Abstract
Control of RNA processing plays a central role in regulating the replication of HIV-1, in particular the 3′ polyadenylation of viral RNA. Based on the demonstration that polyadenylation of mRNAs can be disrupted by the targeted binding of modified U1 snRNA, we examined whether binding of U1 snRNAs to conserved 10 nt regions within the terminal exon of HIV-1 was able to inhibit viral structural protein expression. In this report, we demonstrate that U1 snRNAs complementary to 5 of the 15 regions targeted result in significant suppression of HIV-1 protein expression and viral replication coincident with loss of viral RNA. Suppression of viral gene expression is dependent upon appropriate assembly of a U1 snRNP particle as mutations of U1 snRNA that affect binding of U1 70K or Sm proteins significantly reduced efficacy. However, constructs lacking U1A binding sites retained significant anti-viral activity. This finding suggests a role for these mutants in situations where the wild-type constructs cause toxic effects. The conserved nature of the sequences targeted and the high efficacy of the constructs suggests that this strategy has significant potential as an HIV therapeutic.
INTRODUCTION
As is the case for all retroviruses, replication of HIV-1 is critically dependent upon the controlled processing of its RNA. From a single 9 kb viral transcript, 15 proteins are generated through the process of suboptimal splicing that generates over 30 viral mRNAs (1,2). Control of splicing is achieved in part through the suboptimal nature of the 3′ splice sites (ss) present (3–5) as well as the presence of both exon splicing enhancers (ESEs) and exon splicing silencers (ESSs) (5–11). In the course of analyzing the role of the ESE within the terminal exon of HIV-1 RNA, we observed that, in addition to promoting the use of the adjacent 3′ ss, the ESE was also required for the 3′ end cleavage and polyadenylation of the incompletely spliced viral RNAs (12). Deletion of this ESE resulted in loss of gp160 expression due to a failure to transport the incompletely spliced viral RNA to the cytoplasm (12,13). The correlation of the block to viral RNA export with a reduction in 3′ end formation suggested that targeting this step in the viral life cycle may provide an additional strategy to control HIV-1 replication.
The formation of the 3′ end of RNA polymerase II transcripts is determined in large part by two sequences within the RNA; the consensus AAUAAA polyadenylation signal and the downstream enhancing sequence consisting of either a G or G/U-rich region (14,15). In addition to these basic signals, evidence has also emerged for regulation by trans-acting factors, one of which is U1 snRNA. In several systems studied to date [HIV-1 polyadenylation near the start site of transcription, polyadenylation of the late transcripts of bovine papillomavirus (BPV)], interaction of U1 snRNP via base-pairing close to the polyadenylation signal results in either inhibition of the cleavage or polyadenylation portion of the 3′ end processing reaction (16–20). More recently, this effect has been exploited to suppress other genes by the expression of variants of U1 snRNA in which the first 10 nt have been substituted with a sequence complementary to the gene of interest (21–23). Provided that the targeted sequence is present within the terminal exon of the gene of interest and not present within secondary structure (i.e. is accessible for base pairing with U1 snRNA), a significant inhibition of gene expression can be achieved.
To explore the utility of U1 snRNA variants to inhibit HIV-1 gene expression, we identified highly conserved 10 nt sequences within the terminal exon of the M and O clades of HIV-1 and prepared U1 snRNAs expression vectors with complementarity to 15 of these regions. Several of these constructs induced a dramatic suppression of the expression of HIV-1 p55/p24 upon cotransfection with a HIV-1 proviral expression vector. Little or no effect on expression of these proteins was detected upon overexpression of U1 snRNAs that had no complementarity to HIV-1 and the anti-HIV-1 constructs had no effect on other co-transfected reporter vectors. Combination of anti-HIV constructs leads to an even greater degree of inhibition. The loss of viral protein expression was accompanied by a dramatic decrease in levels of viral mRNAs. Furthermore, the conserved nature of the sequences targeted suggests that this approach may provide broad resistance to a range of HIV-1 strains.
MATERIALS AND METHODS
Bioinformatics
A computation-based selection algorithm was devised for the selection of potential anti-HIV target sites in the 3′-terminal exon of HIV transcripts. Consensus sequences of the env and nef regions of all M and O clades of HIV-1 were retrieved from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/) and were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/) followed by manual adjustment of the multiple sequence alignment (MSA) using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) or JalView (http://www.ebi.ac.uk/~michele/jalview/). A set of well-conserved continuous 10 nt sites was identified by inspection of the MSA. The number of BLAST hits returned for each potential target site against the HIV-1 or human subset of GenBank sequences was recorded. The SR-value of a 10 bp site is defined as the number of BLAST hits returned for the oligo against GenBank HIV-1 sequences. The number of returned BLAST hits for the same oligo against GenBank human sequences was assigned a value N, the number of hits for a non-HIV directed oligo [anti-CAT, (21)] was designated M. The SF-value of an oligo was calculated using its associated SR, N and M values according to the formula SF = SR/(N/M). Multiple sites that satisfy the condition SF > SR were selected for the construction of U1 targeting constructs described.
Plasmid constructs
HIV-1 Hxb2 R-/RI- was provided by E. Cohen (Institut de recherche cliniques de Montreal). Generation of U1 targeting vectors has been previously described (21). The mutant U1 anti-HIV snRNA expression constructs pU1 αHIV-1#1-15 were derived from pUCB U1 [provided by H. Schaal (Institute of Virology, Heinrich Heine University Dusseldorf, Germany)] by PCR (21) or using quickchange mutagenesis (Stratagene). Targeted sequences are indicated in Table 1. Figure 1 illustrates the relative positions of five of the target sites in the HIV-1 3′ LTR proximal region. The following system of nomenclature is employed in this report regarding U1 targeting constructs: pU1 αHIV-1 #1 or pU1 αCAT refers to the plasmid encoding the 5′ end mutated U1 snRNA sequence while U1 αHIV-1 #1 or U1 αCAT refers to the inhibiting U1 snRNAs expressed from those plasmids.
Table 1.
Summary of targeted sites in HIV-1 provirus and their effect on viral p24 expression
| U1 snRNA name | Targeted sequence | Location | Reading frame | Suppression of p24 expression | Conservationa |
|---|---|---|---|---|---|
| 1 | CTTCAGCTAC | 8519 | Rev exon 2 | ++ | 66% |
| 2 | ATAGCAGTAG | 8682 | Env | − | 67% |
| 3 | TTTGCTATAA | 8786 | Env | ++ | 56% |
| 4 | CAGGTACCTT | 9013 | Nef | − | 37% |
| 5 | CATCCGGAGT | 9391 | LTR | + | 71% |
| 6 | TGGGGTACCT | 6344 | Env | − | N/A |
| 7 | GCCTGTGTAC | 6441 | Env | + | N/A |
| 8 | GAAAAGGGGG | 9073 | Nef | − | 92% |
| 9 | GACTGGAAGG | 9083 | LTR | − | 56% |
| 10 | ACACACAAGG | 9143 | LTR | − | 78% |
| 11 | AACTACACAC | 9172 | LTR | − | 74% |
| 12 | GGGGGACTGG | 9079 | LTR | + | 95% |
| 13 | TTTTAAAAGA | 9065 | Nef | − | 91% |
| 14 | GGCAGGGATA | 8350 | Env | − | 85% |
| 15 | TTAGGCAGGG | 8347 | Env | − | 87% |
Position of targeted sequences is stated relative to the reference HIV-1 provirus HXB2 (K03455).
a% of time the complete target sequence is found in a database of 1005 viral genomes belonging to the M and O clades of HIV-1.
++-High activity, suppression of viral p24 expression to background level.
+-Moderate activity, >50% reduction in p24 production expression.
−-No significant change in viral p24 expression observed.
Figure 1.
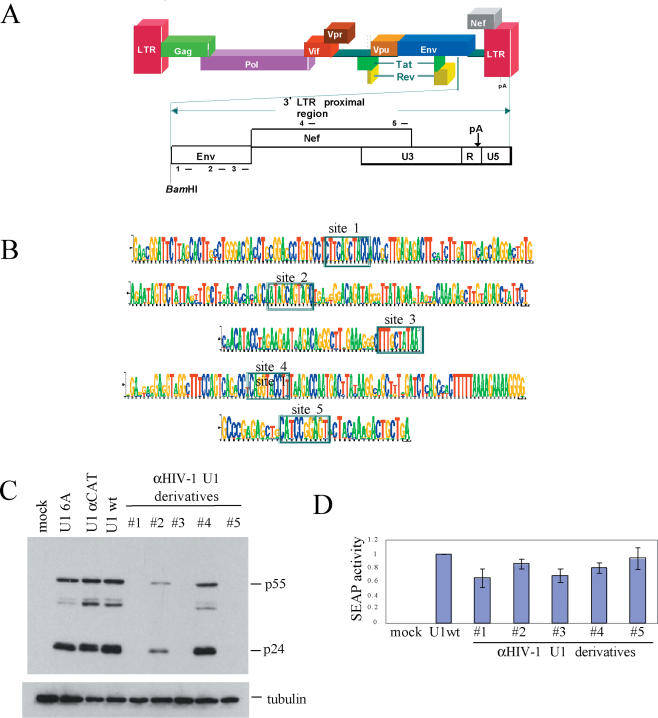
U1 snRNA variants targeted to HIV-1 are able to suppress HIV-1 gene expression. (A) Outline of HIV-1 genome structure. Sites targeted by U1 snRNA variants are indicated by numbered lines above and below reading frames. (B) Sequence logos showing the conservation of sequences targeted by five U1 snRNA variants. Selected 10 nt sequences identified by MSA are boxed, the height of the individual base reflecting the degree of conservation at that position among HIV-1 strains. (C) Effect of anti-HIV-1 U1 snRNAs on viral protein expression. Cells were transfected with a plasmid containing HIV-1 proviral DNA (HIV-1 Hxb2 R-/RI-), CMV PLAP and one of the indicated U1 expression constructs. Forty-eight hours post-transfection, cells were harvested and level of p24 production assessed by western blot. Effect on secreted alkaline phosphatase (SEAP) expression (D) was performed in parallel as outlined in Materials and Methods and data are shown.
To investigate the requirement for the various U1 snRNP protein components to achieve suppression of HIV-1 expression, a number of mutants of the αHIV-1 #1 and #5 expression vector were generated by quickchange (Stratagene) using the following primer pairs: ΔSL1, 5′-CGAAGATCTCAACTCCGGATGGCAGGGCCCAGGGCGAGGCTTATC-3′, 5′-GATAAGCCTCGCCCTGGGCCCTGCCATCCGGAGTTGAGATCTTCG-3′; SL1mut, 5′-GCAGGGGAGATACCAACTAGTGCTTCCTGGTTTTCCCAGGGC-3′, 5′-GCCCTGGGAAAACCAGGAAGCACTAGTTGGTATCTCCCCTGC-3′; ΔSL2, 5′-GGGAAATCGATGGAAAACCACCTTCGTGA-3′, 5′-GGTGGTTTTCCATCGATTTCCCCAAATGT-3′; SL2mut, 5′-GGGCGCGGCTTATCCATTACGCGGATGTGCTGACCCC-3′, 5′-GGGGTCAGCACATCCGCGTAATGGATAAGCCTCGCCC-3′; ΔSm, 5′-CACTACCACCTCGAGTGCAGTCGAGTTTG-3′, 5′-TCGACTGCACTCGAGGTGGTAGTG-3′. All constructs were confirmed by sequencing.
Transfection and western blots
293T cells were transfected by calcium phosphate precipitation with 1.0 μg of HxBruR−/RI− and a maximum of 4 μg of pUCB U1 or its derivatives (24). At 48 h post-transfection, cells were harvested and lysed in whole cell lysis buffer (10 mM Na2HPO4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.2% Na azide, 0.5% Na deoxycholate and 1 mM Na orthovanadate) for 20 min. For p24 blots, protein lysates were fractionated on 12% SDS–PAGE then transferred onto polyvinylidene difluoride (PVDF) membranes. p24 blots were blocked in 1× phosphate-buffered saline (PBS), 5% milk, 0.05% Tween-20 then probed with mouse anti-p24 hybridoma supernatant overnight at 4°C. Donkey anti-mouse horseradish peroxidase (HRP)-conjugated antibody was applied to the blots after washing at a 1:1000 dilution for 1 h, then washed four times prior to development with western lightning (Perkin–Elmer). To normalize for protein loading, blots were stripped and reprobed with mouse anti-tubulin antibody (Sigma).
Northern blot analysis
293T cells (3 × 106) were transfected with 4 μg of HxBruR−/RI− and a total of 12 μg of pUCB U1 or its derivatives. At 48 h post-transfection cells were harvested and lysed in GT (4 M guanidinium thiocyanate, 25 mM sodium citrate (pH 7), 0.5% sarcosyl, 0.1M β-mercaptoethanol). Total RNA was isolated (25) and analyzed by northern blotting. Blots were probed with 32P labeled DNA to HIV-1 LTR (generated by PCR using the primers 5′-CTAATTCACTCCCAACGAAGA-3′ and 5′-TGCTAGAGATTTTCCACACTG-3′), end labeled oligo to GAPDH (5′-AAAGGTGGAGGAGTGGGTGTCGCTGTTGAA-3′), or end labeled oligos specific for each of the U1 snRNA variants (anti-HIV #1 5′-CCTGCCTTCAGCTAC-3′; anti-HIV #5 CCTGCCATC CGGAGT-3′).
Viral challenge assays
To assess the ability of the anti-HIV U1 constructs to suppress HIV-1 replication, HeLa CD4+ cells lines were generated expressing vector alone, U1 6A, or anti-HIV #5. Following selection for stably transduced cells (using puromycin), pooled clones were subsequently used in the viral challenge assay. For HIV-1 infection, 0.5 × 106 cells were plated into tissue culture 6-well, flat-bottom, dishes. After culture for 24 h, cells were infected with 0.3 moi of HIV-1IIIB virus for 1hr at 37oC as previously described (26). The cells were then washed three times with PBS lacking Ca2+/Mg2+ and left in 2 ml of media. On day three of culture, 0.5 ml aliquots were collected and assayed for production of virus by using HIV-1 p24 antigen ELISA kit (Beckman-Coulter Inc.).
RESULTS
Computation-based selection of anti-HIV target sites in the 3′-terminal exon of viral transcripts
U1 snRNP mediated inhibition of polyadenylation requires localization of the modified U1 snRNP particle to the 3′-terminal exon of the target transcript (22). This is accomplished by base pairing the 5′ end of the U1 snRNA in its associated RNP complex to a complementary region in the target pre-mRNA. To achieve selective targeting of U1 snRNA to sequences in HIV-1, we not only identified conserved sequences in the virus but also eliminated the U1 sequences having a significant number of interactions with host cell mRNAs. Due to the 10 nt length of the targeted sequence, binding sites are expected to occur approximately once in 106 bases, corresponding to ∼3000 occurrences in the human genome. However, since exons constitute only 2% of the human genome (27) and inhibition by U1 snRNP particles requires base-pairing to unstructured regions in the 3′-terminal exon of transcripts containing AAUAAA type poly(A) signals (22), the specificity of inhibition can be substantially improved with judicious selection of target sites. In light of the few published studies on U1 snRNA-mediated reduction of target gene expression (21,22), rational design of anti-HIV U1 constructs based on the characteristics of known effective target sites cannot be achieved at this point. To facilitate the discovery of HIV sequences susceptible to U1-mediated inhibition that would provide broad coverage for all HIV-1 strains, a MSA of consensus sequences of the env and nef regions of all M and O group HIV-1 subtypes was generated and a number of highly conserved 10 bp sequences identified in the MSA. The number of BLAST hits returned for a given 10 bp sequence against HIV-1 sequences in GenBank was determined and defined as the Raw Score (SR-value). The magnitude of the SR-value is proportional to the extent of conservation observed in the MSA. This value was then scaled according to the number of BLAST hits in the human genome returned for each sequence to derive the Final Score (SF-value). Sequences with the least hits upon scanning of the human genome were deemed suitable for targeting and placed in a list of candidate sites. Selection of target sites within highly conserved regions of the HIV-1 genome allows the identification of universal targets common to all subtypes. In addition, the conservation of these sites among HIV-1 subtypes suggests that mutations within these regions would have detrimental effects to the virus. Using this computational strategy, multiple anti-HIV targets were identified in the 3′ LTR proximal region of HIV-1 of which five were chosen for initial evaluation. The sequence and position of a subset of the selected target sites are shown in Figure 1A and B. All selected sites reside in the env or nef region and are present in the 3′-terminal exon of all viral transcripts. Selection of these target sites ensures that the expression of all viral proteins will be affected.
Targeting of the HIV-1 3′-terminal exons with anti-HIV U1 constructs inhibits viral gene expression at both the protein and RNA level
Having selected a number of sites within the terminal exon of HIV-1 that met the desired criteria, five U1 expression vectors were generated in which the first 10 nt of the transcript was able to base pair to one of the sites. To measure their anti-viral capacity, U1 snRNA expression plasmids were co-transfected with plasmid HxBruR−/RI− that contains a HIV-1 provirus in which the RT and IN genes have been deleted. In addition to the five anti-HIV U1 variants, parallel transfections were performed with vectors expressing unmodified U1 snRNA (U1 wt), a point mutant of U1 (U1 6A) that abolished activity in splicing, and a U1 variant targeted to chloramphenicol acetyl transferase (CAT) (22). Effect of U1 snRNA expression on HIV-1 gene expression was monitored by western blot for p24. As shown in Figure 1C, three of the five anti-HIV constructs tested (#1, #3 and #5) resulted in a significant reduction in HIV-1 gene expression. In contrast, U1 6A and U1 α-CAT had little to no effect relative to U1 wt (see Figure 1C). Parallel examination of the effect of these vectors on a non-target RNA (secreted alkaline phosphatase, SEAP) failed to show significant levels of inhibition (Figure 1D). The failure to see any large changes in SEAP expression suggests that the viral inhibition observed cannot be attributed to gross toxicity to the cell. In light of this success, we continued to evaluate additional sites within the HIV-1 provirus for their susceptibility to this inhibitory strategy. As detailed in Table 1, 15 sites in total were targeted of which only five yielded significant reduction on viral p24 expression.
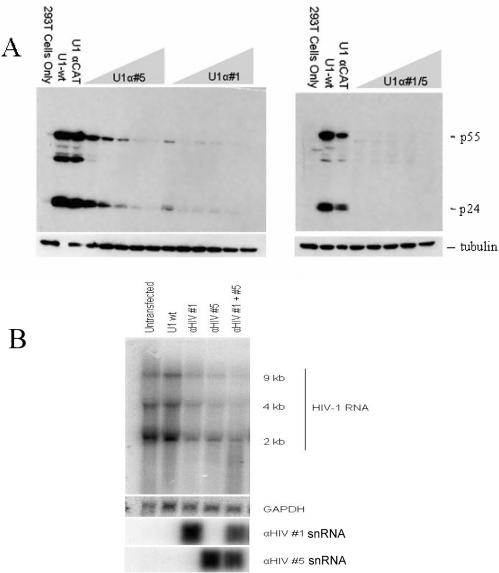
Having established the potency of a subset of the U1 αHIV-1 constructs, we tested the relative efficacy of the anti-HIV U1 targeting vectors alone or in combination. Two U1 αHIV-1 vectors were selected to assess their potency (U1 αHIV-1 #1 and U1 αHIV-1 #5). The two anti-HIV U1 targeting vectors were co-transfected individually or in combination with the proviral clone HxBruR−/RI− at increasing concentrations. The level of viral protein and RNA accumulation at 48 h post-transfection was assessed by western and northern blots, respectively. As shown in Figure 2A, the anti-HIV U1 constructs differed in the amount of plasmid required to suppress viral structural protein (p24 and p55) expression, anti-HIV #5 requiring 2 μg of transfected plasmid while anti-HIV #1 achieved the same degree of suppression at lower levels (0.5 μg of transfected plasmid). Combination of the two anti-HIV constructs (#1 and #5) was able to suppress viral gene expression at all concentrations tested. To explore the basis for the observed response, the effect of the U1 constructs on viral RNA levels was subsequently explored (Figure 2B). Cotransfection with vectors expressing anti-HIV #1 or #5 alone led to significant reduction in viral RNA levels. Further reduction was achieved when the two constructs were added in combination. In contrast, little or no change in viral RNA levels was detectable upon cotransfection with the control vectors (U16A, U1 anti-CAT, data not shown).
Figure 2.
Suppression of HIV-1 gene expression occurs at the level of both viral protein and RNA. Cells were transfected with proviral DNA (pHxb2 R-/RI-) and 4 μg of U1 wt/αCAT expression vector or increasing amounts of αHIV #1/#5 (0.25, 0.5, 1, 2 or 4 μg) alone or in combination. Forty-eight hours post-transfection, cells were harvested and either (A) used to examine HIV-1 p24 expression or (B) RNA extracted and levels of RNA determined by northern blot. Representative gels are shown indicating the position of the viral RNAs (upper), GAPDH (middle) and U1 snRNA variants (lower).
Inhibitory activity is dependent on proper assembly of a U1 snRNP particle
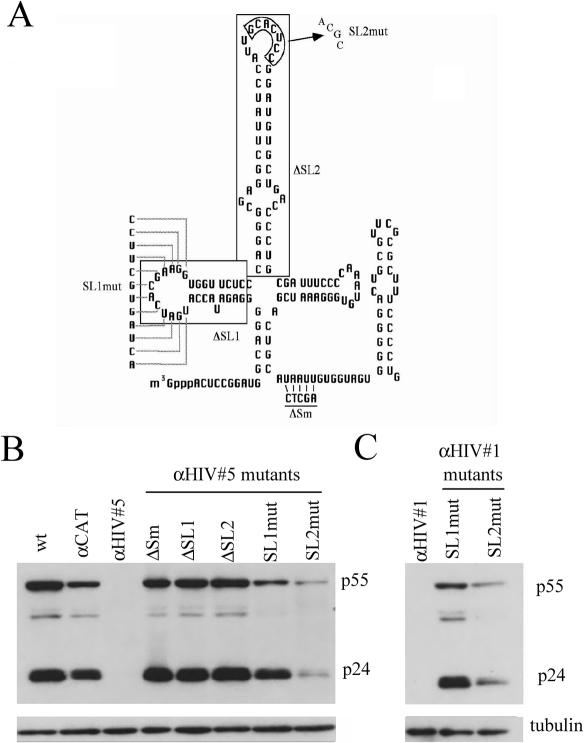
Modified U1 snRNAs have been used in the past to suppress HIV-1 gene expression, but in that instance more extensive regions of base-pairing (61–68 bases) were used along with deletion of stem–loop 1 (ΔSL1) of the resultant RNA (28). Loss of HIV-1 expression in that study was attributed to RNA degradation due to an anti-sense mechanism. To test whether the inhibition seen in this study was operating through the same or another mechanism, we examined whether the response was dependent upon proper assembly of the U1 snRNP particle. If mutants in the various protein assembly domains of U1 snRNA do not alter the response, then only base-pairing potential would be deemed necessary for the loss of HIV-1 expression (suggestive of an anti-sense mechanism). Five variants of αHIV #5 U1 snRNA were generated to probe the requirement of the various regions of the U1 snRNA for function: ΔSL1 or substitutions in stem–loop 1 that results in loss of U1 70 K binding (SL1mut), deletion of stem–loop 2 (ΔSL2) or substitutions in stem–loop 2 that results in loss of U1A binding (SL2 mut), and mutations within the Sm binding region (ΔSm) that abolish assembly of the Sm core (Figure 3A) (29). Cotransfection of these mutant U1 snRNAs with proviral DNA revealed four of the five mutants had no inhibitory activity; expression of HIV-1 p24 was similar to that seen with wt U1 snRNA (Figure 3B). Only αHIV #5 SL2mut retained some inhibitory activity although significantly less than that of the unmodified variant. To assess whether this phenomenon was limited to only one construct or was valid for all U1 variants, both SL1mut and SL2mut mutations were introduced in the context of anti-HIV #1. As shown in Figure 3C, the same spectrum of effects was observed, mutation of SL1 leading to abolition of anti-HIV activity while SL2 mutants retained low but significant anti-viral activity.
Figure 3.
HIV-1 Suppression is dependent upon the integrity of the U1 snRNP. (A) Schematic of U1 snRNA structure with the location and nature of U1 mutants indicated. Shown are the regions of α-HIV U1 #1 and #5 that were mutated to identify the protein components of the snRNP required to suppress viral gene expression. Mutations were as follows: deletion of stem–loop 1 (ΔSL1), deletion of stem–loop 2 (ΔSL2), substitutions in stem–loop 1 (SL1mut), substitutions in the loop region of stem–loop 2 (SL2mut) and mutations in the Sm binding region that abolishes assembly of the Sm core complex (ΔSm). (B and C) Effect of anti-HIV-1 #5/#1 mutation on Suppression of HIV-1 Gene Expression Cells were transfected with a plasmid containing HIV-1 proviral DNA (HIV-1 Hxb2 R-/RI-) and mutants of anti-HIV #5 (B) or #1 (C). Forty-eight hours post-transfection, cells were harvested and level of p24 production assessed by western blot.
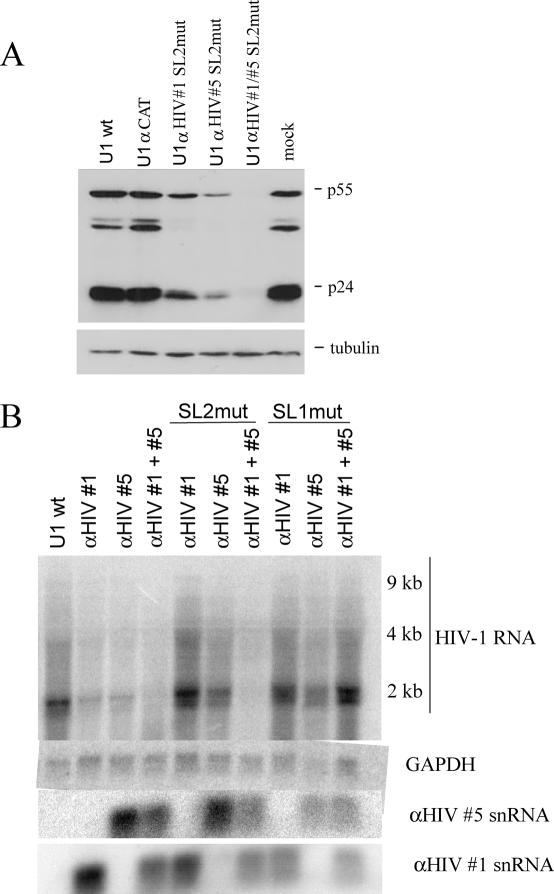
The residual anti-viral activity of the SL2 mutants raised the possibility that these constructs may serve as alternatives should the unmodified anti-viral U1 RNAs have deleterious effects on the cell. To probe this issue further, we assessed the anti-viral activities of the SL2mut constructs alone or in combination (Figure 4). Consistent with their reduced activity, higher amounts of each plasmid were required to repress HIV-1 gene expression (data not shown). However, when used in combination, significant inhibition of viral p24 expression (to background) was achievable, albeit using higher amounts of plasmid. To test whether this effect was attributable to a similar effect on viral RNA expression, Northern blots were also carried out. As shown in Figure 4B, when used in combination SL2 mutants of anti-HIV #1 and #5 led to a significant reduction in HIV-1 RNA levels. A similar response is not evident upon expression of the SL1 variants of the same constructs (Figure 4B).
Figure 4.
SL2mut variants of anti-HIV U1 snRNAs retain anti-viral activity. (A) Cells were transfected with HIV-1 proviral DNA (HIV-1 Hxb2 R-/RI-) and vector expressing anti-HIV #1/#5 SL2mut snRNA either alone or in combination. Forty-eight hours post-transfection, cells were harvested and lysates analyzed for p24 expression by western blot. (B) Cells were transfected with HIV-1 proviral DNA (HIV-1 Hxb2 R-/RI-) and vector expressing anti-HIV #1/#5 wild-type, SL2mut, or SL1mut snRNAs either alone or in combination. Total RNA was subsequently extracted and levels of viral RNA, and GAPDH RNA assayed by northern blot. Shown is representative gel of the results obtained. Identity of viral RNAs, GAPDH RNA and U1 snRNA variants (lower) are as indicated.
Cells stably expressing anti-HIV U1 snRNAs show reduced capacity to support HIV replication
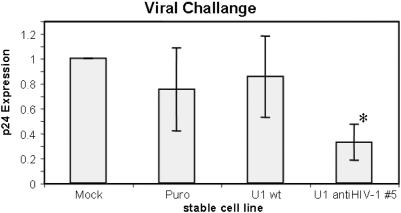
While the experiments to date have clearly established the anti-HIV properties of the U1 constructs generated, we were interested in examining their properties under conditions that more accurately mimicked the in vivo situation. Toward this end, stable cells expressing one of the anti-viral U1 constructs was generated and challenged with virus. As shown in Figure 5, monitoring of p24 production in the media following viral challenge revealed a reduction in HIV-1 replication in only the cell line expressing the anti-HIV U1 snRNA variant. Little to no change in p24 levels was seen with cells expressing either of the control vectors (U1 wt, U16A). Although the inhibition of HIV-1 replication seen in the context of the stable cell lines is less than that observed in transient assays (see Figure 1), subsequent analysis has revealed that expression of the U1 constructs is considerably below that obtained in the transient assays (data not shown). Current efforts are directed at improving the expression of the modified U1 snRNAs.
Figure 5.
Cells stably expressing anti-HIV #5 U1 snRNA show reduced capacity to support HIV-1 replication. HeLa CD4+ cells lines were isolated following transfection with pBABE vectors expressing without insert (puro), U1wt, or anti-HIV #5. Following selection for stable integrants, the pooled clones were challenged with HIV-1 and p24 production assayed five days post-infection by ELISA. Shown are the averages of two independent experiments and a total of four independent assays. Asterisk indicates values that are different from control cells (Mock) at a P-value <0.05.
DISCUSSION
Maturation of pre-mRNA in eukaryotes involves a set of factors that modify the primary transcript into an export competent mRNP (messenger ribonucleoprotein) complex, the formation of which signals its release from the site of transcription and export into the cytoplasm for translation (30). These processing events include capping, splicing, 3′ end cleavage and polyadenylation. Proper 3′ end formation promotes transcription termination (15) and nuclear export of the mRNA (31) and enhances the stability and translation of the mature transcript in the cytoplasm (32,33). Recent evidence implicates the existence of extensive coupling between the gene expression machinery engaged in mRNA synthesis, processing, export and surveillance (34–36) suggesting that disruption of RNA processing will compromise proper gene expression.
Binding of U1 snRNP upstream of the AAUAAA signal in the 3′-terminal exon inhibits the polyadenylation step in 3′ end formation by disrupting the protein–protein interactions in the polyadenylation machinery (20). In contrast, interaction between U1 snRNP and the major splice donor site downstream of the HIV-1 5′ LTR poly(A) site inactivates the cleavage step in a U1 70 K dependent manner and is thought to be important for production of full-length genomic RNA (29). Here we demonstrated, for the first time, that the same component of the splicing machinery recruited by HIV for its survival can be re-programmed to direct its demise.
The target site selection algorithm employed in this study does not account for possible local secondary structure formation which might explain the failure of several of the αHIV constructs to elicit a significant reduction in viral protein production. However, dramatic reduction of viral structural protein expression by five of the constructs (#1, #3, #5, #7 and #12) was observed in this study with little or no effect on expression of the co-transfected SEAP vector. Synergistic inhibition of HIV gene expression by U1 targeting was likely achieved at two levels. The sites targeted are common to all viral transcripts, so expression of all viral proteins is suppressed by U1 targeting. Hence, reduction of viral RNA is not only achieved by direct destabilization of the RNA but also by loss of Tat and the transactivation of the HIV-1 LTR (37). It should be noted that maximum possible inhibition of viral gene expression was likely not observed in this study. The assembly of functional U1 snRNP particles takes at least 16 h (22,38). Therefore, a delay exists between the initiation of gene expression from the co-transfected provirus and when significant nuclear accumulation of anti-HIV U1 snRNPs has occurred.
In addition to establishing the anti-viral activity of the modified U1 snRNAs, we also demonstrate that activity of the αHIV U1 snRNAs is dependent upon proper assembly of the U1 snRNP. Mutations which affected any component of this RNP abolished function, the sole exception being point mutants within stem–loop 2 that affect U1A binding. Assembly of the Sm core is a requisite for import of U1 snRNA (38) into the nucleus while U1 70K has been shown to be required for inhibiting the cleavage/polyadenylation reaction (18,29). These observations suggest that it is unlikely that these constructs are functioning solely in an anti-sense manner. The partial activity of the SL2mut variant of αHIV #1 and #5 raises the possibility that they could be used in gene therapy protocols should the intact form of the snRNA prove to have deleterious side effects.
The high activity of U1 snRNA variants targeted to HIV-1 and their distinct mechanism of action raises the possibility that they could be used in a complementary fashion with RNAi to achieve a significant barrier to HIV-1 replication (39–42). While the U1 snRNA based strategy would affect the nuclear metabolism of the viral RNA, RNAi could be used to reduce the concentration of the same RNA in the cytoplasm, the location of the RISC complex. Future work will focus on expanding the number of U1 snRNA variants with anti-HIV-1 activity and exploration of the durability of the response to multiple viral strains and detection of resistant HIV-1 variants following serial passages of virus on cells expressing these constructs. The demonstrated efficacy of this approach for HIV-1 suggests it may prove of equal value in the suppression of replication of other viruses which display a similar reliance on the host cell polyadenylation machinery for replication.
Acknowledgments
K.A. was supported by a scholarship from CIHR while A.C. is an OHTN scientist. Research was supported by a grant from OHTN. Funding to pay the Open Access publication charges for this article was provided by OHTN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schwartz S., Felber B.K., Benko D.M., Fenyo E.-M., Pavlakis G.N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell D., Martin M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Reilly M.M., McNally M.T., Beemon K.L. Two strong 5′ (splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 4.Staffa A., Cochrane A. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 1994;68:3071–3079. doi: 10.1128/jvi.68.5.3071-3079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Si Z., Amendt B.A., Stoltzfus C.M. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amendt B., Si Z., Stoltzfus C.M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amendt B.A., Hesslein D., Chang L.-J., Stoltzfus C.M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilodeau P.S., Domsic J.K., Mayeda A., Krainer A.R., Stoltzfus C.M. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 2001;75:8487–8497. doi: 10.1128/JVI.75.18.8487-8497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caputi M., Mayeda A., Krainer A., Zahler A. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si Z.-H., Rauch D., Stoltzfus M. The exon splicing silencer in the human immunodeficiency virus type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staffa A., Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaren M., Asai K., Cochrane A. A novel function for Sam68: enhancement of HIV-1 RNA 3′ end processing. RNA. 2004;10:1119–1129. doi: 10.1261/rna.5263904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pongoski J., Asai K., Cochrane A. Positive and negative modulation of human immunodeficiency virus type 1 rev function by cis and trans regulators of viral RNA splicing. J. Virol. 2002;76:5108–5120. doi: 10.1128/JVI.76.10.5108-5120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 16.Ashe M.P., Pearson L.H., Proudfoot N.J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furth P.A., Choe W.T., Rex J.H., Byrne J.C., Baker C.C. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson S.I., Polycarpou-Schwarz M., Mattaj I.W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 19.Kato K., Hitomi Y., Imamura K., Esumi H. Hyperstable U1snRNA complementary to the K-ras transcripts induces cell death in pancreatic cancer cells. Br. J. Cancer. 2002;87:898–904. doi: 10.1038/sj.bjc.6600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagner S., Ruegsegger U., Gunderson S.I., Keller W., Mattaj I.W. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA. 2000;6:178–188. doi: 10.1017/s1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckley S.A., Liu P., Stover M.L., Gunderson S.I., Lichtler A.C., Rowe D.W. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell. Biol. 2001;21:2815–2825. doi: 10.1128/MCB.21.8.2815-2825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortes P., Cuevas Y., Guan F., Liu P., Pentlicky S., Jung S.P., Martinez-Chantar M.L., Prieto J., Rowe D., Gunderson S.I. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu P., Gucwa A., Stover M.L., Buck E., Lichtler A., Rowe D. Analysis of inhibitory action of modified U1 snRNAs on target gene expression: discrimination of two RNA targets differing by a 1 bp mismatch. Nucleic Acids Res. 2002;30:2329–2339. doi: 10.1093/nar/30.11.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriegler M. Gene Transfer and Expression: A Laboratory Manual. 1st edn. NY: Stockton Press; 1990. [Google Scholar]
- 25.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Branch D.R., Valenta L.J., Yousefi S., Sakac D., Singla R., Bali M., Sahai B.M., Ma X.Z. VPAC1 is a cellular neuroendocrine receptor expressed on T cells that actively facilitates productive HIV-1 infection. AIDS. 2002;16:309–319. doi: 10.1097/00002030-200202150-00001. [DOI] [PubMed] [Google Scholar]
- 27.Makalowski W. The human genome structure and organization. Acta Biochim. Pol. 2001;48:587–598. [PubMed] [Google Scholar]
- 28.Liu D., Donegan J., Nuovo G., Mitra D., Laurence J. Stable human immunodeficiency virus type 1 (HIV-1) resistance in transformed CD4+ monocytic cells treated with multitargeting HIV-1 antisense sequences incorporated into U1 snRNA. J. Virol. 1997;71:4079–4085. doi: 10.1128/jvi.71.5.4079-4085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashe M.P., Furger A., Proudfoot N.J. Stem–loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA. 2000;6:170–177. doi: 10.1017/s1355838200991957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y., Carmichael G.C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ford L.P., Bagga P.S., Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preiss T., Hentze M.W. From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Gen. Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 35.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell. Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 36.Vasudevan S., Peltz S.W. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 37.Tang H., Kuhen K.L., Wong-Staal F. Lentivirus replication and regulation. Annu. Rev. Gen. 1999;33:133–170. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- 38.Will C.L., Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 39.Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee N.S., Rossi J.J. Control of HIV-1 replication by RNA interference. Virus Res. 2004;102:53–58. doi: 10.1016/j.virusres.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Michienzi A., Castanotto D., Lee N., Li S., Zaia J.A., Rossi J.J. RNA-mediated inhibition of HIV in a gene therapy setting. Ann. N. Y. Acad. Sci. 2003;1002:63–71. doi: 10.1196/annals.1281.008. [DOI] [PubMed] [Google Scholar]
- 42.Boden D., Pusch O., Silbermann R., Lee F., Tucker L., Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]