) and 27 amino acid (exon 15) (cf. Figure 1A). Only this latter amino acids stretch appears to have putative RNA binding properties and is encountered solely in the Isoe isoform able to bind specifically the G-quartet structure. This 27 amino acid stretch encoded by FXR1P exon 15 being in close proximity to the RGG box of FXR1P encoded by sequences of exon 14 (Figure 1A), it may contribute to the binding to G-quartet mRNA structures together with the RGG box. Alternatively, the presence of this additional sequence in the FXR1P-Isoe, as compared to the other shorter isoforms, may alter the structure of the C-terminal portion of FXR1P, thereby allowing the binding. In a similar way, a different affinity for RNA was also shown for different FMRP isoforms. Indeed, ISO18, a minor isoform of FMRP lacking a small portion of exon 17 (29), is still able to bind G-quartet RNA (36), like ISO1 and ISO7 (13,30). In addition, ISO 18 is able to bind the 3′-UTR of FMR1 mRNA (36), that, conversely, is not bound by ISO1 and ISO7 (13,30).
In adult muscle, where both FMRP and FXR2P are absent, the FXR1P isoforms encountered both contain exon 15 sequences and correspond to a doublet of 82 (Isog) and 84 kDa (Isoe) (20,21,37). Their ability to bind G-quartet RNA structure suggests that, in this tissue, specific RNAs might be recognized by FXR1P via the interaction with G-quartet forming structure. Since FXR1P absence has a strong impact during muscle development (8,9), its RNA binding capacities are critical per se, independently from FMRP's fonction. Indeed, a recent knock-down analysis for xFxr1 produced a list of putative FXR1P target RNAs. Interestingly, several of their human homologues harbor a putative G-quartet structure [(9), and our unpublished data]. In addition, very little is known concerning the precise function of FXR1P in muscle, and it is possible that muscle-specific FXR1P interacting proteins might modulate its affinity for RNA.
Our analysis to dissect the binding capacities to G-quartet RNA FXR1P and its heterodimer with FMRP yielded unexpected findings. We observed a dramatic effect of the FMRP/FXR1P heterodimer on the dynamics of complex formation with G-quartet RNA. This effect was not observed when FMRP and its other partner MSP58 were mixed together, suggesting that MSP58 can probably compete for the same binding site as FMRP. In addition, the interaction of FMRP with FXR1P-Isoa or Isod strongly reduced FMRP specificity for G-quartet RNA. These different behaviors of the two FMRP-interacting proteins illustrate the complexity of the functions and interactions that take place in FMRP-containing mRNPs and in different tissues. FXR1P-Isod and Isoa are the FXR1P isoforms with the highest expression in brain, suggesting that in neurons FMRP interacts mostly with these two isoforms that might regulate negatively its action. Since in brain and cerebellum FXR1P-Isoe mRNA is expressed at a low level as revealed by RT–PCR (our unpublished data), probably only a very small portion of FMRP may be regulated by FXR1P-Isoe. Conversely, FMRP and FXR1P-Isoe and Isog isoforms are co-expressed in myoblasts and in myotubes, suggesting a particular regulation of G-quartet containing target mRNAs during muscle differentiation but not in adult muscle, where FMRP is not expressed anymore (21). The present study highlights the functional differences between FXR1P isoforms and therefore emphasizes the importance of the extensive tissue-specific alternative splicing undergone by FXR1 mRNA. In view of these results, it is clear that in each mRNP the ratio between FMRP and FXR1P different isoforms becomes important to precisely regulate FMRP function. The modulation of the affinity and/or of the dynamics observed for the FXR1P/FMRP heterodimer may reflect a regulation of the exchange of mRNAs between mRNPs or trafficking granules and polyribosomes.
The interaction domain of the two FXR proteins is localized in the N-terminal region of both proteins. This domain mediates the interaction between FMRP and several other proteins (FXR2P, CYFIP1, CYFIP2, NUFIP and 82-FIP) (10,33). On the other hand, despite the high level of homology, the N-terminal region of FXR1P seems to interact only with CYFIP2 and FXR2P (18). CYFIP2, together with CYFIP1, that only interacts with FMRP, belongs to a small family of proteins linking FMRP to the Rac pathway (18,33,38). We have previously proposed that the CYFIP proteins might modulate the ability of the FXR family members to homo and/or heterodimerize (18).
FXR1P and FXR2P are believed to have distincts but overlapping function in conjunction with FMRP, with the possibility to partially compensate for its absence. Our results show here a completely different function for two different FXR1P isoforms, which modulate the action of FMRP. This data reveals how a full understanding of FMRP function may be achieved through the deciphering of the global action of FMRP-containing mRNP complexes.
Acknowledgments
The authors are grateful to Alexandra Charlesworth for help. B.B. is supported by CNRS, Fondation J. Lejeune, GIS Maladies Rares, Fondation pour la Recherche Médicale, FRAXA Foundation and ‘Contrat d'interface’ from INSERM and CHU de Nice. E.B. is recipient of a fellowship from Fondation pour la Recherche Médicale. E.L. is supported by CNRS, Fondation pour la Recherche Médicale and ARC. L.D. is supported by the FRAXA Foundation and by a short term fellowship from the Boeringher Ingelheim Fund and the Conquer Fragile X Foundation. E.W.K. is supported by the NSERC. The authors also thank the support of the French Embassy in Canada for their Travelling Exchange Programm. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bardoni B., Mandel J.L., Fisch G.S. FMR1 gene and fragile X syndrome. Am. J. Med. Genet. 2000;97:153–163. doi: 10.1002/1096-8628(200022)97:2<153::aid-ajmg7>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Bardoni B., Davidovic L., Bensaid M., Khandjian E.W. The fragile X syndrome: exploring its molecular basis and seeking a treatment. Expert Rev. Mol. Med. 2006;8:1–16. doi: 10.1017/S1462399406010751. [DOI] [PubMed] [Google Scholar]
- 3.Siomi M.C., Siomi H., Sauer W.H., Srinivasan S., Nussbaum R.L., Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., O'Connor J.P., Siomi M.C., Srinivasan S., Dutra A., Nussbaum R.L., Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakker C.E., Verheij C., Willemsen R., Vanderhelm R., Oerlemans F., Vermey M., Bygrave A., Hoogeveen A.T., Oostra B., Reyners E., et al. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 6.Morales J., Hiesinger P.R., Schroeder A.J., Kume K., Verstreken P., Jackson F.R., Nelson D.L., Hassan B.A. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 7.Bontekoe C.J., McIlwain K.L., Nieuwenhuizen I.M., Yuva-Paylor L.A., Nellis A., Willemsen R., Fang Z., Kirkpatrick L.L., Bakker C.E., McAninch R., et al. Knockout mouse model for Fxr2: a model of mental retardation. Hum. Mol. Genet. 2002;11:487–498. doi: 10.1093/hmg/11.5.487. [DOI] [PubMed] [Google Scholar]
- 8.Mientjes E.J., Willemsen R., Kirkpatrick L.L., Nieuwenhuizen I.M., Hoogeveen-Westerveld M., Verwzeij M., Reis S.A., Bardoni B., Hoogeveen A.T., Oostra B., et al. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet. 2004;13:1291–1302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 9.Huot M.E., Bisson N., Davidovic L., Mazroui R., Labelle Y., Moss T., Khandjian E.W. The RNA-binding protein fragile X-related 1 regulates somite formation in Xenopus laevis. Mol. Biol. Cell. 2005;14:835–844. doi: 10.1091/mbc.E05-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos A., Hollingworth D., Adinolfi S., Castets M., Kelly G., Frenkiel T.A., Bardoni B., Pastore A. The structure of the N-terminal domain of the Fragile X Mental retardation protein: a platform for protein–protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Kirkpatrick L.L., McIlwain K.L., Nelson D.L. Comparative genomic sequence analysis of FXR gene family: FMR1, FXR1 and FXR2. Genomics. 2001;78:169–177. doi: 10.1006/geno.2001.6667. [DOI] [PubMed] [Google Scholar]
- 12.Khandjian E.W., Bechara E., Davidovic L., Bardoni B. Fragile X Mental retardation Protein: many partners and multiple targets for a promiscuous function. Current Genomics. 2005;6:512–522. [Google Scholar]
- 13.Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B. Kissing complex RNAs mediate interaction between the Fragile X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Yun S.-W., Seto J., Liu W., Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequenses. Neuroscience. 2003;120:1005–10017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- 16.Garnon J., Lachance C., Di Marco S., Hel Z., Marion D., Ruiz M.C., Newkirk M.M., Khandjian E.W., Radzioch D. Fragile X-related Protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J. Biol. Chem. 2005;280:5750–5763. doi: 10.1074/jbc.M401988200. [DOI] [PubMed] [Google Scholar]
- 17.Khandjian E.W., Huot M.E., Tremblay S., Davidovic L., Mazroui R., Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl Acad. Sci. USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schenck A., Bardoni B., Moro A., Bagni C., Mandel J.L. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displayng selective interaction with FMRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidovic L., Bechara E., Gravel M., Jaglin X.H., Tremblay S., Sik A., Bardoni B., Khandjian E.W. The nuclear MicroSpherule protein 58 is a novel RNA-binding protein that interacts with fragile mental retardation protein in polyribosomal mRNPs from neurons. Hum. Mol. Genet. 2006;9:1–14. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick L.L., McIlwain K.L., Nelson D.L. Alternative splicing in the murine and human FXR1 genes. Genomics. 1999;59:193–202. doi: 10.1006/geno.1999.5868. [DOI] [PubMed] [Google Scholar]
- 21.Khandjian E.W., Bardoni B., Corbin F., Sittler A., Giroux S., Heitz D., Tremblay S., Pinset C., Montarras D., Rousseau F., et al. Novel isoform of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- 22.Bardoni B., Schenck A., Mandel J.L. A novel RNA binding protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet. 1999;8:2557–2566. doi: 10.1093/hmg/8.13.2557. [DOI] [PubMed] [Google Scholar]
- 23.Mazroui R., Huot M.E., Tremblay S., Boilard N., Labelle Y., Khandjian E.W. Fragile X mental retardation protein determinants required for its association with polyribosomal ribonucleoparticles. Hum. Mol. Genet. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- 24.Devys D., Lutz Y., Rouyer N., Bellocq J.P., Mandel J.L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:783–789. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 25.Castets M., Schaeffer C., Bechara E., Schenck A., Khandjian E.W., Luche S., Moine H., Rabilloud T., Mandel J.L., Bardoni B. FMRP interferes with Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 26.Miyashiro K.Y., Beckel-Mitchener A., Purk T.P., Becker K.G., Barret T., Liu L., Carbonetto S., Weiler I.J., Greenough W.T., Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Brown S.J. BinDN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acids sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terribilini M., Lee J.H., Yan C., Jernigan R.L., Honovar V., Dobbs D. Prediction of RNA binding sites in proteins from amino acids sequence. RNA. 2006;12:1450–1462. doi: 10.1261/rna.2197306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sittler A., Devys D., Weber C., Mandel J.L. Alternative splicing of exon 14 determines nuclear or cytoplasmic localization of fmr1 protein isoforms. Hum. Mol. Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 30.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 31.Gabus C., Mazroui R., Tremblay S., Khandjian E.W., Darlix J.L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Iacoangeli A., Lin D., Denman R.B., Hellen C.U., Tiedge H. Dendritic BC1 RNA in translational control mechanisms. J. Cell. Biol. 2005;171:811–821. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castets M., Mandel J.L., Bardoni B. FMRP Interacting proteins: from a complex to a pathway. In: Sung Y.J., Denman R.B., editors. The Molecular Basis of Fragile X Syndrome. Kerala, India: Research Signpost; 2005. pp. 117–127. [Google Scholar]
- 34.Darnell J.C., Mostovetsky O., Darnell R.B. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 35.Zanotti J., Lackey P.E., Evans G.L., Milailescu M.R. thermodynamics of the fragile X mental retardation protein RGG box interaction with quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 36.Rackham O., Brown C.M. Visualization of RNA–protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dube M., Huot M.E., Khandjian E.W. Muscle specific fragile X related protein 1 isoforms are sequestred in the nucleus of undifferentiated myoblast. BMC Genet. 2000;1:4. doi: 10.1186/1471-2156-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bardoni B., Mandel J.L. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev. 2002;12:284–293. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]