Abstract
We have previously reported that the CDX1 homeoprotein interacts with the TATA-box binding protein (TBP) on the promoter of the glucose-6-phosphatase (G6Pase) gene. We show here that CDX1 interacts with TBP via the homeodomain and that the transcriptional activity additionally requires the N-terminal domain upstream of the homeodomain. CDX1 interacting with TBP is connected to members of the TFIID and Mediator complexes, two major elements of the general transcriptional machinery. Transcription luciferase assays performed using an altered-specificity mutant of TBP provide evidence for the functionality of the interaction between CDX1 and TBP. Unlike CDX1, CDX2 does not interact with TBP nor does it transactivate the G6Pase promoter. Swapping experiments between the domains of CDX1 and CDX2 indicate that, despite opposite functional effects of the homeoproteins on the G6Pase promoter, the N-terminal domains and homeodomains of both CDX1 and CDX2 have the intrinsic ability to activate transcription and to interact with TBP. However, the carboxy domains define the specificity of CDX1 and CDX2. Thus, intra-molecular interactions control the activity and partner recruitment of CDX1 and CDX2, leading to different molecular functions.
INTRODUCTION
Homeobox-containing genes encode a large family of transcription factors involved in body plan organization during embryogenesis and in tissue homeostasis in many adult organs (1,2). Mutations within these genes cause malformations and metabolic diseases. In addition, several lines of evidence indicate that changes in homeobox gene expression are involved in malignant tumor progression (3,4). Although homeobox genes are thought to control large genetic programs, their molecular mechanisms of action are not fully elucidated.
The three mammalian homeobox genes of the CDX family, related to the Drosophila gene caudal, are major components of the network that defines the antero-posterior axis in embryos. At the adult stage, CDX1 and CDX2 remain selectively active in the continuously renewing intestinal epithelium (5,6). Although CDX2 is expressed in most gut epithelial cells, CDX1 expression is limited to crypts cells, which belong to the proliferating cell compartment (7). The two genes have both common and specific cellular effects. Indeed, whereas they both trigger cell differentiation, CDX1 stimulates while CDX2 decreases proliferation in cell cultures (8–10). They are also differently targeted by regulatory pathways as for example, CDX1 is directly upregulated by Wnt/β-catenin signaling (11), which is mostly active in the crypt compartment, whereas CDX2 is indirectly downregulated by this pathway (12). Transgenic models have provided further evidence that the CDX1 and CDX2 genes exert both common and specific effects in relation to pathological conditions. Indeed, on the one hand the ectopic expression of either CDX1 or CDX2 causes intestinal-type metaplasia of the gastric epithelium (13). On the other hand, CDX2 has been shown to play a tumor suppressor role in murine models of sporadic and genetic colorectal cancers (14,15), whereas CDX1 was reported to mediate the hyperproliferative condition linked to Rb and p130 deficiency in the gut (16). Outside the gut, recent studies have reported that the CDX2 and CDX1 proteins have different effects on the promoters of genes involved in early embryonic development (17). On this basis, it is therefore important to delineate the specific molecular functions of the CDX1 and CDX2 proteins.
One mechanism to account for the different effects exerted by the CDX1 and CDX2 proteins postulates that they could interact with different functional partners. The CDX1 and CDX2 proteins exhibit extensive sequence similarity in their homeodomains responsible for DNA-binding (96.6%), whereas the percentage of identity is lower in the N-terminal (35.5%) and C-terminal domains (36.5%) flanking the homeodomain. We have recently shown that the promoter of the glucose-6-phosphatase (G6Pase) gene is selectively activated by CDX1, whereas CDX2 counteracts this stimulatory effect (18). Both homeoproteins bind to the G6Pase promoter at the level of the TATA-box, but only CDX1 was found to interact with the TATA-box binding protein (TBP) by means of co-immunoprecipitation. In the present study, we have analyzed the functionality of the interaction between TBP and CDX1, and we have used hybrid molecules between CDX1 and CDX2 to investigate the differential effects of these homeoproteins on the G6Pase promoter.
MATERIALS AND METHODS
Plasmids
The plasmids encoding CDX1, CDX2, HA-CDX1, HA-CDX2, CDX1-HD2, CDX2-HD1, TBP, HA-TBP and TBP-spm3 were described previously (8,18,19). To simplify the nomenclature, CDX1-HD2 and CDX2-HD1 were renamed into ND1-HD2-CD1 and ND2-HD1-CD2, respectively, where ND represents the N-terminal domain, HD the homeodomain, and CD the C-terminal domain of these homeoproteins. Hybrid forms with glutathione S-transferase (GST) fused upstream of CDX1 or CDX2 were constructed in the pBC vector (20). For this purpose, the plasmids pCB6-CDX1S and pCB6-CDX2S modified with a NheI site inserted immediately downstream of the translation start-site (18) were cut with NheI, filled in with T4 DNA polymerase and cut with BamHI. The resulting fragments were introduced into the EcoRV/BamHI sites of pBC. Plasmids encoding truncated forms of HA-CDX1 were constructed as follows (see also Figure 3). The plasmid coding for the mutant HA-ND1-HD1, deleted of the C-terminal domain downstream of the homeodomain, was obtained by introducing a stop codon in pCB6-HA-CDX1 (18) using the Gene Editor™ in vitro site directed mutagenesis system (Promega) and the oligonucleotide 5′-AAAGTAAACAAGAAGAGCTAGCAGCAGCAGCAGCCC-3′. The plasmid encoding HA-ND1 corresponding to the N-terminal domain and deleted of the homeodomain and the C-terminal domain was constructed by PCR using the primers 5′-CACCATGCTAGCATATCCCTATGACGTCCCAGACTA-3′/5′-ACCGGTTGTGTAGACCACACGTGACTTGTC-3′ and pCB6-HA-CDX1 as template; the resulting PCR fragment was cloned into the vector pcDNA3.1/V5-His by TOPO-TA cloning (Invitrogen). The plasmid encoding the mutant form having the homeodomain linked to the C-terminal domain, HA-HD1-CD1, was constructed similarly from pCDX2-S using the PCR primers 5′-CACCATGCTAGCATATCCCTATGACGTCCCAGACTATGCGCTAGCTCGGCGCAGCGTGGCGGCTGCAGGC-3′/5′-ACCGGTGGGTAGAAACTCCTCCTTGACGGG-3′. High fidelity Platinum Taq DNA polymerase (Invitrogen) was used for PCR. To construct plasmids encoding the proteins HA-ND1-HD2-CD2 and HA-ND2-HD1-CD1, we first changed the GTGGTCTACACAGAC sequence of pCB6-HA-CDX1 into GTGGTGTACACAGAC to create a Bsrg1 restriction site upstream of the homeobox without changing the encoded protein sequence. Then, the resulting plasmid and the plasmid pCB6-HA-CDX2 were cut with NheI and Bsrg1, and the corresponding Nhe1/Bsrg1 restriction fragments were swapped between both plasmids. The reporter luciferase plasmids pG6Pase-Luc containing the −36 to +60 nt of the G6Pase promoter (18) and pTGTA-Luc (19) were described previously. The plasmid pTGTA-G6Pase-Luc was constructed by changing the TATA-box of pG6Pase-Luc into TGTAAAA using the oligonucleotide 5′-ACAGACTGAATCCAGGGCATGTAAAATGGGCAAGGCACAGA-3′ and the Gene Editor™ in vitro site directed mutagenesis system (Promega). All plasmids were confirmed by sequencing.
Cell transfections, protein extractions, in vitro translation and luciferase assays
HCT116 and HepG2 cells were cultured in DMEM with 10 and 20% FBS, respectively, at 37°C in humidified atmosphere under 5% CO2. Cells at 70–80% confluence were co-transfected using JET-PEI (Polyplus Transfection) with the indicated plasmids and either pEGFP-C1 (Clontech) or the Renilla luciferase control vector pRL-null (Promega) to monitor transfection efficiency. For biochemical analyses, cells were scraped 48 h after transfection in ice-cold NP-40 lysis buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 1% Igepal) and a cocktail of protease inhibitors (Sigma), put 10 min on ice and centrifuged in an Eppendorf 5417R, Rotor F45-30-11 at 10 000 g for 10 min at 4°C. The supernatant was removed and assayed for protein concentration using Bradford reaction. For luciferase assays, cells were lysed in Passive Lysis Buffer (Promega), and firefly luciferase and Renilla luciferase activities were measured sequentially using the dual-luciferase assay reporter system (Promega). Firefly luciferase activity was normalized with Renilla luciferase activity and the data were expressed as fold of induction in the experimental condition compared to the control.
For in vitro production of the full-length HA-CDX1 protein or the truncated form HA-HD1, PCR fragments were respectively produced using pCB6-HA-CDX1 and pCB6-HA-HD1-CD1 as templates and the forward primer containing the T7 promoter 5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGCTAGGATATCCCTATGACGTC-3′ together with the specific reverse primers 5′-30x(T)AGGGTA-GAAACTCCTCCTT GACCGGGCACTG-3′ or 5′-30x(T)ATACTTTGCGCTCCTTGGCCCGGCGGTTCT-3′, respectively. In vitro transcription/translation were performed by adding 500 ng of PCR fragment to the synthesis mixture (TNT-T7 Quick coupled Transcription/Translation System, Promega) containing 40 μCi of [35S]Methionine (1000 Ci/mmol and 10 μCi/mL, Amersham), as recommended by the supplier.
GST-pulldown and co-immunoprecipitation assays
GST-pulldown was performed using glutathione–Sepharose-4B beads (Amersham Biosciences), on either protein extracts of cells transfected with the plasmids encoding GST, GST-CDX1 or GST-CDX2, or on protein extracts of bacteria transformed with the plasmid pGST-TBP (21). After 3 h incubation under gentle agitation at 4°C, the beads were washed in lysis buffer and proteins were analyzed by SDS–PAGE. Proteins from cells transfected with the plasmids encoding HA-tagged CDX1, CDX2, TBP or TBP-spm3 were subjected to immunoprecipitation using anti-HA antibody matrix (3F10, Roche). Alternatively, immunoprecipitation was performed using rabbit anti-CDX1 [C1C, (15)], mouse anti-CDX2 (392 M, Biogenex) or mouse anti-TBP [3G3, (22)] antibodies and subsequent incubation overnight at 4°C with protein A-agarose or protein G-agarose beads (Roche). Then, the beads were washed in lysis buffer and the proteins were analyzed by SDS–PAGE and western blot, as described (23). Briefly, they were transferred on to nitrocellulose filters and identified using the primary antibodies raised against GST (G7781 Sigma, dilution 1:5000), HA (3F10, Roche, dilution 1:1000), CDX1 [C1C (15), dilution 1:2000], CDX2 (392M, Biogenex, dilution 1:5000), MED7 (dilution 1:10, kindly provided by Dr M. Meisterernst), TBP (3G3, dilution 1:2000), TAF7 (19TA, dilution 1:1000), TAF15 (16TA, dilution 1:1000) and TAF12 [22TA, dilution 1:1000 (22)]. Immunodetection used sheep anti-mouse κ-chain immunoglobulins (dilution 1:5000) or donkey anti-rabbit immunoglobulins (dilution 1:5000) or sheep anti-rat immunoglobulins (dilution 1:5000) coupled to horseradish peroxydase (HRP) (Amersham Biosciences). The filters were developed using ECL chemiluminescence (Amersham Biosciences).
Immunocytofluorescence
Cells cultured in eight chamber slides (Lab-Tek) were taken 24 h after transfection, fixed in 4% paraformaldehyde for 10 min and incubated 1 h in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 5% normal goat serum. Primary polyclonal anti-HA antibody (Sigma H6908, dilution 1:1000) was added for overnight incubation at 4°C and the primary antibody was detected by fluorescence using goat anti-rabbit immunoglobulins coupled to Alexa 488 (Molecular Probes, dilution 1:1000) for 1 h in the dark. Cell nuclei were stained with DAPI and slides were mounted in Glycerol/PBS/Phenylenediamine for observation using an Olympus AX60 microscope.
RESULTS
Selective interaction of TBP with the homeoprotein CDX1 but not with CDX2
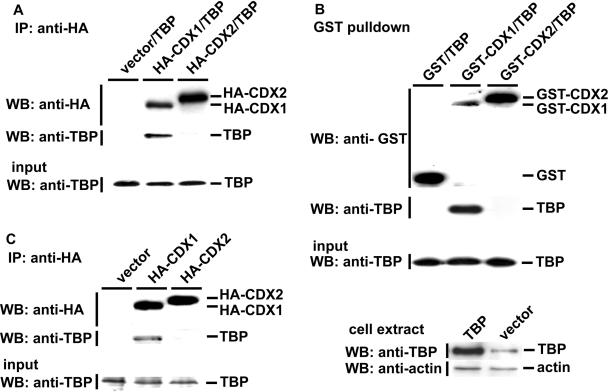
We have used two methods to examine the selective interaction of CDX1 with TBP. When introduced by co-transfection into HCT116 cells, overexpressed TBP was co-immunoprecipitated by HA-tagged CDX1, but not by HA-CDX2 (Figure 1A). To provide evidence that the interaction between TBP and HA-CDX1 is unrelated to the HA-tag and to demonstrate the interaction with a method not dependent on antibodies, we used plasmids encoding GST-fusion versions of CDX1 and CDX2 to perform GST-pulldown assays. After co-transfection with the corresponding plasmids, the results confirmed the selective interaction of TBP with CDX1, and the absence of interaction with CDX2 (Figure 1B). We then investigated if endogenous TBP can be trapped by GST-CDX1 or HA-CDX1. For this purpose, HCT116 or HepG2 cells were transfected only with the plasmids encoding the GST-fusion or HA-tagged forms of CDX1. HA co-immunoprecipitation (Figure 1C) or GST-pulldown (data not shown) indicated that endogenous TBP was recovered together with CDX1. In contrast, no TBP was retained in similar experiments conducted with HA-tagged or GST-fusion forms of CDX2, demonstrating the selectivity of the interaction between CDX1 and endogenous TBP (Figure 1C). Since CDX2 does not interact with TBP, we checked the possibility of an interaction with the TBP-related protein TLF/TRF2. Like for TBP, no interaction was detected between CDX2 and TLF/TRF2. In addition, we also failed to detect any interaction between CDX1 and TLF/TRF2, which strengthens the specificity of the CDX1/TBP interaction (data not shown).
Figure 1.
Interaction of TBP with CDX1 but not with CDX2. (A) Specific co-immunoprecipitation of TBP and HA-CDX1. HCT116 cells were co-transfected with the plasmids encoding TBP and either HA-CDX1, HA-CDX2 or the control empty vector, as indicated. Proteins immunoprecipitated with anti-HA antibody were detected by western blots using anti-HA to control the immunoprecipitation step and anti-TBP antibody to detect the co-immunoprecipitation of TBP. TBP was recovered in the fraction immunoprecipitated with HA-CDX1 but not with HA-CDX2. The presence of TBP in the cell extracts prior to HA-immunoprecipitation was controlled by western blot using anti-TBP antibody (Input). (B) Specific GST-pulldown of overexpressed TBP with GST-CDX1. HCT116 cells were co-transfected with the plasmids encoding TBP and either GST-CDX1, GST-CDX2 or the control GST vector, pBC. GST-pulldown extracts were analyzed by western blots using anti-GST antibody and assayed for the presence of TBP using anti-TBP antibody. TBP was recovered in the fraction containing GST-CDX1 but not GST-CDX2. The presence of TBP in the cell extracts prior to GST-pulldown was controlled by western blot using anti-TBP antibody (Input). The lower panel demonstrates overexpression of TBP in total extracts of HCT116 cells transfected with the TBP-encoding plasmid as compared to cells transfected with the empty vector. (C) Specific co-immunoprecipitation of endogenous TBP by HA-CDX1. HepG2 cells were transfected with the plasmids encoding HA-CDX1, HA-CDX2 or the control plasmid. Immunoprecipitation was performed as described in A with anti-HA antibody followed by western blot analysis with anti-HA or anti-TBP. HA-CDX1, but not HA-CDX2, was able to interact with endogenous TBP. The presence of TBP in the cell extracts prior to HA-immunoprecipitation was controlled by western blot using anti-TBP antibody (Input).
Characterization of the CDX1–TBP interaction
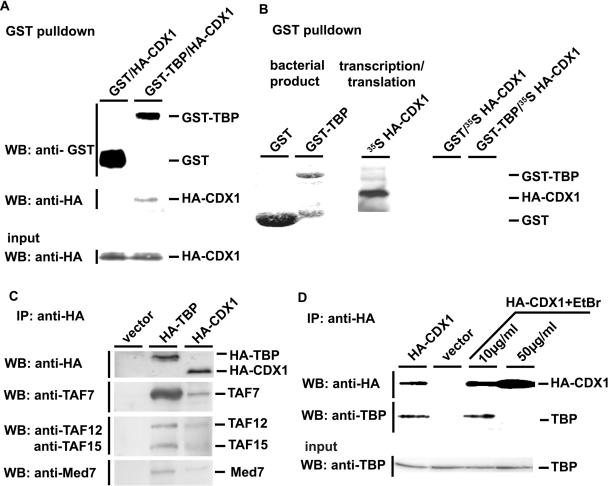
To further address the nature of the interaction between CDX1 and TBP, GST-TBP was produced in bacteria and HA-CDX1 in transfected HCT116 cells. After binding of GST-TBP by affinity to glutathione–Sepharose-4B beads, the beads coupled to GST-TBP were mixed with the extracts of HA-CDX1 transfected cells. Subsequently, GST-pulldown and western blot analysis with anti-HA antibody demonstrated that CDX1–TBP interaction occurred in vitro (Figure 2A). Then we synthesized HA-CDX1 by in vitro transcription/translation and the resulting translation product was mixed with the glutathione–Sepharose-4B beads coupled to GST-TBP. In this condition, no interaction between CDX1 and TBP was observed (Figure 2B).
Figure 2.
Characterization of the CDX1/TBP interaction. (A) Interaction of bacterially-produced GST-TBP and cellular HA-CDX1. GST or GST-TBP produced in bacteria was bound to glutathione–Sepharose-4B beads [see also the first two lanes in (B)] and mixed with HCT116 cells extracts containing HA-CDX1. After GST-pulldown, the presence of GST and GST-TBP was revealed using anti-GST antibody, and the presence of HA-CDX1 was detected using anti-HA antibody. The presence of HA-CDX1 in the cell extracts prior to GST-pulldown was control by western blot with anti-HA antibody (Input). (B) Lack of interaction between bacterially-produced GST-TBP and in vitro translated HA-CDX1. GST (first lane, Coomasie blue staining) and GST-TBP (second lane, Coomasie blue staining) were produced in bacteria and bound to glutathione–Sepharose-4B beads. 35S-Methionine-labeled HA-CDX1 was synthesized by in vitro transcription/translation (third lane, autoradiography). By mixing GST and HA-CDX1 (fourth lane) or GST-TBP and HA-CDX1 (fifth lane), no interaction was detected after GST-pulldown and autoradiography. (C) CDX1 interaction with components of the TFIID and Med complexes. HCT116 cells were transfected with the control empty vector or with the plasmids encoding HA-TBP or HA-CDX1. Proteins immunoprecipitated with anti-HA antibody were detected by western blots using the antibodies raised against TAF7, TAF12, TAF15 and Med7. D. Effect of ethidium bromide (EtBr) on the CDX1/TBP interaction. HCT116 were transfected with the plasmid encoding HA-CDX1 and the proteins were immunoprecipitated with anti-HA antibody in the presence of increasing amount of ethidium bromide. Western blot with anti-HA antibody was used to control the step of immunoprecipitation and anti-TBP was used to detect the co-immunoprecipitated TBP. The presence of TBP in every cell extract prior to co-immunoprecipitation was controlled using anti-TBP antibody (Input).
The lack of interaction between TBP and in vitro translated CDX1, in contrast to HCT116 cells-produced CDX1, could result from an inappropriate folding of the protein when translated in vitro. Alternatively, the TBP–CDX1 interaction could depend on additional factors missing in the in vitro translation system. Since TBP associates with many factors, the TAFs (TBP-associated factors), within the TFIID multiprotein complex (24), we checked whether additional members of TFIID can be co-immunoprecipitated with HA-CDX1. As control, HCT116 cells transfected with HA-TBP were used to demonstrate that endogenous TAFs, namely TAF7 (55 kDa), TAF15 (15 kDa) and TAF12 (20 kDa), actually co-immunoprecipitated with HA-tagged TBP in our conditions (Figure 2C). Then we conducted a similar experiment with HA-CDX1 transfected cells. As shown in Figure 2C, TAF7, TAF15 and TAF12 were also recovered by co-immunoprecipitation with HA-CDX1, although less efficiently than with HA-TBP. In addition to TFIID, recruitment and/or binding of TBP onto the TATA-box is facilitated by the Mediator complex (25). Several Mediator complexes have been described in mammalian cells, all containing the Med7 factor (also called Med34). As expected, endogenous Med7 was co-immunoprecipitated using anti-HA antibody in HA-TBP transfected HCT116 cells; strikingly, Med7 was also co-immunoprecipitated with HA-CDX1 in HA-CDX1 transfected cells (Figure 2C). Together, these data indicate that CDX1 is tightly associated with the TFIID multiprotein complex including TBP, as well as with the Mediator complex.
We have previously shown by electrophoretic mobility shift assay (EMSA) that CDX1 and TBP bind together on the DNA sequence containing the G6Pase TATA-box (18). To address the importance of DNA in the CDX1–TBP interaction, HA-immunoprecipitation experiments were performed in the presence of increasing amount of the DNA intercalary agent, ethidium bromide. In cells co-transfected with HA-CDX1, the interaction with endogenous TBP was not altered by ethidium bromide at 10 μg/ml, a concentration that has been shown to strongly inhibits the interaction between the transcription factor Oct1 and its partners (26). However, at the higher dose of 50 μg/ml the CDX1–TBP interaction was prevented, suggesting that, although not required, the DNA may facilitate the interaction. Yet, the addition of DNA containing the G6Pase TATA-box was unable to restore this interaction in the experiments in which bacterially-produced GST-TBP was mixed to in vitro translated HA-CDX1 (data not shown).
Collectively, these results indicate that CDX1 interacts with two major multiprotein complexes of the transcriptional machinery, the TBP-containing TFIID complex and the Med complex, and that the interaction is facilitated by DNA, indicating that CDX1, TBP and DNA form a ternary complex. However, these experiments do not formally demonstrate the direct physical interaction between CDX1 and TBP. In the following, protein interaction between CDX1 and TBP will be used by means of co-immunoprecipitation or GST-pulldown experiments.
Requirement of different domains of CDX1 for TBP-binding and transactivation
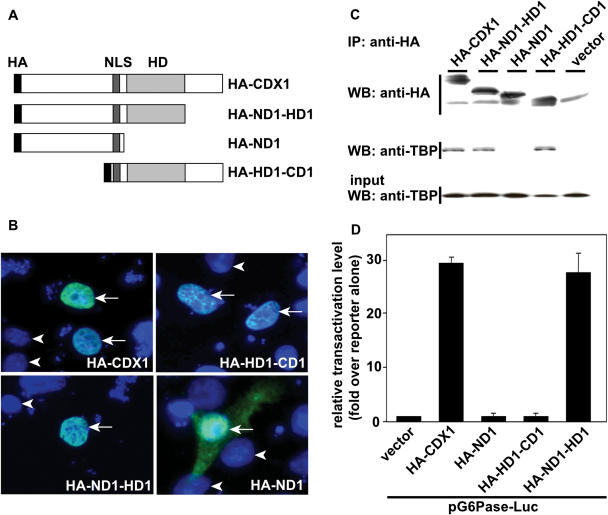
To map the domain of CDX1 responsible for interacting with TBP, we constructed a series of truncated mutants of this homeoprotein, all having the nuclear localization signal of CDX1 and the HA epitope (Figure 3A). The N-terminal domain upstream of the homeodomain and the C-terminal domain downstream of the homeodomain were deleted in the mutants HA-HD1-CD1 and HA-ND1-HD1, respectively. The homeodomain and the C-terminal domain were deleted in the mutant HA-ND1, leaving only the N-terminal domain linked to the nuclear localization signal. Immunofluorescence staining with anti-HA antibody in transfected HCT116 (Figure 3B) and in HepG2 cells (data not shown) revealed that HA-HD1-CD1 and HA-ND1-HD1 accumulated exclusively in the nucleus, like the full-length HA-CDX1. The truncated protein HA-ND1 containing the N-terminal domain linked to the nuclear localization sequence was also present in the nucleus, but a diffuse fluorescence labeling was also detected in the cytoplasm, indicative of less efficient import or retention in the nucleus.
Figure 3.
Requirement of different domains of CDX1 for TBP-binding and transactivation. (A) Schematic representation of the truncated forms of CDX1. HA: hemagglutinin epitope, NLS: nuclear localization signal, HD: homeodomain. (B) Intracellular distribution of the recombinant proteins. HCT116 cells were transfected with the plasmids encoding the indicated proteins and immunolabeled with primary rabbit anti-HA antibody followed by secondary anti-rabbit immunoglobulins coupled to Alexa 488. Cell nuclei were stained with DAPI. Arrows denote cells expressing the mutant forms of CDX1; arrowheads indicate nuclei of non-transfected cells labeled only with DAPI. (C) Co-immunoprecipitation of TBP with the truncated forms of CDX1. HCT116 cells were co-transfected with the plasmids encoding TBP and either HA-CDX1, HA-ND1-HD1, HA-ND1, HA-HD1-CD1 or the control empty vector. Proteins immunoprecipitated with anti-HA antibody were detected by western blots using anti-HA and co-immunoprecipitation of TBP was assayed using anti-TBP. TBP was recovered in every fraction except with HA-ND1. The presence of TBP in the cell extracts prior to HA-immunoprecipitation was controlled by western blot using anti-TBP antibody (Input). (D) Transcriptional activity of the truncated forms of CDX1. HepG2 cells were co-transfected with the reporter plasmid pG6Pase-Luc and with each of the expression vectors encoding HA-CDX1, HA-ND1-HD1, HA-ND1 or HA-HD1-CD1, together with the control reporter pRL-null. The data obtained in triplicate ±SD are representative of three independent experiments.
The different HA-tagged CDX1 mutants were then checked for their ability to co-immunoprecipitate endogenous TBP, using anti-HA antibody. Figure 3C shows that the HA-ND1-HD1 and HA-HD1-CD1 mutants, which have the nuclear localization signal and the homeodomain in common, were both able to interact with TBP as efficiently as wild-type CDX1. However HA-ND1, lacking both the homeodomain and the C-terminal domain, no longer interacted with TBP. Taken together, these data strongly suggest that the homeodomain is crucial for the interaction between CDX1 and TBP. We have also created an expression plasmid encoding the truncated form containing only the nuclear localization signal plus the homeodomain of CDX1, but we failed to detect this protein after transfection, suggesting that it is likely degraded. When synthesized in vitro by in vitro transcription/translation, the polypeptide overlapping only the homeodomain of CDX1, HD1, was not able to interact with GST-TBP (data not shown), as observed above for the full-length CDX1 protein synthesized in vitro.
Next, we analyzed the transcriptional activity of the different truncated mutants on the G6Pase promoter (Figure 3D). Co-transfection of full-length HA-CDX1 with the reporter plasmid pG6Pase-Luc led to a nearly 30-fold stimulation of transcriptional activity. A 30-fold stimulation was also obtained with the HA-ND1-HD1 mutant, lacking the C-terminal domain of CDX1. However, it is worth noting that the HA-HD1-CD1 mutant was devoid of transcriptional activity, despite its ability to interact with TBP. The mutant HA-ND1 that did not interact with TBP was also transcriptionally inactive. These data indicate that the TBP-binding and transactivation activities map on discrete domains of CDX1, respectively the homeodomain and the N-terminal domain, and that cooperation between both domains is required for efficient function of CDX1.
Dependence of the TBP-binding activity of the CDX1 and CDX2 homeodomains to regions outside the homeodomains
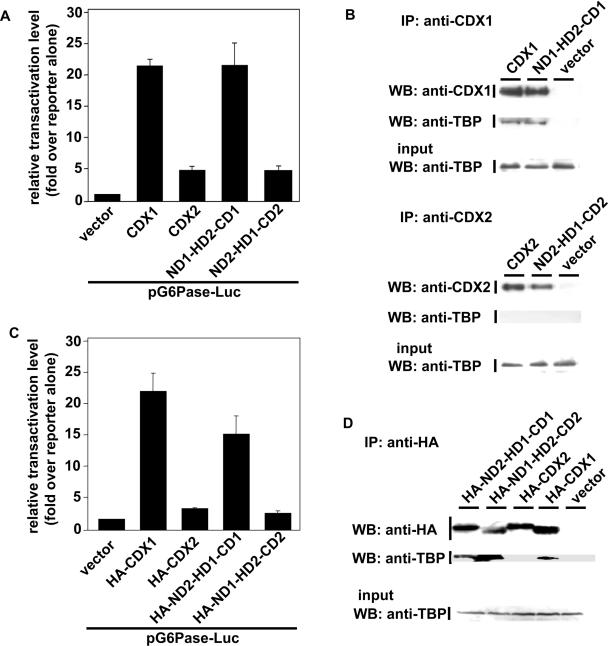
Although CDX2, unlike CDX1, does not interact with TBP, we asked if the CDX2 homeodomain could be turned on to bind TBP in the context of CDX1. Reciprocally, we investigated the TBP-binding activity of the CDX1 homeodomain in the context of CDX2. The homeodomains of CDX1 and CDX2 differ significantly at 2 amino acid, the residues N190 and T194 of CDX1 corresponding to T221 and S225 in CDX2 (18). Changing N190 and T194 of CDX1, respectively into T and S creates the chimeric molecule ND1-HD2-CD1 in which the homeodomain of CDX2 is placed in the context of CDX1 whereas, replacing T221 and S225 of CDX2 with N and T produces the chimeric molecule ND2-HD1-CD2 in which the homeodomain of CDX1 is placed in the context of CDX2. Luciferase assays using the G6Pase promoter indicated that ND1-HD2-CD1 stimulated the transcriptional activity like CDX1, whereas ND2-HD1-CD2 was much less active than CDX1, like CDX2 [Figure 4A, (18)].
Figure 4.
Interaction of TBP with chimeric mutants of CDX1 and CDX2. (A). Transcriptional activity of the swapped mutants of CDX1 and CDX2 at the level of the homeodomain. HepG2 cells were co-transfected with the reporter plasmids pG6Pase-Luc and pRL-null and with the expression plasmids encoding CDX1 or CDX2 or ND1-HD2-CD1 or ND2-HD1-CD2. Relative transactivation levels, obtained from the measurements of luciferase activity, are given ±SD for four independent experiments in triplicate. (B) Cells were transfected with the plasmids encoding CDX1 or ND1-HD2-CD1, or with the plasmids encoding CDX2 or ND2-HD1-CD2. The proteins immunoprecipitated, respectively, with anti-CDX1 or with anti-CDX2 were analyzed by western blot using anti-CDX1 and anti-TBP antibodies, or with anti-CDX2 and anti-TBP antibodies. The presence of TBP in the cell extracts prior to immunoprecipitation was controlled by western blot using anti-TBP antibody (Input). (C) Transcriptional activity of the swapped mutants of CDX1 and CDX2 at the level of the N-terminal domain. HepG2 cells were co-transfected with the reporter plasmids pG6Pase-Luc and pRL-null and with the expression plasmids encoding HA-CDX1 or HA-CDX2 or ND2-HD1-CD1 or ND1-HD2-CD2. Relative transactivation levels, obtained from the measurements of luciferase activity, are given ±SD for four independent experiments in triplicate. (D) Cells were transfected with the plasmids encoding HA-CDX1, HA-CDX2, HA-ND1-HD2-CD2 or ND2-HD1-CD1. Proteins immunoprecipitated with anti-HA antibody were analyzed by western blot using anti-HA and anti-TBP antibodies. The presence of TBP in the cell extracts prior to immunoprecipitation was controlled by western blot using anti-TBP antibody (Input).
Next, we performed co-immunoprecipitation assays to investigate the interaction of the chimeric proteins with TBP. Figure 4B shows that ND1-HD2-CD1 immunoprecipitated with anti-CDX1 antibody was able to interact with endogenous TBP, like wild-type CDX1, whereas ND2-HD1-CD2 immunoprecipitated with anti-CDX2 was unable to interact with TBP, like CDX2. Reciprocally, when TBP was immunoprecipitated with anti-TBP antibody, ND1-HD2-CD1 was recovered into the TBP fraction, like CDX1, whereas CDX2 and the chimeric protein ND2-HD1-CD2 were not recovered (data not shown). Therefore, these results indicate first, that both the CDX1 and the CDX2 homeodomains are able to interact with TBP and second, that the actual binding activity depends on the regions outside the homeodomains, in the sense that the CDX1 context is permissive or stimulatory for TBP-binding whereas the CDX2 context exerts a dominant-negative effect on the homeodomain for its binding to TBP.
Having demonstrated first, that the homeodomain and N-terminal domain of CDX1 are required for TBP interaction and transcriptional activity, respectively (Figure 3), and second that the regions of CDX2 outside the homeodomain can blunt the CDX1 homeodomain for interaction with TBP (Figure 4A and B), we constructed the chimeric protein in which the N-terminal domain of CDX1 was linked to the homeodomain and carboxy domain of CDX2 (HA-ND1-HD2-CD2) as well as the reciprocal chimeric protein in which the N-terminal domain of CDX2 was linked to the homeodomain and carboxy domain of CDX1 (HA-ND2-HD1-CD1). After transfection of the corresponding plasmids in HCT116 cells and co-immunoprecipitation with anti-HA antibody, endogenous TBP was recovered with both HA-ND1-HD2-CD2 and HA-ND2-HD1-CD1 proteins (Figure 4D). However Luciferase assays with the G6Pase-Luc reporter indicated that the HA-ND2-HD1-CD1 protein was transcriptionaly active, although less than CDX1, whereas HA-ND1-HD2-CD2 was inactive, like CDX2 (Figure 4C). This indicates that the N-terminal domain of CDX2 confers transcriptional activity when linked to the CDX1 homeodomain and C-terminal domain, and that the carboxy domain of CDX2 is able to blunt the transcriptional activity of the CDX1 N-terminal domain, yet without compromising TBP interaction with the homeodomain.
Functional interaction and promoter selectivity of the cooperation between CDX1 and TBP
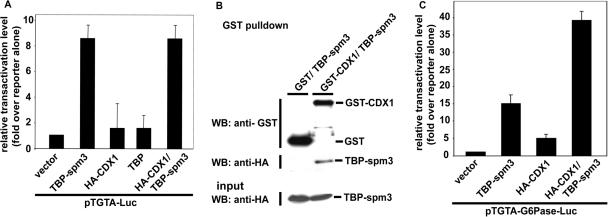
While CDX1 stimulates the G6Pase promoter and interacts with TBP associated with the TFIID complex, the functionality of the interaction between CDX1 and TBP remains to be established. To address this point, we investigated the ability of CDX1 to cooperatively activate a reporter plasmid along with TBP. Co-transfection experiments with TBP and CDX1 to check their co-operativity on the pG6Pase-Luc reporter plasmid gave uninterpretable results because of the presence of the endogenous TBP. To circumvent this problem, we used a previously-described model in which the altered-specificity mutant of TBP, spm3, activates transcription via a TGTA rather than via the canonical TATA DNA-binding site (19,27). The reporter plasmid pTGTA-Luc was co-transfected with the expression plasmids encoding TBP-spm3 and/or HA-CDX1 (Figure 5A). As expected, luciferase activity was stimulated by 8-fold in the presence of TBP-spm3, whereas wild-type TBP was almost inactive on this promoter. HA-CDX1 alone had no effect on this promoter. Moreover, HA-CDX1 did not stimulate the transcriptional effect of TBP-spm3 on the standard pTGTA-Luc reporter plasmid. In parallel experiments, the GST-pulldown recovery of HA-TBP-spm3 together with GST-CDX1 ruled out the possibility that the failure of CDX1 to enhance the stimulatory effect of TBP-spm3 on pTGTA-Luc was due to a loss of interaction between CDX1 and TBP-spm3 (Figure 5B). We therefore created a variant of the G6Pase promoter, pTGTA-G6Pase-Luc, in which the TATAAAA sequence was changed into TGTAAAA. As shown in Figure 5C, the luciferase activity of the pTGTA-G6Pase-Luc reporter was increased 15-fold in the presence of TBP-spm3, whereas CDX1 alone had only a weak effect (4-fold). However important, when TBP-spm3 was coexpressed with CDX1, a much stronger 35-fold stimulation of the pTGTA-G6Pase was observed. This result demonstrates the functionality of the interaction between TBP-spm3 and CDX1, specifically in the context of the TGTA-G6Pase promoter.
Figure 5.
Functional interaction between TBP and CDX1. (A) Lack of activity of CDX1 on the pTGTA-Luc reporter. HepG2 cells were co-transfected with the reporter plasmids pTGTA-Luc and pRL-null, and with the expression plasmids encoding TBP-spm3, HA-CDX1, TBP or with the combination of TBP-spm3 plus CDX1. The data obtained in triplicate ±SD are representative of three independent experiments. (B) Interaction of TBP-spm3 with CDX1. Cells were co-transfected with the plasmid encoding HA-TBP-spm3 and the control vector pBC or the plasmid coding for GST-CDX1. GST-pulldown extracts were analyzed by western blots using anti-GST and anti-HA antibodies, demonstrating the interaction between TBP-spm3 and CDX1. The presence of HA-TBP-spm3 in the cell extracts prior to immunoprecipitation was controlled by western blot using anti-HA antibody (Input). (C) Synergistic effect of TBP-spm3 and CDX1 on the pTGTA-G6Pase-Luc reporter plasmid. HepG2 cells were co-transfected with the reporter plasmids pTGTA-G6Pase-Luc and pRL-null, and with the expression plasmids encoding HA-TBP-spm3, HA-CDX1 or HA-TBP-spm3 plus HA-CDX1. The data obtained in triplicate ±SD are representative of three independent experiments.
DISCUSSION
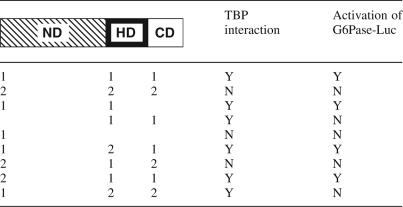
The TBP binds to TAFs within TFIID and also to Mediator, two major complexes of the general transcription machinery, whereas homeodomain proteins are transcription activators acting in a stage- and/or tissue-specific manner. Here we show by means of co-immunoprecipitation and GST-pulldown that CDX1 interacts with TBP in connection to several components of TFIID, TAF7, TAF12 and TAF15, and to Med7, a common component of the Mediator complexes. Unlike CDX1, CDX2 does not interact with TBP. Truncated forms of CDX1 and swapped mutants between CDX1 and CDX2 (Table 1) provide evidence that separate domains of these homeoproteins, namely the N-terminal domain, the central homeodomain and the C-terminal domain, are involved in transcriptional activity, TBP interaction and regulation, and that the combination of these domains within CDX1 and CDX2 dictates their specific function on the G6Pase promoter. Finally, using an altered-specificity mutant of TBP, we demonstrate that the TBP–CDX1 interaction is functional and that co-operativity is selective for G6Pase promoter activation.
Table 1.
Functional organization of the CDX1 and CDX2 homeoproteins
The organization of the N-terminal domain (ND), the homeodomain (HD) and the C-terminal domain (CD) of CDX homeoproteins is schematized. (1) and (2), respectively, designate the CDX1 or CDX2 origin of each domain. Thus, the wild-type CDX1 and CDX2 proteins appear here as ND1-HD1-CD1 and ND2-HD2-CD2, respectively. Interaction with TBP is defined by means of co-immunoprecipitation or GST-pulldown experiments.
Co-immunoprecipitation and GST-pulldown indicate that CDX1 and TBP belong to a common complex with TAFs, although the direct physical interaction between CDX1 and TBP could not be established. Indeed, while GST-TBP interacts with CDX1 produced in intestinal cells, it does not with in vitro translated CDX1 nor with the in vitro translated homeodomain, which indicates either that the TBP–CDX1 interaction is indirect and needs cofactors, or that it is direct but that the conformation of in vitro-synthesized CDX1 or homeodomain is not adequate. However, evidence is provided here for the functionality of the TBP–CDX1 interaction, using the specificity mutant of TBP, spm3 and the TGTA variant of the G6Pase TATA-box. The use of TBP-spm3 together with the TGTA variant of the G6Pase promoter allowed us to bypass the effects of endogenous TBP in cotransfections studies. Interestingly, we found that CDX1 and TBP-spm3 cooperatively activate the pTGTA-G6Pase-Luc reporter, but not the standard pTGTA-Luc vector based on the RARAβ2 proximal promoter, suggesting that CDX1 functions in a context-dependent manner and that sequences outside the TATA-box are also important for controlling CDX1 DNA-binding and/or activity. Recently, it has been shown that the base pair located 3 nt away from the CDX binding site is involved in the mechanism by which CDX2 discriminates among promoters of the UDP-glucuronosyltransferase gene family for DNA-binding and transcriptional activation (28). Introducing the corresponding nucleotide changes within the pTGTA-Luc and pTGTA-G6Pase-Luc plasmids did not modify the response of these promoters to CDX1 (data not shown), which suggests that other cis-elements, still to be elucidated, dictate the differential effects of CDX1 on the pTGTA-G6Pase and pTGTA-Luc reporters.
Although CDX1 and CDX2 have a similar enhancer activity on several intestinal promoters, CDX1 but not CDX2 activates the G6Pase promoter and, moreover, CDX2 blunts the stimulatory effect exerted by CDX1 on the G6Pase promoter (18). CDX1 and CDX2 do not co-immunoprecipitate, ruling out the possibility that CDX2 counteracts CDX1 by trapping it into an inactive heterodimer (data not shown). However, CDX2, like CDX1, is able to bind to the G6Pase TATA-box (18), suggesting that the opposite transcriptional effects of both proteins result from their intrinsic properties. Three domains are generally reported in homeoproteins, with the DNA-binding homeodomain near the centre. Table 1 recapitulates the results obtained in this study with the different truncated and swapped forms of CDX1 and CDX2. It comes out from the truncated mutants that the homeodomain of CDX1 is essential for the interaction with TBP, in addition to its DNA-binding activity. The homeodomain of other homeoproteins has also been involved in protein–protein interactions beside their DNA-binding function (29,30). Despite its TBP-binding activity, the homeodomain of CDX1 is not active on the G6Pase promoter unless it is linked to the N-terminal domain, indicating that this latter domain is crucial for transcriptional activity, as already reported for the N-terminal domain of the CDX2 protein (31). Strikingly, when the CDX2 homeodomain is placed in the context of the CDX1 N-terminal and C-terminal domains, it becomes competent for TBP interaction. Moreover, the CDX2 N-terminal domain becomes transcriptionaly active when associated to the CDX1 homeodomain and C-terminal domain. This suggests that the N-terminal domains and homeodomains of both CDX1 and CDX2 have, respectively, the intrinsic ability to activate transcription and to interact with TBP and therefore, that the opposite effects of CDX1 and CDX2 on the G6Pase promoter depend on their carboxy domains. This conclusion is further supported by additional swapping mutants. Indeed, transcriptional activity (but not TBP interaction) is lost by changing the C-terminal domain of CDX1 into the carboxy domain of CDX2 in the protein built from the CDX1 N-terminal domain and CDX2 homeodomain. Moreover, transcriptional activity and TBP interaction is lost by changing the C-terminal domain of CDX1 into the carboxy domain of CDX2 in the protein built from the CDX2 N-terminal domain and CDX1 homeodomain. Hence, in the context of the G6Pase promoter, the CDX2 C-terminal domain has an inhibitory effect on both CDX1 and CDX2 N-terminal domains responsible for transcription activation, and it also blunts the TBP interaction activity of the CDX1 homeodomain. Alternatively these data may suggest that the CDX1 carboxy domain has a stimulatory effect on the transcriptional and TBP interaction activities of the associated N-terminal domain and homeodomains. Taken together, these data uncover the role of the C-terminal domains of CDX1 and CDX2 as regulators of the functional specificity of these homologous proteins.
This study identifies the domains involved in transcriptional activity, interaction with TBP and regulation within the CDX1 and CDX2 homeoproteins. It suggests that the specific activity of these transcription factors depends on intra-molecular interactions between their domains, with a major role played by the homeodomain for cooperation with TBP and the C-terminal domains for controlling active and inactive conformations of the homeoproteins. Changes between open and closed conformations of HOX and PBX homeoproteins have already been proposed to explain their association with either co-activators or co-repressors (32). The phosphorylation/de-phosphorylation balance is a common way to induce conformational changes and to modify interactions with protein partners. Recently, we have identified a complex phosphorylation site in the carboxy domain of CDX2, that can regulate the half-life and activity of the protein (23). We also have indications that CDX1 is subjected to post-translational modifications (I. Gross, unpublished data). Further studies will investigate if post-translational modifications can alter the conformation of CDX1 and/or CDX2 to change their pattern of interaction with the transcriptional machinery and thereby to modify their downstream genetic program during embryonic development, intestinal homeostasis and/or colorectal cancers.
Acknowledgments
The authors thank L. Tora (IGBMC, INSERM U596, France) for the anti-TPB antibody and for the TBP-expressing plasmids, B. Chatton (CNRS UMR7100, France) for providing the vector pBC, M. Meisterernst (GSF-National Research Center, Munich, Germany) for providing the anti-Med7 antibody and E. Martin for excellent technical assistance. This work was supported by INSERM, the Association pour la Recherche sur le Cancer (ARC, #3286) and the Ligue Contre le Cancer (Comités Départementaux du Bas-Rhin et du Haut-Rhin, France). A.C. is a fellow of the INSERM/Région-Alsace and I.G. was funded by the Association pour la Recherche sur le Cancer and by the Institute National du Cancer (INCa). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellon G., McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays. 1998;20:116–125. doi: 10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 2.Hombria J.C., Lovegrove B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 3.Cillo C., Cantile M., Faiella A., Boncinelli E. Homeobox genes in normal and malignant cells. J. Cell. Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 4.Samuel S., Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur. J. Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freund J.N., Domon-Dell C., Kedinger M., Duluc I. The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell. Biol. 1998;76:957–969. doi: 10.1139/o99-001. [DOI] [PubMed] [Google Scholar]
- 7.Silberg D.G., Swain G.P., Suh E.R., Traber P.G. Cdx1 and Cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 8.Lorentz O., Duluc I., De Arcangelis A., Simon-Assmann P., Kedinger M., Freund J.N. Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J. Cell. Biol. 1997;139:1553–1565. doi: 10.1083/jcb.139.6.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soubeyran P., Andre F., Lissitzky J.C., Mallo G.V., Moucadel V., Roccabianca M., Rechreche H., Marvaldi J., Dikic I., Dagorn J.C., et al. Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology. 1999;117:1326–1338. doi: 10.1016/s0016-5085(99)70283-0. [DOI] [PubMed] [Google Scholar]
- 10.Suh E., Traber P.G. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lickert H., Domon C., Huls G., Wehrle C., Duluc I., Clevers H., Meyer B.I., Freund J.N., Kemler R. Wnt/β-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 12.Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell. Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mutoh H., Sakurai S., Satoh K., Osawa H., Hakamata Y., Takeuchi T., Sugano K. Cdx1 induced intestinal metaplasia in the transgenic mouse stomach: comparative study with Cdx2 transgenic mice. Gut. 2004;53:1416–1423. doi: 10.1136/gut.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoki K., Tamai Y., Horiike S., Oshima M., Taketo M.M. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc(+/Delta716) Cdx2(+/−) compound mutant mice. Nature Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 15.Bonhomme C., Duluc I., Martin E., Chawengsaksophak K., Chenard M.P., Kedinger M., Beck F., Freund J.N., Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haigis K., Sage J., Glickman J., Shafer S., Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J. Biol. Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- 17.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 18.Gautier-Stein A., Domon-Dell C., Calon A., Bady I., Freund J.N., Mithieux G., Rajas F. Differential regulation of the glucose-6-phosphatase TATA box by intestine-specific homeodomain proteins CDX1 and CDX2. Nucleic Acids Res. 2003;31:5238–5246. doi: 10.1093/nar/gkg747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavigne A.C., Gangloff Y.G., Carre L., Mengus G., Birck C., Poch O., Romier C., Moras D., Davidson I. Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of the hTAFII28 histone fold. Mol. Cell. Biol. 1999;19:5050–5060. doi: 10.1128/mcb.19.7.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatton B., Bahr A., Acker J., Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. Biotechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 21.Jacq X., Brou C., Lutz Y., Davidson I., Chambon P., Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 22.Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J.M., Tora L., Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross I., Lhermitte B., Domon-Dell C., Duluc I., Martin E., Gaiddon C., Kedinger M., Freund J.N. Phosphorylation of the homeotic tumor suppressor Cdx2 mediates its ubiquitin-dependent proteasome degradation. Oncogene. 2005;24:7955–7963. doi: 10.1038/sj.onc.1208945. [DOI] [PubMed] [Google Scholar]
- 24.Davidson I. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 2003;28:391–398. doi: 10.1016/S0968-0004(03)00117-8. [DOI] [PubMed] [Google Scholar]
- 25.Wu S.Y., Zhou T., Chiang C.M. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell. Biol. 2003;23:6229–6242. doi: 10.1128/MCB.23.17.6229-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai J.S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl Acad. Sci. USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strubin M., Struhl K. Yeast and human TFIID with altered DNA-specificity specificity for TATA elements. Cell. 1992;68:730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 28.Gregory P.A., Lewinsky R.H., Gardner-Stephen D.A., Mackenzie P.I. Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1alpha and the caudal-related homeodomain protein 2. Mol. Pharmacol. 2004;65:953–963. doi: 10.1124/mol.65.4.953. [DOI] [PubMed] [Google Scholar]
- 29.Bruun J.A., Thomassen E.I., Kristiansen K., Tylden G., Holm T., Mikkola I., Bjorkoy G., Johansen T. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res. 2005;33:2661–2675. doi: 10.1093/nar/gki562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai J., Lin H., Canete-Soler R., Schlaepfer W.W. HoxB2 binds mutant SOD1 and is altered in transgenic model of ALS. Hum. Mol. Genet. 2005;14:2629–2640. doi: 10.1093/hmg/ddi297. [DOI] [PubMed] [Google Scholar]
- 31.Rings E.H., Boudreau F., Taylor J.K., Moffett J., Suh E.R., Traber P.G. Phosphorylation of the serine 60 residue within the cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 32.Saleh M., Rambaldi I., Yang X.J., Featherstone M.S. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]