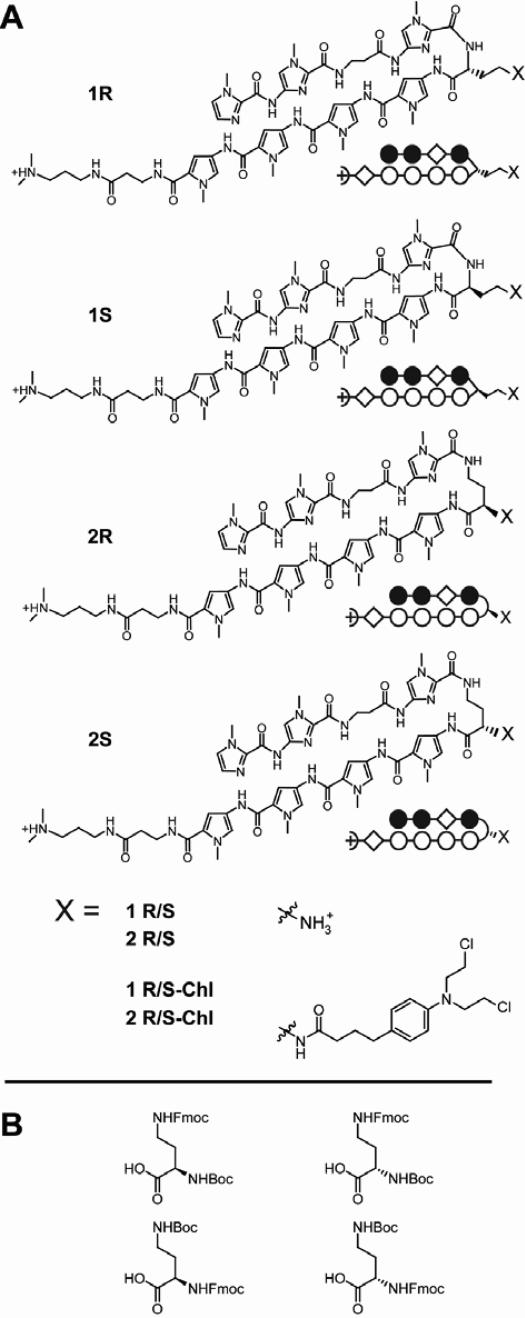
Abstract
Hairpin polyamide–chlorambucil conjugates containing an α-diaminobutyric acid (α-DABA) turn moiety are compared to their constitutional isomers containing the well-characterized γ-DABA turn. Although the DNA-binding properties of unconjugated polyamides are similar, the α-DABA conjugates display increased alkylation specificity and decreased rate of reaction. Treatment of a human colon carcinoma cell line with α-DABA versus γ-DABA hairpin conjugates shows only slight differences in toxicities while producing similar effects on cell morphology and G2/M stage cell cycle arrest. However, striking differences in animal toxicity between the two classes are observed. Although mice treated with an α-DABA hairpin polyamide do not differ significantly from control mice, the analogous γ-DABA hairpin is lethal. This dramatic difference from a subtle structural change would not have been predicted.
INTRODUCTION
The ability to control gene expression through the use of DNA sequence-specific, cell-permeable molecules holds therapeutic promise. Based on the natural product distamycin A, pyrrole–imidazole polyamides are a class of molecules that can be programmed to bind a broad repertoire of DNA sequences with specificities comparable to naturally occurring DNA-binding proteins (1). They have also been shown to permeate living cells, localize to the nuclei (2,3) and regulate gene expression (4,5).
Pairing rules have been established for minor groove recognition, whereby an N-methylpyrrole/N-methylimidazole (Py/Im) pair recognizes C•G, the reverse (Im/Py) recognizes G•C and Py/Py specifies A•T or T•A (1,6). The replacement of Py with β-alanine (β) can enhance binding affinity by allowing the polyamide to reset its curvature with that of the DNA minor groove (7,8). Polyamides may be linked via a turn moiety to give a hairpin structure which binds in the ‘forward’ direction N→C with respect to the 5′→3′ direction of the DNA strand (9). In earlier studies, linkage of antiparallel three-ring subunits via a γ-aminobutyric acid moiety, resulting in a turn with three methylene units, was shown to enhance DNA-binding affinity ∼100-fold over unlinked subunits (10). The γ-aminobutyric acid turn demonstrates selectivity for A•T or T•A base pairs over G•C and C•G (11). Use of a glycine amino acid, with one methylene turn unit, resulted in a 3-fold reduction in affinity relative to the γ-aminobutyric acid. Subsequent NMR studies revealed that the glycine amino acid substituted polyamide bound DNA in an extended, rather than a hairpin, conformation (12,13).
Substitution of the γ-aminobutyric acid with the chiral (R)-2,4-diaminobutyric acid [(R)-γ-DABA] was later found to further increase DNA-binding affinity ∼10-fold (14). In contrast, hairpin polyamides with the opposite enantiomer (S)-γ-DABA result in an unfavorable steric clash with the DNA minor groove and decreased binding for the forward orientation. Remarkably, binding affinity to the opposite orientation, in which the polyamide binds C→N with respect to the 5′→3′ direction of the DNA strand, is energetically favorable due to relief of the steric clash.
Polyamides have been used to regulate gene expression by binding to promoter and enhancer elements, but they do not prevent RNA polymerase elongation when bound to the coding region of genes (1). Conjugation of an alkylating moiety to a sequence-specific DNA-binding molecule would be expected to result in a molecule capable of covalent DNA attachment that could arrest transcription (15,16). An early example of this is Tallimustine (FCE 24517), which is a conjugate of distamycin A and the nitrogen mustard chlorambucil (17). When attached to a DNA minor groove binder, chlorambucil is capable of alkylation at the N3 position of an adenine proximal to the binding site (18,19). Hairpin polyamide–chlorambucil conjugates have been demonstrated to target specific sequences of the HIV-1 promoter and simian virus 40 (19,20).
Recently, a hairpin polyamide–chlorambucil conjugate, synthesized at The Scripps Research Institute, was found to alkylate within the coding region of the histone H4c gene of the human colon carcinoma cell line SW620 and to block proliferation of these cells in culture (21). In addition, this conjugate was found to block tumor growth in a SW620 xenograft mouse model without apparent toxicity. When we attempted to scale-up additional conjugate at Caltech, the newly-synthesized molecule was found to be highly toxic in athymic nude mice, contrary to prior experiments (22). Subsequent studies showed that the two laboratories had utilized two different turn units in the synthesis of the conjugate: the conjugate synthesis at Scripps used Boc-D-Dab(Fmoc)-OH, while the conjugate synthesis at Caltech used Fmoc-D-Dab(Boc)-OH. The molecules are therefore constitutional isomers differing in the hairpin turn moiety. The original molecule made at Scripps continues to be called 1R-Chl (22) and the new molecule is designated 2R-Chl. 1R-Chl utilizes an α-DABA turn unit, whereas 2R-Chl employs the standard γ-DABA turn unit (Figure 1). Linkage via the new α-DABA turn unit results in one methylene group in the hairpin, whereas 2R-Chl has three methylene groups. In addition, the linker between the polyamide core and the chlorambucil alkylator in 1R-Chl is lengthened by two methylene groups relative to 2R-Chl.
Figure 1.
Polyamide structures and syntheses. (A) Chemical and ball-and-stick structures for polyamides 1R, 1S, 2R, 2S and their Chl conjugates. (B) Chemical structures of the four turn units used in the syntheses of the parent polyamides and polyamide-chlorambucil conjugates: Boc-D-Dab(Fmoc)-OH (upper left, used in the syntheses of 1R and 1R-Chl); Boc-Dab(Fmoc)-OH (top right, used in the syntheses of 1S and 1S-Chl); Fmoc-D-Dab(Boc)-OH (lower left, used in the syntheses of 2R and 2R-Chl); Fmoc-Dab(Boc)-OH (lower right, used in the syntheses of 2S and 2S-Chl).
Polyamides utilizing the α-DABA turn moiety have not been previously studied and the reasons for the unique biological properties displayed by 1R-Chl are unknown. In the current study, we report the chemical and biological characterization of α-DABA polyamides and compare them to the analogous γ-DABA polyamides. A series of four parent molecules and their corresponding chlorambucil conjugates were synthesized (Figure 1A). Each molecule contains the heterocyclic sequence: ImImβIm-HT-PyPyPyPy-βDp (where HT = hairpin turn and Dp = 3-(dimethylamino)-propylamine). Molecules in series 1 contain the new α-DABA turn and molecules in series 2 utilize the γ-DABA turn. The R and S enantiomers of the parent polyamides and chlorambucil conjugates in each series have been generated for comparison. The DNA-binding affinities of the parent molecules were measured by DNase I footprinting and the alkylation profiles (sequence specificity and reactivity) of the polyamide–chlorambucil conjugates were established using thermal cleavage assays. The effects of polyamide treatment on SW620 cell morphology, proliferation, viability and cell cycle profiles were compared using phase contrast microscopy, cell counting, trypan blue exclusion assay and fluorescence-assisted cell-sorting (FACS) analysis, respectively. Importantly, toxicity effects of the polyamide constitutional isomers on mice were determined.
MATERIALS AND METHODS
Polyamide synthesis and characterization
Polyamides were synthesized on solid phase on Boc-β-Pam resin using Boc-protected monomers and dimers according to protocols described previously (23). The following monomers/dimers were used in generating each of the molecules: Boc-Py-OBt (OBt = benzotriazol-1-yloxy), Boc-β-Im-OH and Im-Im-OH. Boc deprotection was conducted at r.t. for 30 min using 80% TFA/DCM prior to the first coupling reaction and after addition of each heterocycle. Carboxylic acids were activated with N,N-diisopropylethylamine (DIEA) and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) for 30 min at 37°C. Monomer and dimer couplings were allowed to continue for 1 h (OBt esters) or 2 h (activated carboxylic acids). Polyamides were cleaved from resin with 3-(dimethylamino)-propylamine neat at 55°C for ∼18 h and subsequently purified by reverse-phase high-performance liquid chromatography (HPLC).
Conjugation of polyamides to chlorambucil (4 eq) proceeded by activating the carboxylic acid with HBTU (4 eq) and DIEA (excess) for 30 min at r.t., after which the activation mixture was added to the polyamide. After allowing the reaction to continue for 1.5–2 h, products were purified by reverse-phase HPLC and immediately lyophilized.
The purity of all compounds were established by analytical HPLC and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry.
1R: ImImβIm-(R)H2Nα-PyPyPyPy-βDp Synthesized using Boc-D-Dab(Fmoc)-OH. MALDI-TOF C54H71N22O10+ calculated [M + H]+: 1188.28, found:1188.02
1S: ImImβIm-(S)H2Nα-PyPyPyPy-βDp Synthesized using Boc-Dab(Fmoc)-OH. MALDI-TOF C54H71N22O10+ calculated [M + H]+: 1188.28, found: 1187.99
2R: ImImβIm-(R)H2Nγ-PyPyPyPy-βDp Synthesized using Fmoc-D-Dab(Boc)-OH. MALDI-TOF C54H71N22O10+ calculated [M + H]+: 1188.28, found: 1187.95
2S: ImImβIm-(S)H2Nγ-PyPyPyPy-βDp Synthesized using Fmoc-Dab(Boc)-OH. MALDI-TOF C54H71N22O10+ calculated [M + H]+: 1188.28, found:1187.77
1R-Chl: ImImβIm-(R)Chlα-PyPyPyPy-βDp Synthesized from 1R. MALDI-TOF C68H88Cl2N23O10+ calculated [M + H]+:1474.48, found: 1474.98
1S-Chl: ImImβIm-(S)Chlα-PyPyPyPy-βDp Synthesized from 1S. MALDI-TOF C68H88Cl2N23O10+ calculated [M + H]+:1474.48, found: 1475.03
2R-Chl: ImImβIm-(R)Chlγ-PyPyPyPy-βDp Synthesized from 2R. MALDI-TOF C68H88Cl2N23O10+ calculated [M + H]+:1474.48, found: 1474.92
2S-Chl: ImImβIm-(S)Chlγ-PyPyPyPy-βDp Synthesized from 2S. MALDI-TOF C68H88Cl2N23O10+ calculated [M + H]+:1474.48, found: 1474.73
Construction of plasmid pMFST2
Oligonucleotides were purchased from Integrated DNA Technologies. The plasmid pMFST2 was constructed by annealing the two oligonucleotides 5′-AGCTGCGCCCTTTAGGTGTTACGTAGTGGTGCCGTAGGTCTTAGCCGTAGCTGTTGCCGTAAGGGCGAATTCTGC-3′ and 5′-GATCGCAGAATTCGCCCTTACGGCAACAGCTACGGCTAAGACCTACGGCACCACTACGTAACACCTAAAGGGCGC-3′, followed by ligation into the BamHI/HindIII restriction fragment of pUC19 using T4 DNA ligase. The plasmid was then transformed into Escherichia coli JM109 competent cells. Ampicillin-resistant white colonies were selected from 25 ml Luria–Bertani (LB) agar plates containing 50 mg/ml ampicillin treated with XGAL and isopropyl-β-d-thiogalactopyranoside (IPTG) solutions and grown overnight at 37°C. Cells were harvested the following day and purification of the plasmid was performed with a Wizard Plus Midiprep DNA purification kit (Promega). DNA sequencing of the plasmid insert was performed by the sequence analysis facility at the California Institute of Technology.
Preparation of 5′ 32P-end-labeled DNA
The primer 5′-GAATTCGAGCTCGGTACCCGGG-3′ was labeled at the 5′ end and subsequently used with the primer 3′-CAGCCCTTTGGACAGCACGGTC-5′ to amplify plasmid pMFST2 as described previously (24).
DNase I footprint titrations
Polyamide equilibrations and DNase I footprint titrations were conducted on the 5′ end-labeled PCR product of pMFST2 according to standard protocols (24). DNA was incubated with polyamide conjugates or water (control) for 12 h at r.t. prior to reaction with DNase.
Thermal cleavage assays
Thermal cleavage assay experiments were conducted on the 5′ end-labeled PCR product of pMFST2 as described previously (19). DNA was incubated with polyamide conjugates or water (control) for 24 h at 37°C prior to work-up, unless otherwise noted.
Cell culture
The cells used in this work were derived from the human colon adenocarcinoma cell line SW620 (purchased from ATCC) and were maintained in Leibovitz's L-15 medium (Gibco) supplemented with 10% heat-inactivated FBS (Cambrex), 10 mM HEPES buffer (Gibco), 1 mM sodium pyruvate (Gibco) and 1% antibiotic-antimycotic (Invitrogen). Cells were grown in a humidified 5% CO2 atmosphere at 37°C. For polyamide treatment experiments, cells were plated in 25 cm2 cell culture flasks (Corning) at 600 000 cells/flask in 10 ml media. Flasks were incubated at 37°C for 2.5 h before addition of polyamide stock solution or water (control). Three flasks were treated per condition. Cells were incubated for 3 days prior to analysis. Cell morphology was recorded by phase contrast microscopy with 40× magnification (Nikon Diaphot with attached Nikon D100 camera). Cells were then trypsinized for 7 min at 37°C and recombined with their media. An aliquot was removed to determine cell viability and proliferation via trypan blue exclusion and cell count using a Vi-Cell XR cell viability analyzer (Beckman–Coulter). One hundred images were obtained per flask (three flasks/condition). The remaining cells from each treatment condition were then combined, pelleted, washed with Dulbecco's phosphate-buffered saline (PBS) (Gibco) and fixed at −20°C with 70% EtOH for 35 min. The fixed cells were then pelleted, washed with Dulbecco's PBS, re-pelleted and suspended in a solution containing 0.1% Triton X-100, 0.2 mg/ml RNAse A (Sigma) and 0.02 mg/ml propidium iodide (Sigma) in Dulbecco's PBS. Cells were incubated overnight at 4°C in the dark prior to FACS analysis (FACSCalibur; Becton Dickinson). Parameters for FL2 were adjusted to give histograms with G0/G1 peaks centered at 200 for each sample to compensate for differences in cell size.
Animal experiments
Female BALB/c mice were purchased from The Scripps Research Institute Division of Animal Resources. Mice 8–12 weeks of age were given injections of PBS (control) or 100 nmol polyamide/injection via the tail vein on days 0, 2 and 4, with the exception of 2R-Chl. Doses of 2R-Chl were given only on days 0 and 2 due to lethality by day 4. The mice were weighed and their behavior monitored and recorded daily. Experimental protocols were approved by The Scripps Institutional Animal Welfare Committee and were conducted in conformity with institutional guidelines, which are in compliance with national and international laws and policies.
RESULTS AND DISCUSSION
Polyamide synthesis
Four hairpin polyamides were prepared by solid phase synthesis (23). The hairpin polyamides differed in the turn substituents utilized: Boc-D-Dab(Fmoc)-OH (1R, 1R-Chl), Boc-Dab(Fmoc)-OH (1S, 1S-Chl), Fmoc-D-Dab(Boc)-OH (2R, 2R-Chl) and Fmoc-Dab(Boc)-OH (2S, 2S-Chl) (Figure 1B). Conjugates were generated by coupling the HBTU-activated carboxylic acid of chlorambucil to the parent polyamide following reverse-phase HPLC of the free amine and purified similarly.
Plasmid design
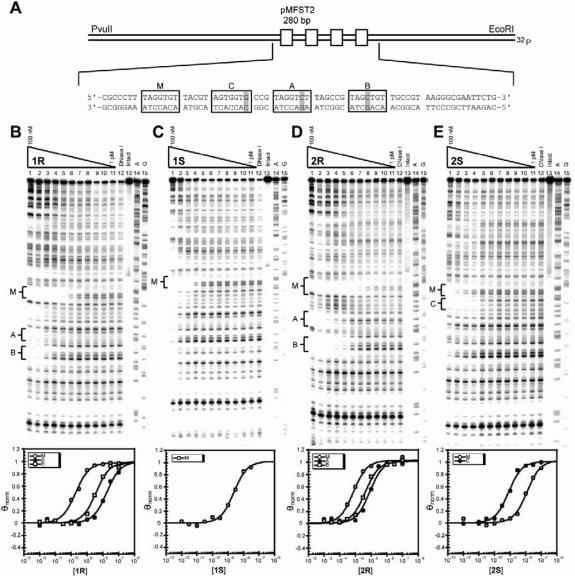
The core polyamide structure was constructed to bind the sequence 5′-WWGGWGW-3′, where W = A or T. In order to probe the affinity and sequence specificity of the parent polyamides, plasmid pMFST2 was designed with an insert containing a single match site (‘M’ = 5′-TAGGTGT-3′) and two single base pair mismatch sites (Figure 2A). The mismatch sites consist of single G•C to C•G replacements at different locations within each binding site (‘A’ = 5′-TAGGTCT-3′, ‘B’ = 5′-TAGCTGT-3′). For the purpose of thermal cleavage assays, each binding site was designed with proximal adenines which the chlorambucil moiety would be expected to alkylate (19). In addition to the designed sites, inherent in the plasmid structure is a single base pair mismatch site (‘C’ = 5′-AGTGGTG-3′) for a polyamide binding in reverse (Supplementary Figure S1).
Figure 2.
Plasmid design and DNA-binding properties of the parent polyamides. (A) Illustration of the designed portion of the EcoRI/PvuII restriction fragment derived from plasmid pMFST2. The four polyamide binding sites are indicated by boxes. Single base pair mismatches are indicated by shaded regions. Site C is a mismatch site for a polyamide binding in reverse. (B–E) Quantitative DNase I footprint titration experiments for polyamides 1R (B), 1S (C), 2R (D), 2S (E) on the 280 bp, 5′ end-labeled PCR product of plasmid pMFST2: lanes 1–11, 100, 30, 10, 3, 1nM; 300, 100, 30, 10, 3 and 1 (M−1) polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. Each footprinting gel is accompanied by its respective binding isotherms for the indicated sites (below).
DNA affinity and sequence specificity
Quantitative DNase I footprint titrations to determine binding site affinities and specificities were conducted for parent polyamides 1R, 1S, 2R and 2S on the 5′-labeled 280 bp PCR product of plasmid pMFST2 (Figure 2 and Table 1). Previous studies have indicated that attachment of the chlorambucil moiety does not significantly alter the DNA-binding affinity of the parent polyamide (19).
Table 1.
Binding affinities (M−1) for parent polyamides
| Polyamide | 5′-TAGGTGT-3′ | 5′-TAGGTCT-3′ | 5′-TAGCTGT-3′ | 5′-AGTGGTG-3′ |
|---|---|---|---|---|
| 1R | 6.7 (±0.1) × 109 | 7.1 (±1.8) × 107 | 2.6 (±0.2) × 108 | — |
| 1S | 4.8 (±0.9) × 108 | — | — | — |
| 2R | 1.6 (±0.3) × 1010 | 1.1 (±0.1) × 109 | 2.9 (±1.0) × 109 | — |
| 2S | 5.8 (±2.1) × 107 | — | — | 1.2 (±0.2) × 109 |
Equilibrium association constants reported are mean values from three DNase I footprint titration experiments; SDs are shown in parentheses.
The α-DABA polyamide 1R bound the match site, M, with Ka = 6.7 × 109 M−1 and it had a 94-fold specificity over single mismatch site A and 26-fold specificity over single mismatch site B (Figure 2B). The enantiomer 1S bound the match site M with Ka = 4.8 × 108 M−1, a 14-fold decrease relative to 1R. Binding affinities at the other sites were too low to be measured and the polyamide was found to coat DNA at high concentrations (Figure 2C).
The γ-DABA polyamides 2R and 2S bound DNA in accordance with the expected orientation of the free amine (14). 2R was shown to bind the match and both single base pair mismatch binding sites (Figure 2D). The affinity of 2R for site M was Ka = 1.6 × 1010 M−1, with 15- and 6-fold preferences over mismatch sites A and B, respectively. The enantiomer, 2S, bound the single base pair mismatch reverse site C with Ka = 1.2 × 109 M−1, a 21-fold enhancement over the forward match site M (Figure 2E).
The binding isotherms for all four molecules were best-fit to a 1:1 binding model. Previous studies have found a glycine amino acid-linked polyamide is able to bind DNA in an extended dimeric conformation that can be observed via a larger binding site in a DNase I footprinting gel. We do not observe evidence of an extended conformation for either 1R or 1S, as the binding sites of both molecules are the same size as those of 2R and 2S. The substituted glycine favors a turn conformation.
The DNase I footprinting experiments reveal that the α-DABA polyamide 1R has a 2-fold reduced binding affinity relative to the γ-DABA polyamide 2R. A contributing factor to the decreased binding observed for 1R and 1S could be a reduced ability of the molecules to fit into the minor groove. The shorter turn linkage between the two subunits in an α-DABA polyamide may reduce its flexibility in conforming to the shape of the groove (Supplementary Figure S2). The energetics of α-DABA hairpin polyamides appear less sensitive than the γ-DABA series to the R and S stereochemistry at the turn.
DNA alkylation specificity and time-dependence
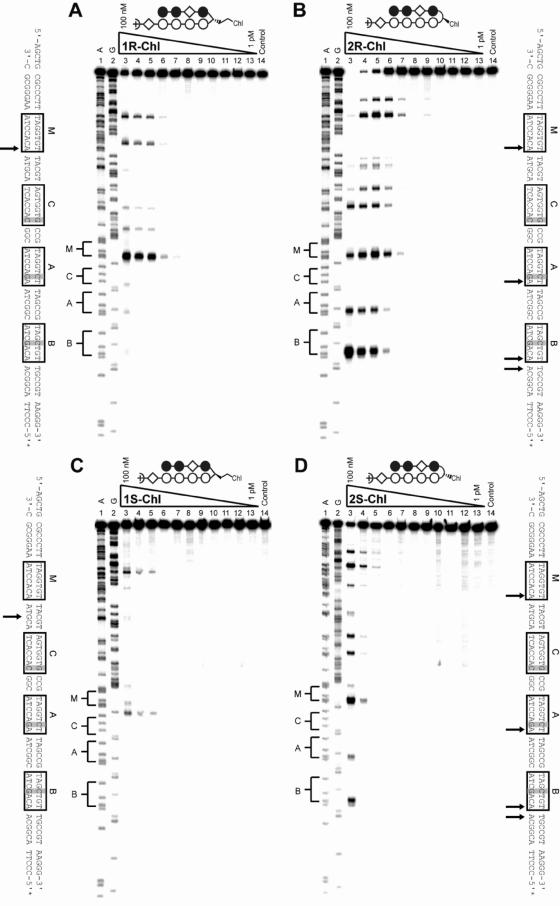
To determine the alkylation properties of the polyamide-chlorambucil conjugates, thermal cleavage assays were performed on the 5′ 32P-labeled 280 bp PCR product of pMFST2 (Figure 3). The alkylation profiles of 1R-Chl and 2R-Chl are dramatically different: 1R-Chl alkylates DNA more specifically than 2R-Chl and appears less reactive. For the designed sequence, 1R-Chl appears to specifically alkylate the adenine proximal to the turn at the match site M at concentrations up to 30 nM (Figure 3A). It only begins to alkylate at the designed mismatch binding sites at 100 nM concentration and alkylation can be observed at concentrations as low as 1 nM. In contrast, 2R-Chl is more promiscuous, alkylating at site M and both mismatch sites, A and B, at concentrations as low as 3 nM (Figure 3B). Only at 1 nM concentration was 2R-Chl observed to specifically alkylate at site M over sites A and B.
Figure 3.
Alkylation specificities of the polyamide conjugates. Thermal cleavage assay experiments with 1R-Chl (A), 2R-Chl (B), 1S-Chl (C) and 2S-Chl (D) on the 280 bp, 5′ end-labeled PCR product of pMFST2: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 100, 30, 10, 3, 1 nM; 300, 100, 30, 10, 3 and 1 pM polyamide, respectively; lane 14, intact DNA. Putative major sites of alkylation on the DNA fragment are indicated by arrows on the sequences adjacent to each gel.
The S enantiomers of the conjugates are similar to their R analogs in their alkylation specificities, but they are muted in activity. In contrast to 1R-Chl, 1S-Chl appears to specifically alkylate the guanine between sites M and C up to a concentration of 30 nM (Figure 3C). Alkylation at N3 of guanine has been observed previously (18,21,25). Based on the DNase I footprinting data for 1S, the observed alkylation is probably due to binding of 1S-Chl at the match site M. The 2S-Chl alkylation profile at 100 nM is similar to that of 2R-Chl at 10 nM, with the polyamide alkylating at the match site and both single base pair mismatch sites (Figure 3D). However, at 30 nM, 2S-Chl is specific for the match site.
Outside of the designed pMFST2 insert, 1R-Chl and 1S-Chl alkylate fewer sites than 2R-Chl and 2S-Chl, respectively, further demonstrating their enhanced alkylation specificity. There are a number of forward-binding and reverse-binding single base pair mismatch sites, as well as one reverse-binding match site, inherent in the labeled fragment of pMFST2 used in these studies. The major sites of alkylation observed for 1R-Chl and 2R-Chl outside of the designed plasmid insert can generally be attributed to foward-binding single base pair mismatch sites, although the gel resolution is often too poor to definitively identify specific alkylation sites. The additional major alkylation sites observed for 2S-Chl can be attributed to a combination of forward-binding and reverse-binding single base pair mismatch sites, while the additional major alkylation site observed for 1S-Chl may correspond to a reverse-binding single base pair mismatch site.
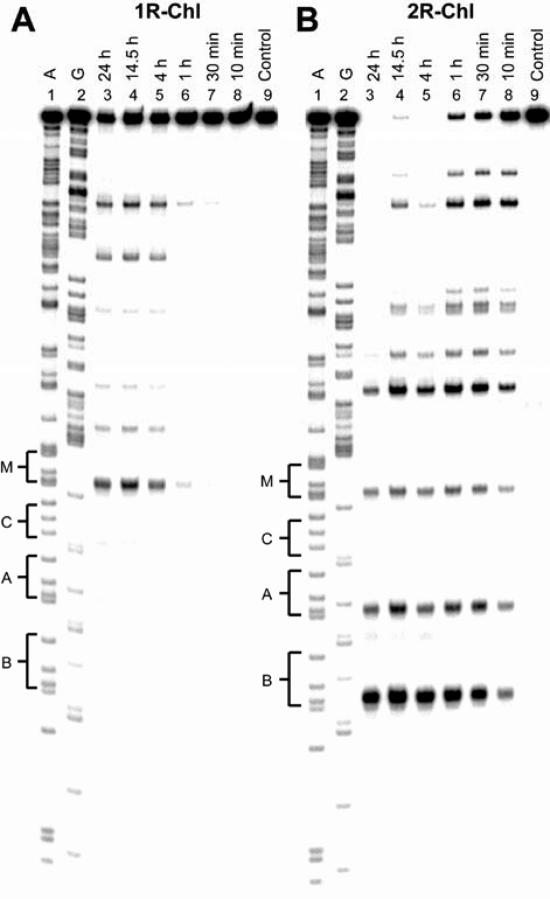
We next studied the time-dependence of alkylation for 1R-Chl and 2R-Chl to quantitate their kinetic parameters (Figure 4). At 500 nM polyamide and 37°C, the half-life of the labeled DNA (i.e. disappearance of full-length DNA) is 20 h for 1R-Chl and 10 min for 2R-Chl. Thus, 2R-Chl alkylates DNA 120 times faster than 1R-Chl under these conditions. Given the minimal structural differences between 1R-Chl and 2R-Chl and the similar binding affinities of their parent molecules, the dramatic difference in alkylation specificity and kinetics between hairpins with a common alkylator moiety would not have been predicted.
Figure 4.
Time-dependence of alkylation for polyamide conjugates. Thermal cleavage assay experiments with 500 nM 1R-Chl (A) and 500 nM 2R-Chl (B) on the 280 bp, 5′ end-labeled PCR product of pMFST2: lane 1, A reaction; lane 2, G reaction; lanes 3–8, equilibrations for 24, 14.5, 4, 1 h; 30 and 10 min, respectively; lane 9, intact DNA.
Effects of polyamides on SW620 cells
1R-Chl treatment of cultured SW620 cells has been shown to induce a morphology change and arrest cell growth in the G2/M phase. In addition, the polyamide was found to be a cytostatic agent, decreasing cell proliferation without significantly affecting cell viability, as determined by trypan blue staining (21). A two-hit mechanism for growth arrest by 1R-Chl was suggested by experiments where a different polyamide-chlorambucil conjugate, which by itself had no effect on SW620 cell proliferation, was found to cause growth arrest after treatment of these cells with an siRNA directed against H4c mRNA (26). Our model for the action of 1R-Chl involves direct alkylation of the H4c gene, leading to a block in transcription and eventual depletion of H4 protein, opening up the genome for massive alkylation by 1R-Chl and G2/M arrest.
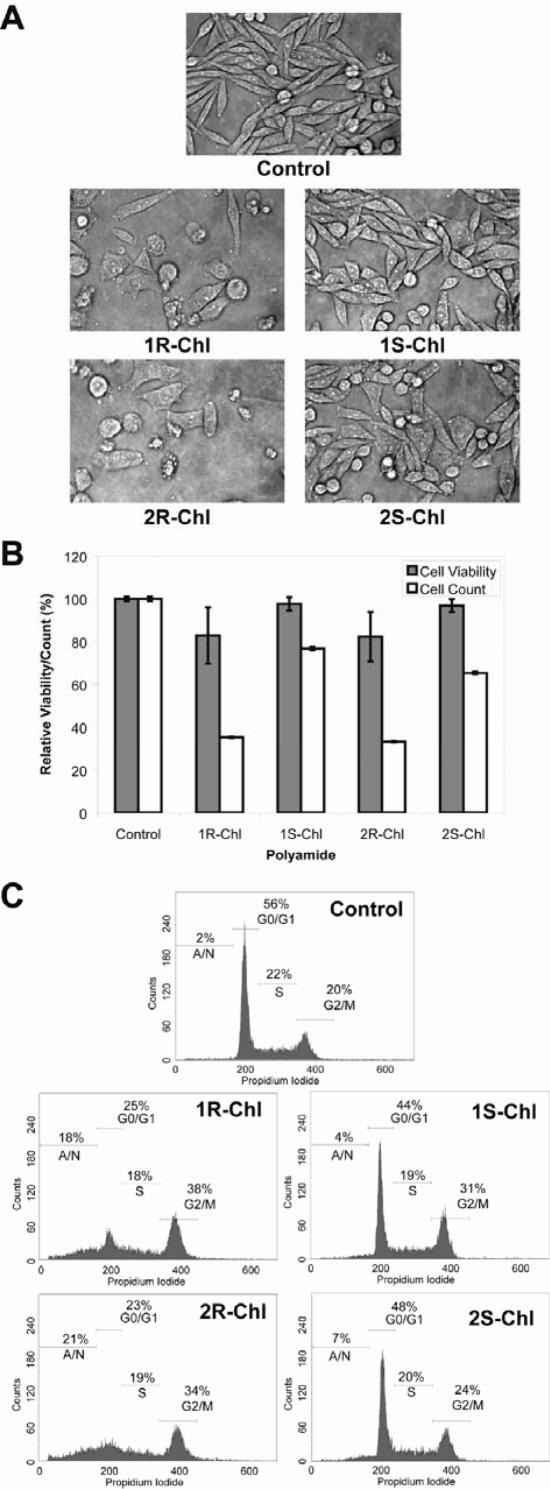
We studied the effects of our polyamide library on SW620 cultured cells to determine whether the differences between the polyamides observed in vitro would translate to disparities in cell morphology, proliferation, viability and cell cycle profile. As expected, no effects were observed for the treatment of cells with the four parent polyamides (Supplementary Figure S3). Cells treated for 3 days with 1R-Chl or 2R-Chl show the same morphological change as established previously (Figure 5A). Each conjugate also decreases cell proliferation to ∼30% relative to control with only a small decrease in cell viability, as measured with trypan blue exclusion staining (Figure 5B). FACS analysis using propidium iodide staining indicates both molecules arrest cells in the G2/M phase of growth (38% for 1R-Chl and 34% for 2R-Chl, compared to 20% for control; Figure 5C).
Figure 5.
Effects of polyamide conjugates on cultured cells. Cultured SW620 cells were treated with 200 nM of the indicated polyamide for 3 days prior to analysis. (A) Representative phase microscopy images. (B) Normalized cell viabililty and proliferation values. Data shown are the averages of three samples; error bars indicate standard deviations. (C) Fluorescence-activated cell-sorting analysis of cells. Plots indicate cell numbers versus propidium iodide staining. Percentages of cells in G0/G1, S, G2/M and apoptotic/necrotic cells (A/N) are indicated.
Interestingly, a less pronounced cellular effect is observed for cells treated with 1S-Chl and 2S-Chl. Although most cells appear identical to control, some of the cells treated with each of the S enantiomers display a morphology change (Figure 5A). Cell viability is not significantly reduced, but proliferation decreases slightly (77% for 1S-Chl and 65% for 2S-Chl relative to control; Figure 5B), indicating a slight cytostatic effect for these conjugates. This is also apparent in the FACS data for 1S-Chl, where 31% of the cells are found to be in the G2/M phase of growth. The greater population of apoptotic/necrotic cells following 2S-Chl treatment may explain why significant growth arrest is not observed with these cells. We find that the trend of similar but muted effects of S relative to R enantiomers observed in the alkylation data appears to correlate to biological effects on SW620 cells. Previous studies have correlated the DNA interstrand crosslinking ability of alkylating agents with biological activity (27,28). Since the S isomers are poor DNA alkylators and have minimal biological activity in SW620 cells, we chose not to investigate them further.
Additional experiments were performed with longer incubation times to determine if cells treated with 1R-Chl or 2R-Chl die by apoptosis (Supplementary Figure S4). SW620 cells were treated with 200 nM polyamide for either 3 or 6 days, with the 6-day experiment involving replacement on the third day with fresh media containing 200 nM polyamide. Comparison of the FACS analyses of these cells using annexin V-fluorescein isothiocyanate (annexin V-FITC) and propidium iodide staining shows that after 6 days, 2R-Chl treatment leads to greater cell death than 1R-Chl treatment (33.7 versus 20.2%, respectively). However, a significant difference in apoptosis (i.e. cells that are annexin V positive and propidium iodide negative) between treatments with 1R-Chl and 2R-Chl is not observed (5.28 versus 6.73%, respectively).
In vivo effects of polyamides on BALB/c mice
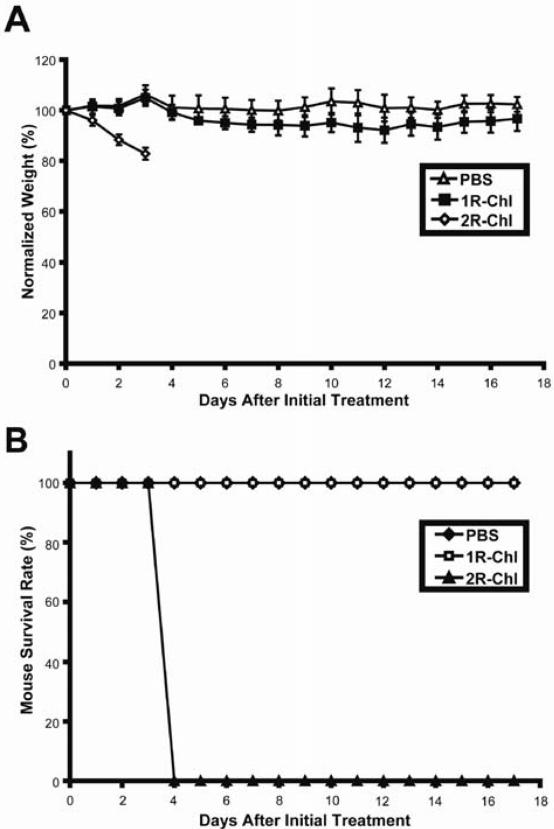
1R-Chl has been shown to arrest cancer growth in a SW620 xenograft nude mouse tumor model, whereas 2R-Chl treatment was toxic to the mice (21). The effects of 1R-Chl and 2R-Chl on a normal mouse strain were tested. Groups of five BALB/c mice were injected with PBS (control), 100 nmols 1R-Chl, or 100 nmols 2R-Chl every other day for up to 5 days. The mice were observed and weighed each day for up to 17 days (Figure 6A). Mice treated with 1R-Chl do not differ significantly from the control group. However, 2R-Chl treatment of mice results in an average weight loss of 17% after 3 days and is found to be lethal to 100% of the mice after 4 days (Figure 6B).
Figure 6.
Effects of polyamide conjugates on BALB/c mice. Female BALB/c mice were treated with PBS (control), 100 nmols 1R-Chl, or 100 nmols 2R-Chl at 0, 2 and 4 (PBS and 1R-Chl only) days. Five mice were treated for each experimental condition. (A) Average mouse weights, normalized to weight at day 0, over time. Error bars indicate standard deviations. (B) Survival rates for treated mice.
CONCLUSION
We have shown that changing the turn unit of a hairpin pyrrole–imidazole polyamide–chlorambucil conjugate from the standard γ-DABA to an α-DABA can result in dramatically different chemical and biological properties. Although unconjugated parent hairpin polyamides 1R and 2R bind DNA with comparable binding affinities, their corresponding chlorambucil conjugates have different alkylation profiles. The polyamide conjugate 1R-Chl, which contains the (R)-α-DABA turn unit, alkylates DNA much more specifically and at a decreased rate relative to 2R-Chl, which contains the (R)-γ-DABA turn. Strikingly, treatment of a common mouse strain with 1R-Chl did not appear to have adverse effects on the mice, whereas treatment with 2R-Chl was lethal. Although it is possible the difference in toxicities between the two molecules is due to variance in absorption, distribution, metabolism or excretion, given their structural similarities, it is not unreasonable to speculate the origin is the dramatic difference in alkylation specificity and reactivity. These results would not have been predicted a priori and indicate that hairpin polyamides containing the α-DABA turn unit may be an important class of DNA-binding small molecules with interesting biological properties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank David Alvarez for helpful discussions; David M. Chenoweth for molecular modeling; Kathy Klingensmith, Pastorcito Nieto and Antonio Reyes from TSRI ICND Vivarium; and Dr Kent Osborn DMV and Leslie Nielsen from TSRI Pathology lab for helpful discussions and assistance on animal care and protocols. The authors are grateful to the National Institutes of Health for research support (CA107311) and for a predoctoral NRSA training grant to M.E.F. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health (CA107311).
Conflict of interest statement. None declared.
REFERENCES
- 1.Dervan P.B., Poulin-Kerstien A.T., Fechter E.J., Edelson B.S. Regulation of gene expression by synthetic DNA-binding ligands. Top. Curr. Chem. 2005;253:1–31. [Google Scholar]
- 2.Best T.P., Edelson B.S., Nickols N.G., Dervan P.B. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl Acad. Sci. USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edelson B.S., Best T.P., Olenyuk B., Nickols N.G., Doss R.M., Foister S., Heckel A., Dervan P.B. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olenyuk B.Z., Zhang G.J., Klco J.M., Nickols N.G., Kaelin W.G., Dervan P.B. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl Acad. Sci. USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett R., Melander C., Puckett J.W., Son L.S., Wells R.B., Dervan P.B., Gottesfeld J.M. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA•TTC repeats in Friedreich's ataxia. Proc. Natl Acad. Sci. USA. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wemmer D.E., Dervan P.B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 1997;7:355–361. doi: 10.1016/s0959-440x(97)80051-6. [DOI] [PubMed] [Google Scholar]
- 7.Turner J.M., Swalley S.E., Baird E.E., Dervan P.B. Aliphatic/aromatic amino acid pairings for polyamide recognition in the minor groove of DNA. J. Am. Chem. Soc. 1998;120:6219–6226. [Google Scholar]
- 8.Wang C.C.C., Ellervik U., Dervan P.B. Expanding the recognition of the minor groove of DNA by incorporation of beta-alanine in hairpin polyamides. Bioorg. Med. Chem. 2001;9:653–657. doi: 10.1016/s0968-0896(00)00282-0. [DOI] [PubMed] [Google Scholar]
- 9.White S., Baird E.E., Dervan P.B. Orientation preferences of pyrrole-imidazole polyamides in the minor groove of DNA. J. Am. Chem. Soc. 1997;119:8756–8765. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- 10.Mrksich M., Parks M.E., Dervan P.B. Hairpin peptide motif—a new class of oligopeptides for sequence-specific recognition in the minor-groove of double-helical DNA. J. Am. Chem. Soc. 1994;116:7983–7988. [Google Scholar]
- 11.Swalley S.E., Baird E.E., Dervan P.B. Effects of gamma-turn and beta-tail amino acids on sequence-specific recognition of DNA by hairpin polyamides. J. Am. Chem. Soc. 1999;121:1113–1120. [Google Scholar]
- 12.Geierstanger B.H., Mrksich M., Dervan P.B., Wemmer D.E. Extending the recognition site of designed minor groove binding molecules. Nature Struct. Biol. 1996;3:321–324. doi: 10.1038/nsb0496-321. [DOI] [PubMed] [Google Scholar]
- 13.de Clairac R.P.L., Seel C.J., Geierstanger B.H., Mrksich M., Baird E.E., Dervan P.B., Wemmer D.E. NMR characterization of the aliphatic beta/beta pairing for recognition of a A•T/T•A base pairs in the minor groove of DNA. J. Am. Chem. Soc. 1999;121:2956–2964. [Google Scholar]
- 14.Herman D.M., Baird E.E., Dervan P.B. Stereochemical control of the DNA binding affinity, sequence specificity, and orientation preference of chiral hairpin polyamides in the minor groove. J. Am. Chem. Soc. 1998;120:1382–1391. [Google Scholar]
- 15.Shinohara K., Sasaki S., Minoshima M., Bando T., Sugiyama H. Alkylation of template strand of coding region causes effective gene silencing. Nucleic Acids Res. 2006;34:1189–1195. doi: 10.1093/nar/gkl005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bando T., Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006 doi: 10.1021/ar030287f. doi:10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- 17.Pezzoni G., Grandi M., Biasoli G., Capolongo L., Ballinari D., Giuliani F.C., Barbieri B., Pastori A., Pesenti E., Mongelli N., et al. Biological profile of Fce-24517, a novel benzoyl mustard analog of distamycin-A. Br. J. Cancer. 1991;64:1047–1050. doi: 10.1038/bjc.1991.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt M.D., Lee M., Hartley J.A. Alkylation specificity for a series of distamycin analogues that tether chlorambucil. Anti-Cancer Drug Des. 1997;12:49–60. [PubMed] [Google Scholar]
- 19.Wurtz N.R., Dervan P.B. Sequence specific alkylation of DNA by hairpin pyrrole-imidazole polyamide conjugates. Chem. Biol. 2000;7:153–161. doi: 10.1016/s1074-5521(00)00085-5. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y.D., Dziegielewski J., Wurtz N.R., Dziegielewska B., Dervan P.B., Beerman T.A. DNA crosslinking and biological activity of a hairpin polyamide-chlorambucil conjugate. Nucleic Acids Res. 2003;31:1208–1215. doi: 10.1093/nar/gkg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson L.A., Burnett R., Melander C., Edelson B.S., Arora P.S., Dervan P.B., Gottesfeld J.M. Arresting cancer proliferation by small-molecule gene regulation. Chem. Biol. 2004;11:1583–1594. doi: 10.1016/j.chembiol.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Dickinson L.A., Burnett R., Melander C., Edelson B.S., Arora P.S., Dervan P.B., Gottesfeld J.M. Erratum: arresting cancer proliferation by small-molecule gene regulation. Chem. Biol. 2006;13:339. doi: 10.1016/j.chembiol.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Baird E.E., Dervan P.B. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 24.Trauger J.W., Dervan P.B. Footprinting methods for analysis of pyrrole-imidazole polyamide/DNA complexes. Meth. Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 25.Wyatt M.D., Lee M., Garbiras B.J., Souhami R.L., Hartley J.A. Sequence specificity of alkylation for a series of nitrogen mustard-containing analogs of distamycin of increasing binding-site size—evidence for increased cytotoxicity with enhanced sequence specificity. Biochemistry. 1995;34:13034–13041. doi: 10.1021/bi00040a014. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez D., Chou C.J., Latella L., Zeitlin S.G., Ku S., Puri P.L., Dervan P.B., Gottesfeld J.M. A two-hit mechanism for pre-mitotic arrest of cancer cell proliferation by a polyamide-alkylator conjugate. Cell Cycle. 2006;5:1537–1548. doi: 10.4161/cc.5.14.2913. [DOI] [PubMed] [Google Scholar]
- 27.Pawlak K., Pawlak J.W., Konopa J. The mode of action of cyto-toxic and antitumor 1-nitroacridines 4. Cyto-toxic and antitumor-activity of 1-nitroacridines as an after effect of their interstrand DNA cross-linking. Cancer Res. 1984;44:4289–4296. [PubMed] [Google Scholar]
- 28.Sunters A., Springer C.J., Bagshawe K.D., Souhami R.L., Hartley J.A. The cytotoxicity, DNA cross-linking ability and DNA-sequence selectivity of the aniline mustards melphalan, chlorambucil and 4-[bis(2-chloroethyl)amino] benzoic-acid. Biochem. Pharmacol. 1992;44:59–64. doi: 10.1016/0006-2952(92)90038-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.